Abstract
Abscisic acid (ABA) plays important roles in multiple physiological processes, such as plant response to stresses and plant development. The ABA receptors pyrabactin resistance (PYR)/ PYR1-like (PYL)/regulatory components of ABA receptor (RCAR) play a crucial role in ABA perception and signaling. However, little is known about the details regarding PYL family genes in Brassica juncea var. tumida. Here, 25 PYL family genes were identified in B. juncea var. tumida genome, including BjuPYL3, BjuPYL4s, BjuPYL5s, BjuPYL6s, BjuPYL7s, BjuPYL8s, BjuPYL10s, BjuPYL11s, and BjuPYL13. The results of phylogenic analysis and gene structure showed that the PYL family genes performed similar gene characteristics. By analyzing cis-elements in the promoters of those BjuPYLs, several hormone and stress related cis-elements were found. The results of gene expression analysis showed that the ABA receptor homologous genes were induced by abiotic and biotic stress. The tissue-specific gene expression patterns of BjuPYLs also suggested those genes might regulate the stem swelling during plant growth. These findings indicate that BjuPYLs are involved in plant response to stresses and organ development. This study provides valuable information for further functional investigations of PYL family genes in B. juncea var. tumida.
1. Introduction
The plant phytohormone abscisic acid (ABA) is involved in multiple plant physiological processes, such as seed dormancy, seed germination, seedling development, post-germinative growth, stomatal movement, and synthesis of storage proteins and lipids [1,2,3,4,5,6,7]. ABA also plays an essential role in plant responses to adverse environmental stresses, such as abiotic stresses (salinity and low-temperature stresses) and biotic stresses [1,2,8]. So far, the core components of the ABA signaling pathway have been identified in Arabidopsis [9,10,11]. The ABA signaling pathway mainly contains pyrabactin resistance (PYR)/PYR1-like (PYL) regulatory components of ABA receptor (RCAR) protein family (ABA receptors), the co-receptors group A protein type 2C phosphatases (PP2Cs), and sucrose nonfermenting-1-related protein kinase class 2 (SnRK2s) [9,10,11]. In the absence of ABA, PP2Cs interact with SnRK2s and inhibit their kinase activity, which resulted in turning off of ABA signaling pathway. In the presence of ABA, PYR/PYL/RCAR receptors bind the hormone ABA, allowing them to physically associate with PP2Cs and eliminate the inhibitory effect of the phosphatases on SnRK2s, which phosphorylate bZIP (basic-leucine zipper) transcription factors and ion channels to turn on the ABA signaling.
The ABA receptors PYR/PYL/RCAR are responsible for ABA perception and signaling [6,12]. In Arabidopsis, the ABA receptors contain 14 family members, which are PYR1, PYL1–PYL13. Recently, the mechanisms about how these ABA receptors regulate plant responses to ABA, abiotic stresses, and biotic stresses have been characterized. For example, overexpression of NtPYL4 in tobacco hairy roots caused the reprogramming of the cellular metabolism, leading to a decreased alkaloid accumulation, and conferred ABA sensitivity to the production of alkaloids [13]. Overexpression of RSL1 (RING FINGER OF SEED LONGEVITY1) in Arabidopsis resulted in decreased ABA sensitivity of plants by promoting the degradation of PYR1 and PYL4 [14]. OsPYL5, acting as an ABA receptor, can increase rice tolerance to salt and drought stresses [15]. In Brassica napus, the transcript levels of BnPYLs genes were analyzed by qPCR in plant roots under multiple abiotic stresses treatment, and the results showed that the gene expression levels of BnPYR1-3, BnPYL1-2, and BnPYL7-2 were induced by drought, heat, and salinity treatments [16].
B. juncea var. tumida (AABB, 2n = 36), which belongs to Brassicaceae, is an allotetraploid species by a natural cross between Brassica rapa (AA, 2n = 20) and Brassica nigra (BB, 2n = 16) following with subsequent chromosome doubling [17]. B. juncea var. tumida is a kind of vegetable in China and some south-east Asia countries. The swollen stem (the diameter of the stem can swell to more than 20 cm) of B. juncea var. tumida is the raw material for Fuling mustard, which is famous for its special flavor and nutritional value [18]. However, little is known about the regulation mechanism of stem swelling. Tuber mustard always suffers from abiotic stresses (salinity and low-temperature) and biotic stress (Plasmodiophora brassicae), resulting in inhibition of plant growth and huge economic loss. The ABA receptors are key components in the ABA signaling pathway, mediating plant development and response to stresses. Identifying the ABA receptor family genes in B. juncea var. tumida is not only helpful for further understanding the ABA signaling pathway but also provides cues for the plant to defend against stresses. However, the role and function of the ABA receptor family genes in B. juncea var. tumida remains mainly unknown.
