The Capability of O-Acetyl-ADP-Ribose, an Epigenetic Metabolic Small Molecule, on Promoting the Further Spreading of Sir3 along the Telomeric Chromatin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strains
2.2. Purification of Proteins and AAR
2.3. Dot Blotting
2.4. Isothermal Titration Calorimetry (ITC)
2.5. Chromatin Immunoprecipitation on Chip (ChIP-on-chip), and Chromatin Affinity-Precipitation on Chip (ChAP-on-chip)
2.6. Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
2.7. Electron Microscopy (EM)
3. Results and Discussion
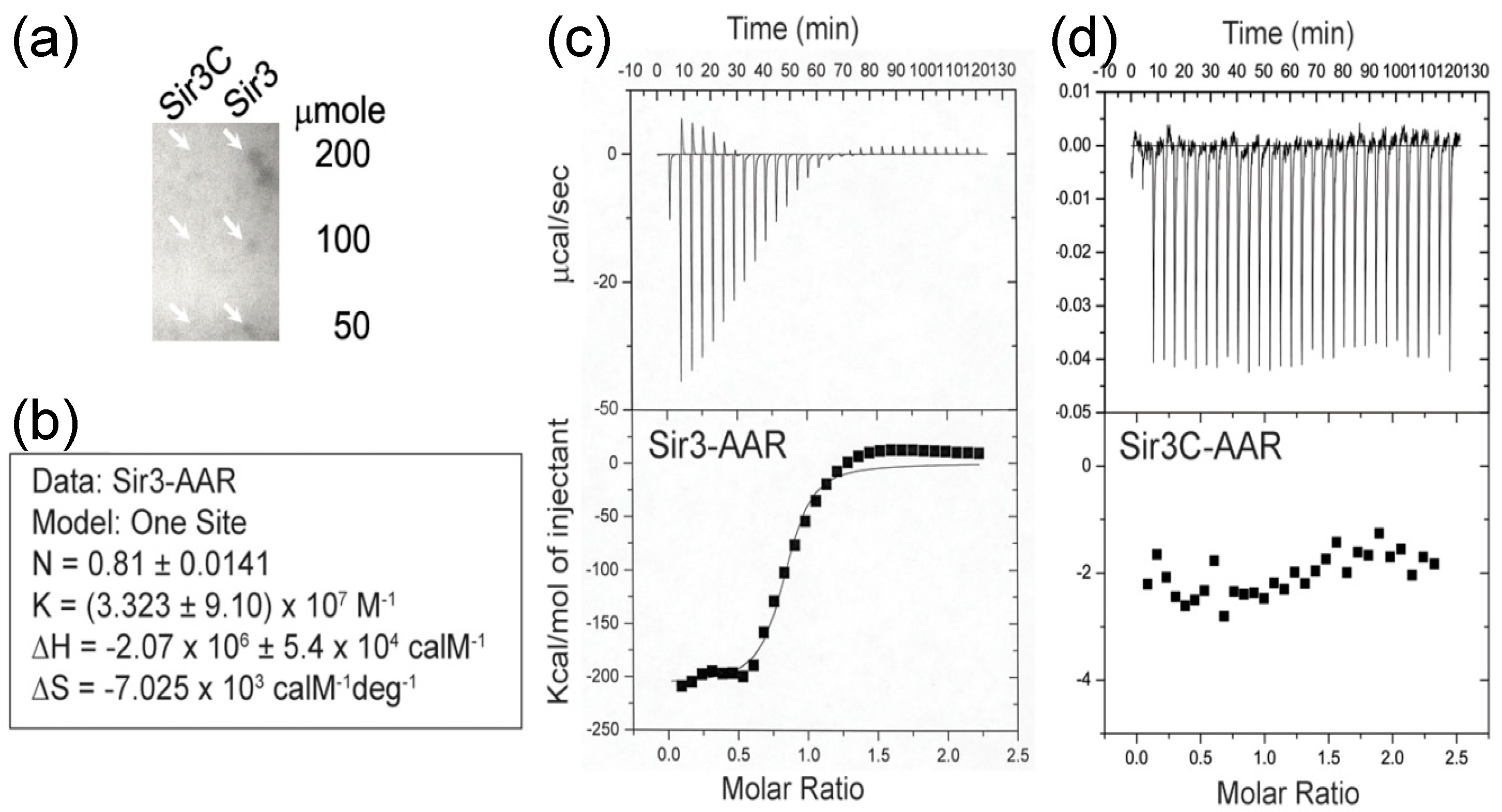
3.1. Interaction of AAR with Sir3
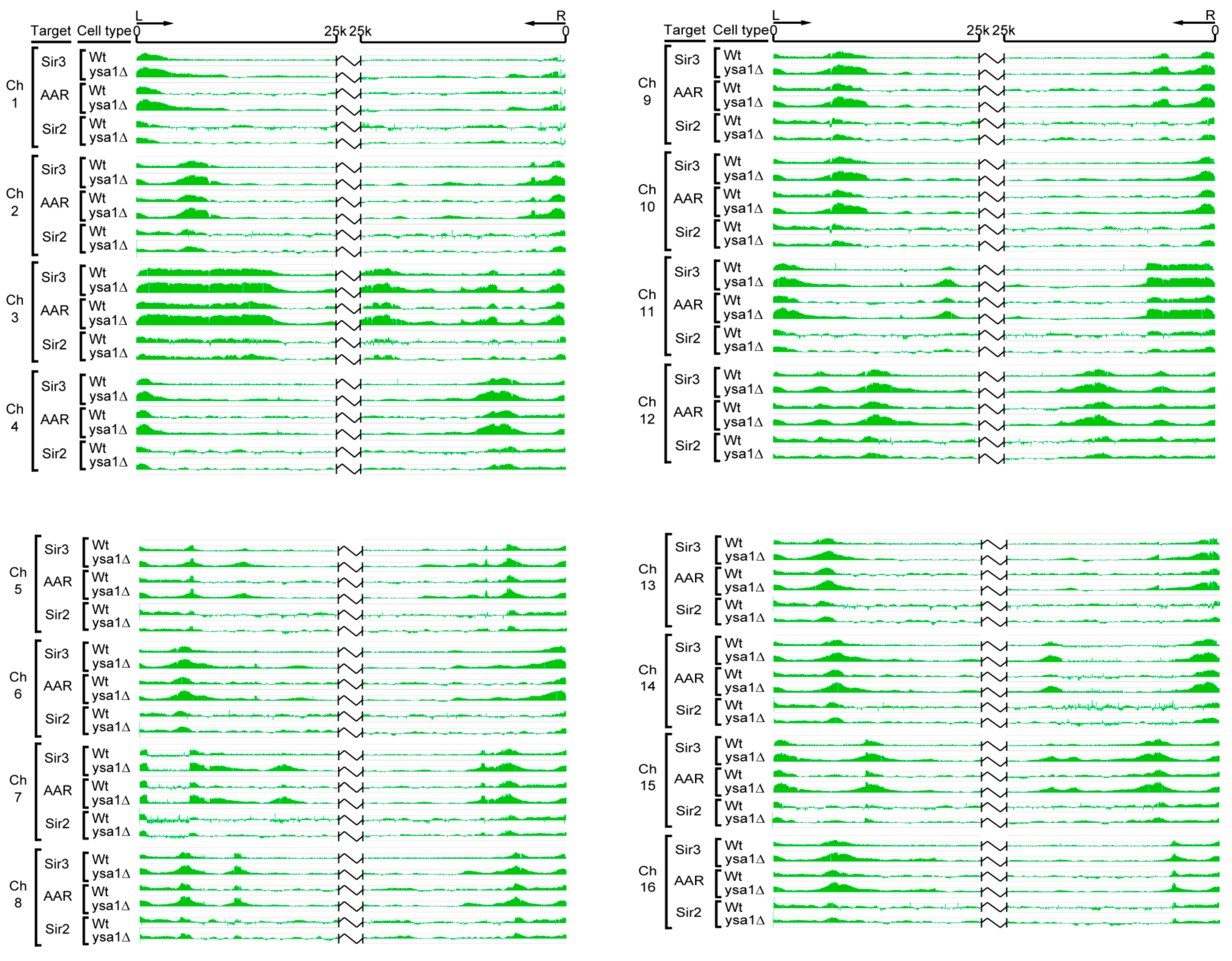
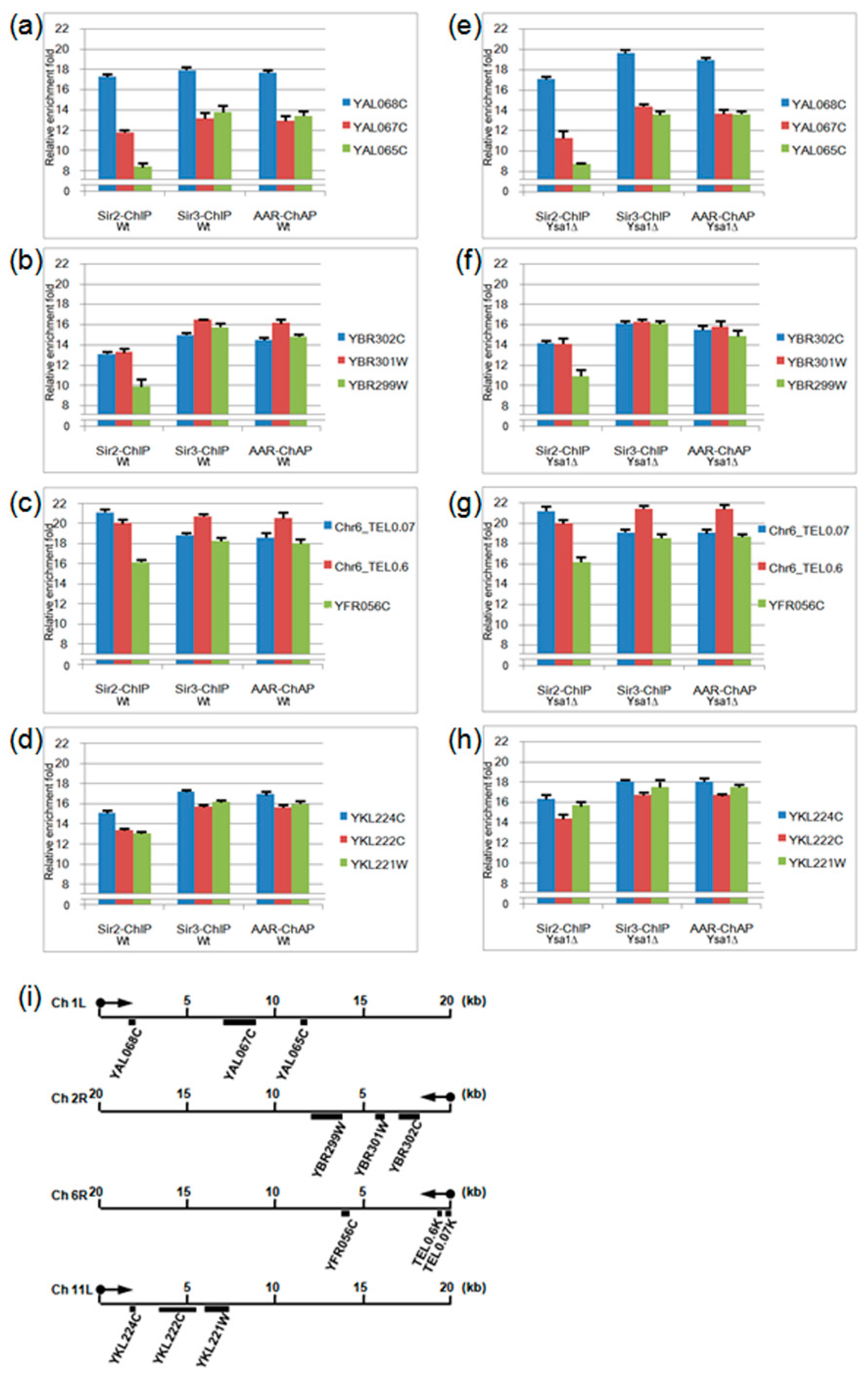
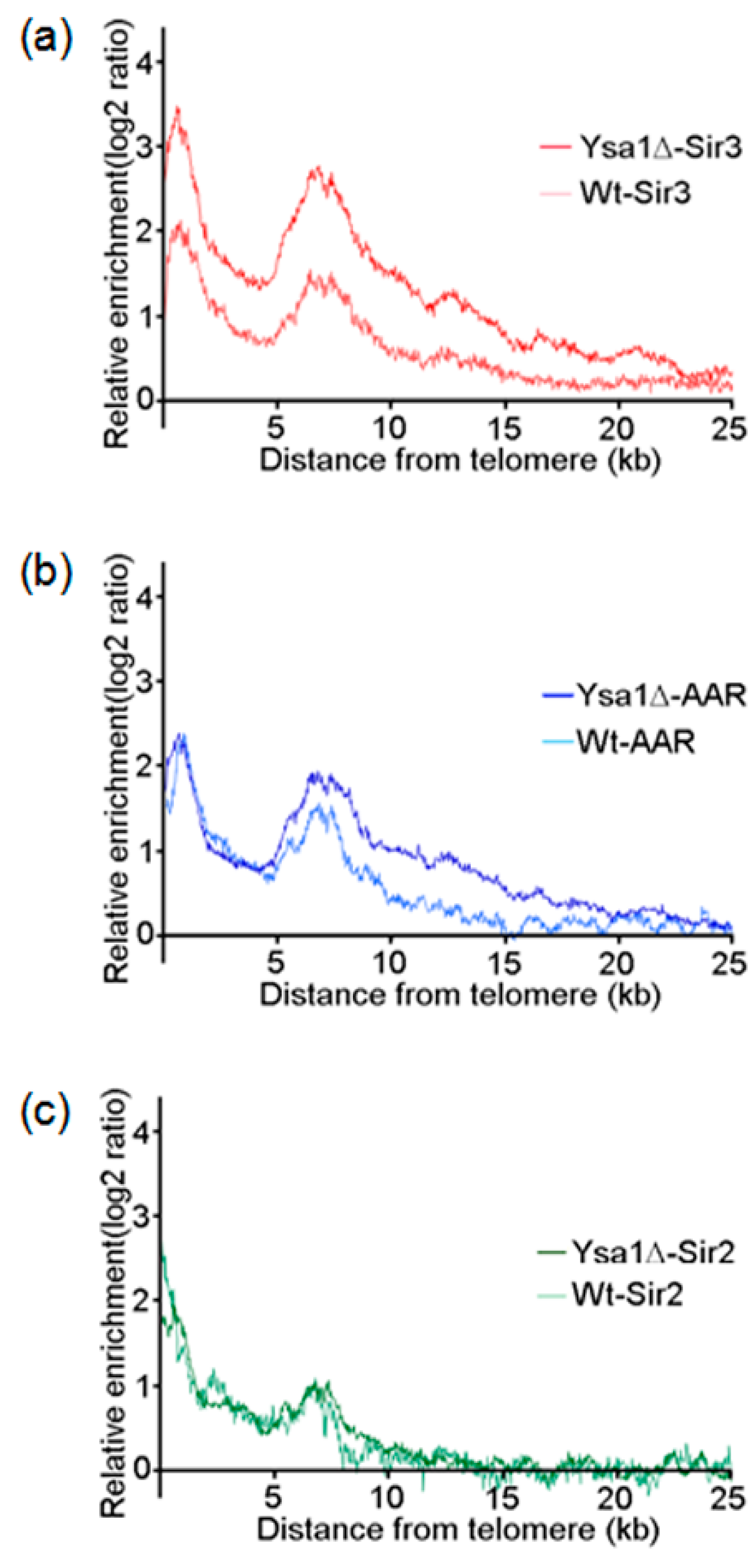
3.2. Occupancies of AAR, Sir2 and Sir3 on Telomeres
3.3. Effect of AAR on Sir2 Spreading and Sir3 Spreading on Telomeres
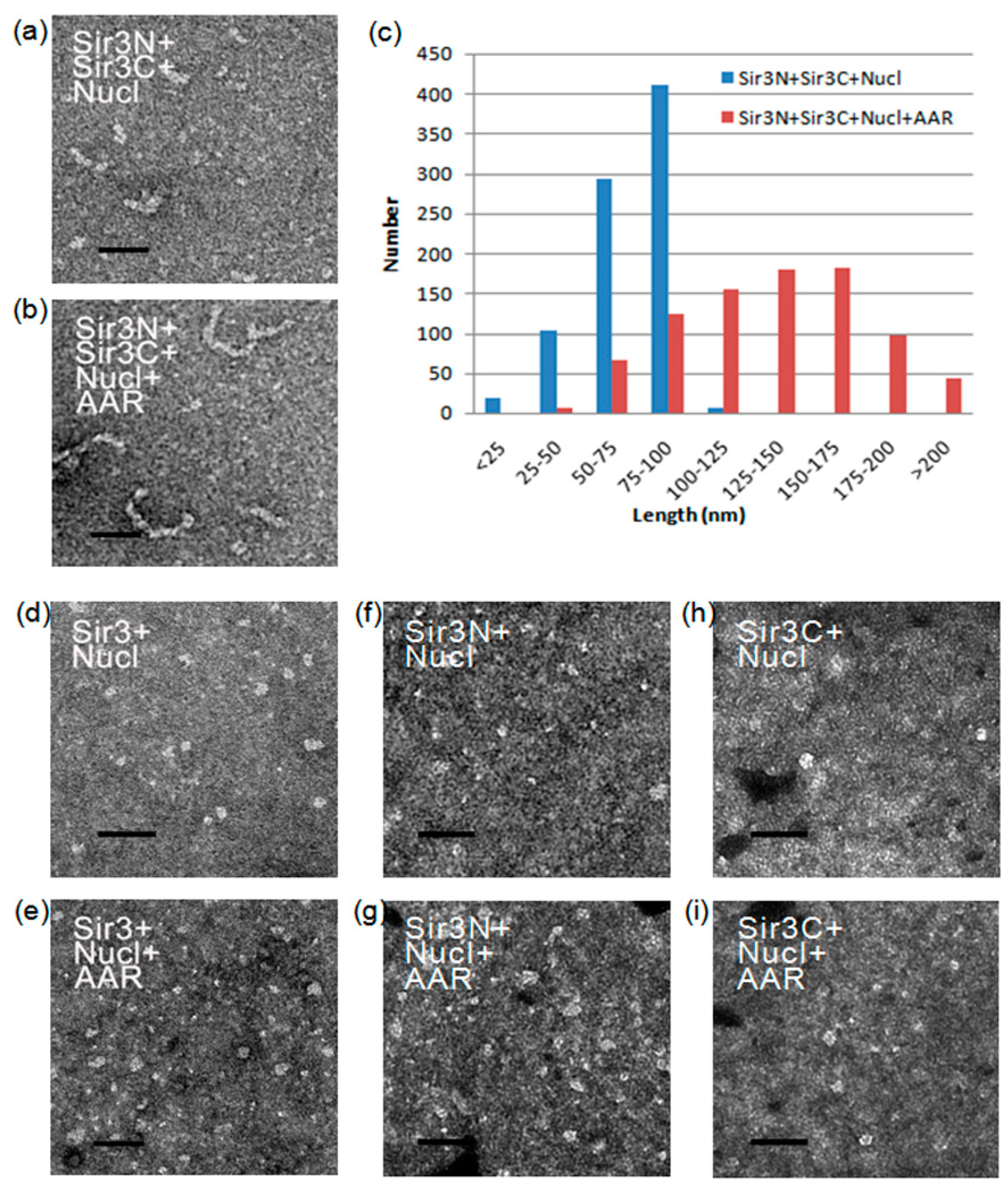
3.4. Effect of AAR on Sir3 Interactions In Vitro
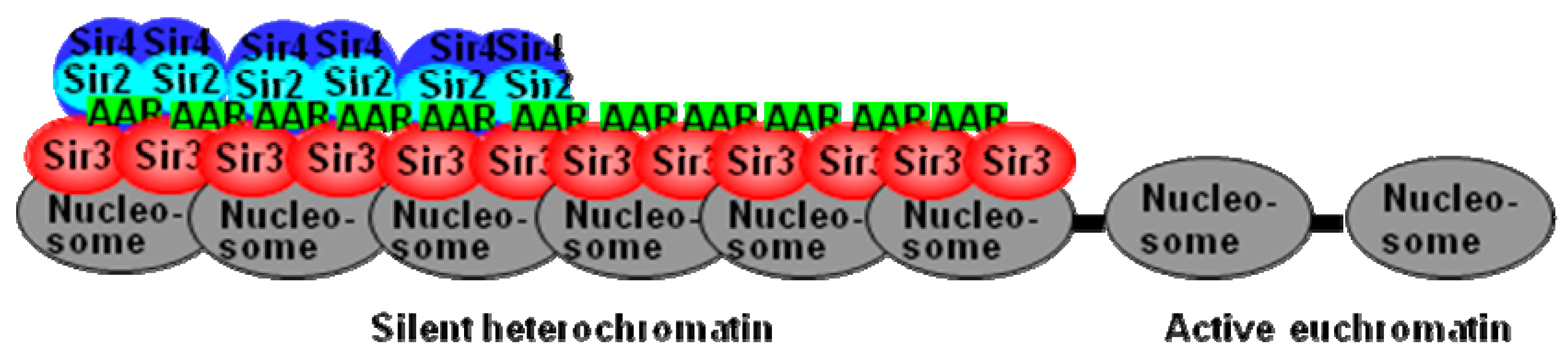
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moazed, D. Common themes in mechanisms of gene silencing. Mol. Cell 2001, 8, 489–498. [Google Scholar] [CrossRef]
- Richards, E.J.; Elgin, S.C. Epigenetic codes for heterochromatin formation and silencing rounding up the usual suspects. Cell 2002, 108, 489–500. [Google Scholar] [CrossRef]