In this study, we identified 25 ABA receptor family genes in the B. juncea var. tumida genome. Based on the analysis of the phylogenic relationship, gene structures, and promoter cis-elements, similar gene characteristics were found between BjuPYLs and AtPYLs. The BjuPYLs expression patterns in different tissues were analyzed by qPCR, and the result showed that the 25 BjuPYLs have a different organ and tissue expression patterns. In addition, we analyzed the genes transcriptional levels of BjuPYLs under abiotic stresses (including NaCl, ABA, and low-temperature) and biotic stress (P. brassicae). We also analyzed the BjuPYLs transcriptional levels during different developmental stages of B. juncea var. tumida (the developmental stages of D1 to D5). The results showed that BjuPYLs were induced by abiotic and biotic stresses in B. juncea var. tumida, and BjuPYLs (BjuPYL4-3, BjuPYL5-3, BjuPYL5-5, BjuPYL6-2, BjuPYL8-2) might play roles in stem swelling.
2. Materials and Methods
2.1. Materials and Growth Conditions
Tuber mustard cultivar Yong’an was used for gene expression analysis. The seeds were surface sterilized and plated on MS medium (Sigma-Aldrich, St. Louis, MO, USA) with 1% sucrose and 8 g/L agar (Sigma-Aldrich, St. Louis, MO, USA) and then cultivated in growth room at 22 °C and 6000 lx under long-day conditions (16 h light/8 h dark) for seven days and then treated with 50 μM ABA, 200 mM NaCl, and at low-temperature (4 °C) for 3 h. For pathogen (P. brassicae) treatment, 2-week-old seedlings of B. juncea var. tumida were irrigated with P. brassicae suspension liquid (OD600 = 0.07) for the indicated time points.
2.2. Bioinformatics Analysis
The gene sequences of AtPYLs and their homologous genes in B. juncea var. tumida were searched in TAIR (http://www.arabidopsis.org/) and Brassica database (http://brassicadb.org/brad/). The phylogenic tree was constructed using the neighbor-joining method with the bootstrap values of 1000 by MEGA5 [19]. The gene structure analysis was done by online software (http://gsds.cbi.pku.edu.cn/). The protein sequences were aligned by ESPript 3.0 online software (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) and WebLogo online software (http://weblogo.berkeley.edu/). The promoter cis-element analysis was performed using online analysis software of PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action= page&page=newplace).
2.3. Gene Expression Analysis
Total RNA of different samples were extracted from B. juncea var. tumida seedlings using TRIzol reagent. The RNA samples were used for cDNA synthesis using a cDNA synthesis Supermix with gDNA remover kit (TransGen Biotech, Beijing, China) following the manufacturer’s instructions. qRT-PCR was carried out using SYBR Green qPCR Supermix (Invitrogen, Carlsbad, CA, USA). The transcript abundance was calculated by the comparative CT (cycle threshold) method, and BjuActin3 was used as the internal control. The qRT-PCR experiments were carried out three times, each with three replicates. The primers used are listed in Table S1.
3. Results
3.1. Genome-Wide Identification and Characterization of BjuPYLs in B. juncea var. tumida
25 BjuPYLs were identified in B. juncea var. tumida genome through BLASTP in Brassica database as homologs of AtPYLs by using nine AtPYLs protein sequences as references. No homologs were found for the proteins of AtPYR1, AtPYL1, AtPYL2, AtPYL9, and AtPYL12 (Table 1). The lengths of these ABA receptor genes ranged from 486 bp to 1238 bp with 1–3 exons in each sequence. The protein lengths of BjuPYLs ranged from 162 (BjuPYL11-1) to 221 (BjuPYL6-3) amino acid (aa) residues. The relative molecular weights of these proteins varied from 18.05 kD (BjuPYL11-1) to 24.25 kD (BjuPYL6-3), and the isoelectric point (PI) was 5.01–9.12 (Table 1). The 25 BjuPYLs genes were distributed in 11 of 18 chromosomes of B. juncea var. tumida. Each of the chromosomes A04, A06, B06 contained one gene; A01, A02, A10, B02 contained two genes; A03, B01, B05 contained three genes; the other four genes were all located in B08 (Figure 1).