- Rusche, L.N.; Kirchmaier, A.L.; Rine, J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003, 72, 481–516. [Google Scholar] [CrossRef] [PubMed]
- Bryk, M.; Briggs, S.D.; Strahl, B.D.; Curcio, M.J.; Allis, C.D.; Winston, F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 2002, 12, 165–170. [Google Scholar]
- Smith, J.S.; Boeke, J.D. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997, 11, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Moazed, D.; Johnson, D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 1996, 86, 667–677. [Google Scholar] [CrossRef]
- Moazed, D.; Kistler, A.; Axelrod, A.; Rine, J.; Johnson, A.D. Silent information regulator protein complexes in Saccharomyces cerevisiae: A SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 1997, 94, 2186–2191. [Google Scholar] [CrossRef]
- Moretti, P.; Freeman, K.; Coodly, L.; Shore, D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994, 8, 2257–2269. [Google Scholar] [CrossRef]
- Strahl-Bolsinger, S.; Hecht, A.; Luo, K.; Grunstein, M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997, 11, 83–93. [Google Scholar] [CrossRef]
- Aparicio, O.M.; Billington, B.L.; Gottschling, D.E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 1991, 66, 1279–1287. [Google Scholar] [CrossRef]
- Gottschling, D.E.; Aparicio, O.M.; Billington, B.L.; Zakian, V.A. Position effect at S. Cerevisiae telomeres: Reversible repression of pol II transcription. Cell 1990, 63, 751–762. [Google Scholar] [PubMed]
- Klar, A.J.S.; Fogel, S.; Macleod, K. MAR1-a regulator of the HMa and HMα loci in Saccharomyces cerevisiae. Genetics 1979, 93, 37–50. [Google Scholar] [PubMed]
- Rine, J.; Herskowitz, I. Four genes responsible for a position effect on expression from HML and HMR in Saccnaromyces cerevisiae. Genetics 1987, 116, 9–22. [Google Scholar] [PubMed]
- Blander, G.; Guarente, L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 2004, 73, 417–435. [Google Scholar] [CrossRef] [PubMed]
- Borra, M.T.; O’Neill, F.J.; Jackson, M.D.; Marshall, B.; Verdin, E.; Foltz, K.R.; Denu, J.M. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem. 2002, 277, 12632–12641. [Google Scholar] [CrossRef] [PubMed]
- North, B.J.; Marshall, B.L.; Borra, M.T.; Denu, J.M.; Verdin, E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 2003, 11, 437–444. [Google Scholar] [CrossRef]
- Shore, D. The Sir2 protein family: A novel deacetylase for gene silencing and more. Proc. Natl. Acad. Sci. USA 2000, 97, 14030–14032. [Google Scholar] [CrossRef] [Green Version]