Table 1.
The BjuPYLs family members in B. juncea var. tumida.
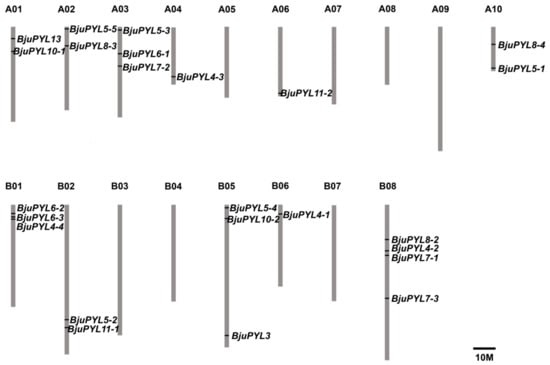
Figure 1.
The distribution of BjPYLs in B. juncea var. tumida chromosomes. Twenty-five identified BjuPYLs were mapped to the 11 of 18 chromosomes. The chromosome name is at the top of each bar. The scale of the chromosome is in millions of bases (Mb).
3.2. The Phylogenic Analysis and Gene Structures of BjuPYLs
To analyze the evolutionary relationships between BjuPYLs and AtPYLs, a phylogenetic tree was constructed by MEGA5 software with the neighbor-joining method using the protein and genomic sequences (Figure 2, Figure S1). According to the phylogenic tree, 25 BjuPYLs with 14 AtPYLs were identified. The BjuPYL genes were named following their homologs in Arabidopsis (Figure 2, Table 1, Figure S1). To understand the gene structures of BjuPYLs, the gene exon-introns were identified using the online software of GSDS2.0 server. According to the result, BjuPYL3, BjuPYL4-1 to BjuPYL4-4, BjuPYL5-1 to BjuPYL5-5, BjuPYL6-1 to BjuPYL6-2, BjuPYL11-1 to BjuPYL11-2, and BjuPYL13 all had one exon; BjuPYL6-3 had two exons; BjuPYL7-1 to BjuPYL7-3, BjuPYL8-1 to BjuPYL8-4, and BjuPYL10-1 to BjuPYL10-2 all contained three exons (Figure 2). Almost all the AtPYLs had the same gene structures with their homologs genes in B. juncea var. tumida, except BjuPYL6-3, which had two exons, while AtPYL6 had one exon. These results indicated that the BjuPYLs shared similar gene structures with their homologs in Arabidopsis.
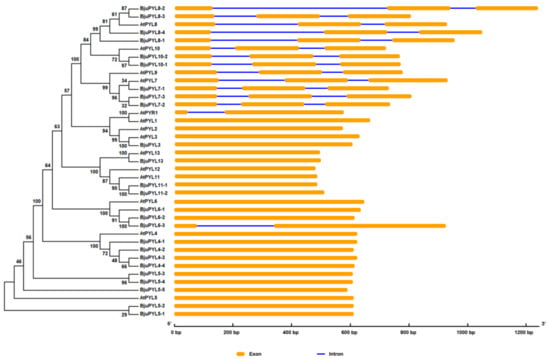
Figure 2.
The phylogenic tree and gene structures of BjuPYLs and AtPYLs. The phylogenic tree was built with the neighbor-joining (NJ) method using the protein sequences, and the exon-intron structure of pyrabactin resistance (PYR)1-like (PYL) homologs was drawn according to their phylogenic relationships. The orange boxes and blue lines denote exons and introns, respectively.
3.3. The Alignment of PYL Proteins and Motif Analysis
The PYL protein sequences were aligned by ESPript 3.0 software (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) [20]. The results showed that the peptide sequences of these PYLs were conserved (Figure S2). The protein sequence identities among PYL3s, PYL4s, PYL5s, PYL6s, PYL7s, PYL8s, PYL10s, PYL11s, and PYL13s were more than 83.33%, 83.81%, 82.84%, 76.78%, 78.14%, 81.68%, 82.70%, 80.98%, and 79.88%, respectively (Figure S2). Moreover, the SGLPA (gate) and HRL (latch) sequences were invariant among the PYL family members, indicating that the gate and latch mechanism was likely to be a common feature of these receptors and they might be typical ABA receptors (Figure 3A,B) [21].