- Tanner, K.G.; Landry, J.; Sternglanz, R.; Denu, J.M. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. USA 2000, 97, 14178–14182. [Google Scholar] [CrossRef]
- Tung, S.-Y.; Hong, J.-Y.; Walz, T.; Moazed, D.; Liou, G.-G. Chromatin affinity-precipitation using a small metabolic molecule: Its application to analysis of O-acetyl-ADP-ribose. Cell Mol. Life Sci. 2012, 69, 641–650. [Google Scholar] [CrossRef]
- Wang, S.-H.; Tung, S.-Y.; Su, K.-C.; Shen, H.-H.; Hong, J.-Y.; Tsai, M.-S.; Liou, G.-G. Enhancer role of a native metabolite, O-acetyl-ADP-ribose, on the Saccharomyces cerevisiae chromatin epigenetic gene silencing. Genes Cells 2019. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.-G.; Tanny, J.C.; Kruger, R.G.; Walz, T.; Moazed, D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 2005, 121, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Martino, F.; Kueng, S.; Robinson, P.; Tsai-Pflugfelder, M.; van Leeuwen, F.; Ziegler, M.; Cubizolles, F.; Cockell, M.M.; Rhodes, D.; Gasser, S.M. Reconstitution of yeast silent chromatin: Multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol. Cell 2009, 33, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Onishi, M.; Liou, G.