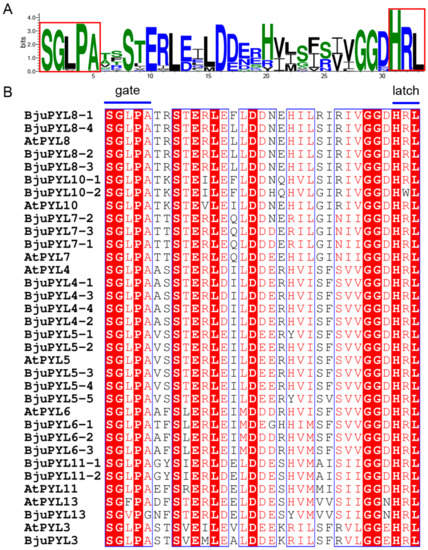
Figure 3.
The alignment of PYL proteins and protein domain prediction. (A) The PYLs protein sequences were aligned by WebLogo online software, and the gate and latch residues are boxed. (B) The PYLs protein sequences were aligned by ESPript online software. Conserved residues are highlighted, and the gate and latch residues are noted.
3.4. The Promoter cis-Acting Regulatory Elements Prediction of BjuPYLs
To further understand the potential roles of BjuPYLs in B. juncea var. tumida and how the genes’ expression is regulated, we chose the 2000 bp DNA fragment upstream of the ATG start code as the promoter sequences and performed the promoter cis-elements analysis using online software of PlantCARE and PLACE. According to the result, the promoters of BjuPYLs contained hormone-related elements, such as ABRE (ACGTG, responsive to Abscisic acid) [22], ARFAT (TGTCTC, responsive to auxin) [23], GMSAUR (CATATG, responsive to auxin) [24], ASF1MOTIFCAMV (TGACG, responsive to auxin and salicylic acid) [25], and ABREATRD22 (RYACGTGGYR, responsive to Abscisic acid) [26] (Figure 4). In addition, the BjuPYLs promoters also contained stressed-related elements, such as MYCCONSENSUSAT (CANNTG, responsive to dehydration stress) [27], MYB1AT (WAACCA, responsive to dehydration stress) [27], MYBATRD22 (CTAACCA, responsive to dehydration stress) [28], CBFHV (RYCGAC, responsive to dehydration stress) [29], GT1GMSCAM4 (GAAAAA, responsive to pathogen and salt stress) [30], GCCCORE (GCCGCC, responsive to pathogen) [31], MYB1LEPR (GTTAGTT, responsive to defence) [32], CRTDREHVCBF2 (GTCGAC, responsive to low-temperature) [33], and LTRECOREATCOR15 (CCGAC, responsive to low-temperature) [34] (Figure 4). Together, the promoters of BjuPYLs contained diversities of cis-elements responsive to ABA, auxin, SA (salicylic acid), dehydration stress, pathogen, salt stress, and low-temperature, indicating that the BjuPYLs genes might be involved in the regulation of the response of B. juncea var. tumida to hormone and stresses.
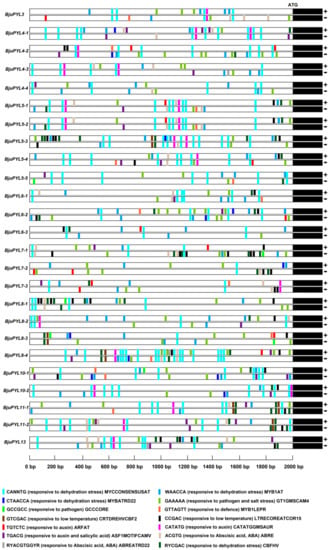
Figure 4.
The promoter cis-elements analysis of BjuPYLs. The 2 kb DNA fragments upstream of the ATG staring code of BjuPYLs were analyzed using online analysis software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace).