-G.; Buchberger, J.R.; Walz, T.; Moazed, D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol. Cell 2007, 28, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Tung, S.-Y.; Wang, S.-H.; Lee, S.-P.; Tsai, S.-P.; Shen, H.-H.; Chen, F.-J.; Wu, Y.-Y.; Hsiao, S.-P.; Liou, G.-G. Modulations of SIR-nucleosome interactions of reconstructed yeast silent pre-heterochromatin by O-acetyl-ADP-ribose and magnesium. Mol. Biol. Cell 2017, 28, 381–386. [Google Scholar] [CrossRef]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999, 9, 27–43. [Google Scholar] [PubMed]
- Hoppe, G.J.; Tanny, J.C.; Rudner, A.D.; Gerber, S.A.; Danaie, S.; Gygi, S.P.; Moazed, D. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell Biol. 2002, 22, 4167–4180. [Google Scholar] [CrossRef]
- Luo, K.; Vega-Palas, M.A.; Grunstein, M. Rap1–Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002, 16, 1528–1539. [Google Scholar] [CrossRef]
- Gartenberg, M.R.; Smith, J.S. The nuts and bolts of transcriptionally silent chromatin in Saccharomyces cerevisiae. Genetics 2016, 203, 1563–1599. [Google Scholar] [CrossRef]
- Rusche, L.N.; Kirchmaier, A.L.; Rine, J. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 2002, 13, 2207–2222. [Google Scholar] [CrossRef]
- Rudner, A.D.; Hall, B.E.; Ellenberger, T.; Moazed, D. A nonhistone protein-protein interaction required for assembly of the SIR complex and silent chromatin. Mol. Cell Biol. 2005, 25, 4514–4528. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Tong, L.; Denu, J.M. Quantification of endogenous sirtuin metabolite O-acetyl-ADP-ribose. Anal. Biochem. 2008, 383, 174–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hecht, A.; Strahl-Bolsinger, S.; Grunstein, M. Spreading of transcriptional represser SIR3 from telomeric heterochromatin. Nature 1996, 383, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Renauld, H.; Aparicio, O.M.; Zierath, P.D.; Billington, B.L.; Chhablani, S.K.; Gottschling, D.E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength and by SIR3 dosage. Genes Dev. 1993, 7, 1133–1145. [Google Scholar] [CrossRef]
- Sperling, A.; Grunstein, M. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc. Natl. Acad. Sci. USA 2009, 106, 13153–13159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tung, S.-Y.; Lee, K.-W.; Hong, J.-Y.; Lee, S.-P.; Shen, H.-H.; Liou, G.-G. Changes in the genome-wide localization pattern of Sir3 in Saccharomyces cerevisiae during different growth stages. Comput. Struct. Biotechnol. J. 2013, 7, e201304001. [Google Scholar] [CrossRef] [PubMed]