3.5. The Tissue-Specific Expression Pattern Analysis of BjuPYLs
To investigate the tissue-specific expression patterns of BjuPYLs, we analyzed the genes’ expression levels at different growth stages and tissues (root, stem, swollen stem, leaf, pod, and flower) using qRT-PCR. The results showed that BjuPYL4-1 and BjuPYL6-1 highly expressed in pod; BjuPYL4-1 and BjuPYL5-2 highly expressed in leaf; BjuPYL5-2, BjuPYL5-5, BjuPYL6-1, and BjuPYL7-3 highly expressed in swollen stem; BjuPYL5-2, BjuPYL6-1, and BjuPYL7-3 highly expressed in stem; BjuPYL4-1, BjuPYL5-2, and BjuPYL7-3 highly expressed in root, indicating that different BjuPYLs might be involved in different growth and development stages, and the expression patterns of BjuPYLs were existence of space-time specificity (Figure 5). In contrast, the expression levels of BjuPYL4-3, BjuPYL4-4, BjuPYL5-4, BjuPYL6-2, BjuPYL6-3, BjuPYL8-1, BjuPYL8-2, BjuPYL8-4, and BjuPYL11-2 were very low, with nearly no expression in all the tissues, indicating that these BjuPYLs genes had limited function during plant growth and development (Figure 5). Interestingly, we found that the expression level of BjuPYL5-5 was much higher in the swollen stem than that in the stem, suggesting that BjuPYL5-5 might play a role in regulating stem swelling (Figure 5).
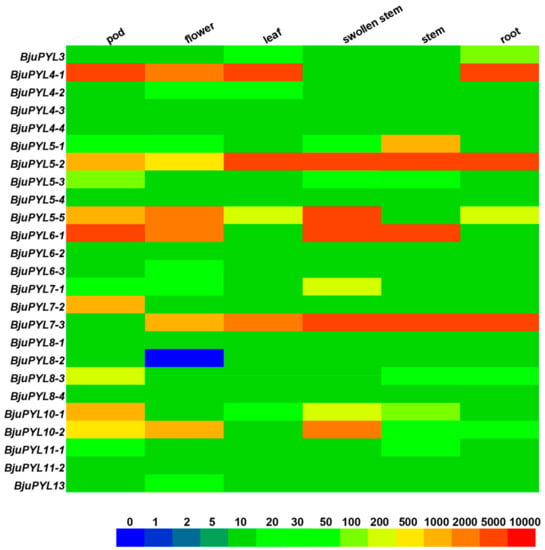
Figure 5.
Expression levels of BjuPYLs in different tissues. Tissue-specific expression pattern of BjuPYLs was analyzed by qPCR. BjuActin3 was used as internal control. The boxes display the gene expression levels, and different colors represent different expression levels.
3.6. The Gene Expression Levels of BjuPYLs in B. juncea var. tumida Under Abiotic Stress
To further explore the expression levels of BjuPYLs in B. juncea var. tumida under abiotic stresses treatment, qRT-PCR was performed using the 7-day-old seedlings treated with 200 mM NaCl, 50 μM ABA, and at low-temperature (4 °C) for 3 h. Under low-temperature stress condition, the transcript levels of BjuPYL3, BjuPYL4-1, BjuPYL5-5, BjuPYL6-1, BjuPYL7-3, BjuPYL8-3, and BjuPYL8-4 were induced significantly; however, there were no obvious expression differences between CK (control check) and low-temperature treatment of other BjuPYLs, indicating that BjuPYL3, BjuPYL4-1, BjuPYL5-5, BjuPYL6-1, BjuPYL7-3, BjuPYL8-3, and BjuPYL8-4 regulated the response of B. juncea var. tumida to low-temperature stress (Figure 6). BjuPYL4-1, BjuPYL5-4, BjuPYL5-5, BjuPYL6-1, BjuPYL6-2, BjuPYL6-3, BjuPYL7-3, and BjuPYL8-3 were induced significantly after ABA treatment; in contrast, the gene expression levels of other BjuPYLs were stable, suggesting BjuPYL4-1, BjuPYL5-4, BjuPYL5-5, BjuPYL6-1, BjuPYL6-2, BjuPYL6-3, BjuPYL7-3, and BjuPYL8-3 might be involved in ABA signaling pathway (Figure 6). Under NaCl treatment, BjuPYL5-4, BjuPYL5-5, BjuPYL6-2, BjuPYL8-3, and BjuPYL8-4 were highly induced by salt stress, indicating that these BjuPYLs play roles in plant response to salt stress (Figure 6). Taken together, the expression patterns of BjuPYLs changed under NaCl, ABA, and low-temperature treatments, indicating that BjuPYLs in B. juncea var. tumida might be important candidates for regulating plant tolerance to abiotic stresses.