- Guidi, M.; Ruault, M.; Marbouty, M.; Loïodice, I.; Cournac, A.; Billaudeau, C.; Hocher, A.; Mozziconacci, J.; Koszul, R.; Taddei, A. Spatial reorganization of telomeres in long-lived quiescent cells. Genome Biol. 2015, 16, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, L.; Lee, S.; Denu, J.M. Hydrolase regulates NAD+ metabolites and modulates cellular redox. J. Biol. Chem. 2009, 284, 11256–11266. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef]
- Connelly, J.J.; Yuan, P.; Hsu, H.C.; Li, Z.; Xu, R.M.; Sternglanz, R. Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain. Mol. Cell. Biol. 2006, 26, 3256–3265. [Google Scholar] [CrossRef] [PubMed]
- Oppikofer, M.; Kueng, S.; Keusch, J.J.; Hassler, M.; Landurner, A.G.; Gut, H.; Gasser, S.M. Dimerization of Sir3 via its C-erminal winged helix domain is essential for yeast heterochromatin formation. EMBO J. 2013, 32, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Swygert, S.G.; Manning, B.J.; Senapati, S.; Kaur, P.; Lindsay, S.; Demeler, B.; Peterson, C.L. Solution-state conformation and stoichiometry of yeast Sir3 heterochromatin fibers. Nat. Commun. 2014, 5, 4751. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tung, S.-Y.; Wang, S.-H.; Lee, S.-P.; Tsai, S.-P.; Su, K.-C.; Shen, H.-H.; Hong, J.-Y.; Tsai, M.-S.; Liou, G.-G. The Capability of O-Acetyl-ADP-Ribose, an Epigenetic Metabolic Small Molecule, on Promoting the Further Spreading of Sir3 along the Telomeric Chromatin. Genes 2019, 10, 577. https://doi.org/10.3390/genes10080577
Tung S-Y, Wang S-H, Lee S-P, Tsai S-P, Su K-C, Shen H-H, Hong J-Y, Tsai M-S, Liou G-G. The Capability of O-Acetyl-ADP-Ribose, an Epigenetic Metabolic Small Molecule, on Promoting the Further Spreading of Sir3 along the Telomeric Chromatin. Genes. 2019; 10(8):577. https://doi.org/10.3390/genes10080577
Chicago/Turabian StyleTung, Shu-Yun, Sue-Hong Wang, Sue-Ping Lee, Shu-Ping Tsai, Kuan-Chung Su, Hsiao-Hsuian Shen, Jia-Yang Hong, Ming-Shiun Tsai, and Gunn-Guang Liou. 2019. "The Capability of O-Acetyl-ADP-Ribose, an Epigenetic Metabolic Small Molecule, on Promoting the Further Spreading of Sir3 along the Telomeric Chromatin" Genes 10, no. 8: 577. https://doi.org/10.3390/genes10080577
APA StyleTung, S.-Y., Wang, S.-H., Lee, S.-P., Tsai, S.-P., Su, K.-C., Shen, H.-H., Hong, J.-Y., Tsai, M.-S., & Liou, G.-G. (2019). The Capability of O-Acetyl-ADP-Ribose, an Epigenetic Metabolic Small Molecule, on Promoting the Further Spreading of Sir3 along the Telomeric Chromatin. Genes, 10(8), 577. https://doi.org/10.3390/genes10080577