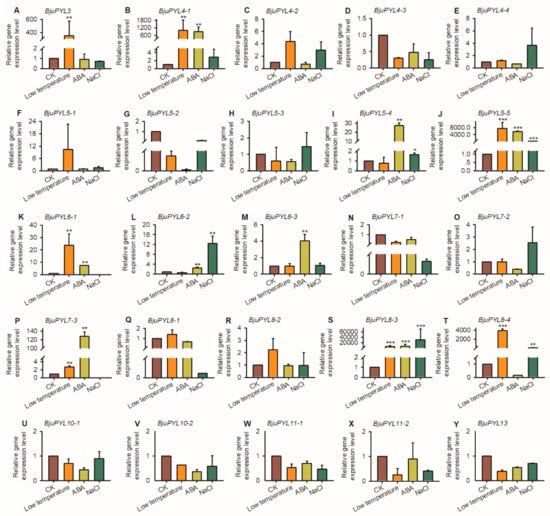
Figure 6.
The expression patterns of BjuPYLs under abiotic stresses. Total RNA was extracted from 7-day-old seedlings treated with 200 mM NaCl, 50 μM ABA, and at low-temperature (4 °C) for 3 h. Data were normalized to the expression level of BjuActin3. The values are means ± standard error. Three independent biological repeats were performed. CK: Control Check; ABA: Abscisic acid.
3.7. The Gene Expression Levels of BjuPYLs in B. juncea var. tumida under Pathogen Treatment
P. brassicae is a main and serious pathogen of B. juncea var. tumida, which usually results in the formation of clubroot and restricts the growth and development of crucifer plants. To investigate the function of BjuPYLs during plant response to P. brassicae, 2-week-old seedlings were treated with P. brassicae (OD600 = 0.07) for 0 d, 0.25 d, 0.5 d, 1 d, 3 d, 5 d, 7 d, and 9 d. qRT-PCR assay was performed, and the result showed that gene expression levels of BjuPYL3, BjuPYL4s, BjuPYL5s, BjuPYL6s, BjuPYL7s, BjuPYL8s, and BjuPYL13 were highly induced by P. brassicae, especially at 1 d and 3 d after pathogen treatment; in contrast, the other BjuPYLs showed similar expression levels after P. brassicae treatment (Figure 7). Taken together, BjuPYLs induced by pathogen (P. brassicae) treatment might be important candidates for regulating plant response to P. brassicae.
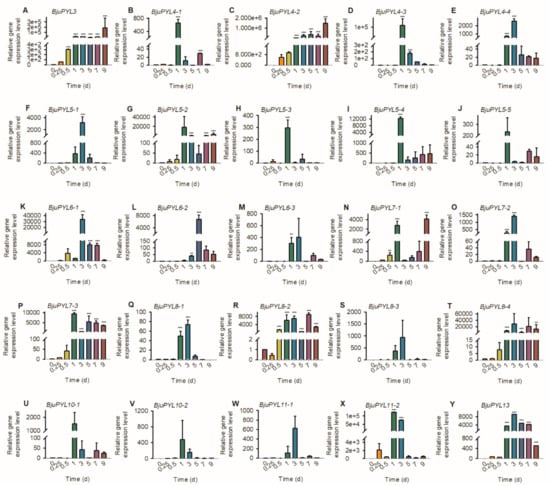
Figure 7.
The expression patterns of BjuPYLs under pathogen treatment. Total RNA was extracted from 2-week-old seedlings treated with P. brassicae for the indicated time points. Data were normalized to the expression level of BjuActin3. The values are means ± standard error. Three independent biological repeats were performed.
3.8. The Expression Patterns of BjuPYLs in B. juncea var. tumida During Stem Swelling Stages
To further explore the roles of BjuPYLs in regulating the stem swelling of B. juncea var. tumida, the qRT-PCR assay was performed. We collected the samples of B. juncea var. tumida, which were grown in the field at different growth stages, and named the samples as D1 (the stems of 1-month-old seedlings, six leaf stage), D2 (the stems of 2-month-old seedlings, primary stage of stem swelling), D3 (the stems of 3-month-old seedlings, early stage of stem swelling), D4 (the stems of 4.5-month-old seedlings, fast-growing stage of stem swelling), and D5 (the stems of 5-month-old seedlings, last stage of stem swelling). The qPCR result showed that BjuPYL4-3, BjuPYL5-3, BjuPYL5-5, BjuPYL6-2, and BjuPYL8-2 were induced with the stem swelling, and at D4 stage (fast-growing stage of stem swelling), the expression levels of these BjuPYL genes were highest, and at D5 stage (last stage of stem swelling), the expression levels decreased, indicating that BjuPYL4-3, BjuPYL5-3, BjuPYL5-5, BjuPYL6-2, and BjuPYL8-2 might be involved in regulating stem swelling of B. juncea var. tumida (Figure 8).
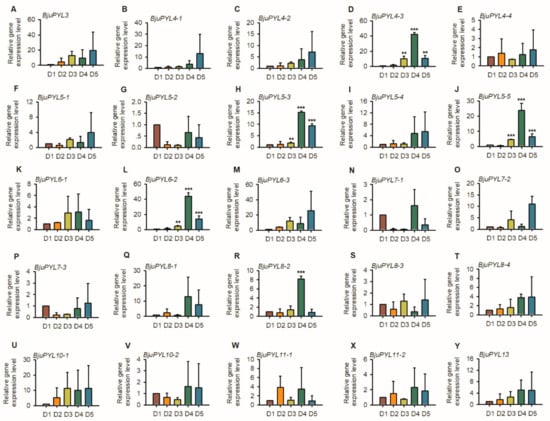
Figure 8.
The expression patterns of BjuPYLs at different stages of stem swelling. Total RNA was extracted from the seedlings of D1, D2, D3, D4, and D5 stages. Data were normalized to the expression level of BjuActin3. The values are means ± standard error. Three independent biological repeats were performed.
4. Discussion
The ABA receptors PYR/PYL/RCAR are core regulatory components of the ABA signaling pathway, which functions for ABA perception and signaling [6,12,16,35]. In our study, we identified 25 ABA receptor family genes in the B. juncea var. tumida genome and analyzed the functions of BjuPYLs in the regulation of B. juncea var. tumida responding to abiotic stresses, biotic stresses, and stem swelling. We noticed that 25 PYL homologous genes to 14 Arabidopsis PYL genes were found in B. juncea var. tumida, and the phylogenetic analysis using the PYL protein sequences was consistent with that of the PYL genomic sequences, indicating that the PYL family genes were conserved in Arabidopsis and B. juncea var. tumida (Figure 2, Figure S1). Most of the PYLs had more than two homologs; however, PYR1/PYL1/PYL2/PYL9/PYL12 did not have homologous genes, and PYL3/PYL13 only had one homologous gene in B. juncea var. tumida genome. The loss or not duplication of homologs suggests that these homologous genes may perform a functional redundancy or divarication during the evolutionary process. The expansion and loss of some PYL family genes in the B. juncea var. tumida genome suggest their possible functional differentiation in response to environmental conditions. The number of PYL genes was possibly sufficient for B. juncea var. tumida against stress from the outer environment during the long evolutionary process [36]. The losses of genes during the genome duplication event also frequently exist in other species, such as the PYLs in rice, PYLs in Gossypium, and the chitinase family genes in B. rapa [36,37,38].
B. juncea var. tumida is an allotetraploid species resulted from hybridization between B. rapa and B. nigra following with genome duplication [17]. In Arabidopsis thaliana, 14 PYLs genes were identified [6,10,11,12]. According to our results, 25 BjuPYLs were found in the genome of B. juncea var. tumida. PYL4, PYL5, and PYL8 had four homologous genes located in A sub-genome and B sub-genome; PYL6 and PYL7 had three homologous genes located in A sub-genome and B sub-genome; PYL10 and PYL11 had two homologous genes located in A sub-genome and B sub-genome; PYL3 and PYL13 only had one homologous gene located in B sub-genome B05 and A sub-genome A01, respectively (Figure 1, Table 1). The comparable homologous gene numbers in A sub-genome and B sub-genome indicated that the B. juncea var. tumida genome experienced co-linearity gene duplication [36].
The roles of PYLs in regulating plant response to abiotic stresses have been investigated in many plants. In rice, a total of 13 OsPYLs were identified, and expressions of most OsPYLs were detected in all tissues. OsPYL2 and OsPYL9 expressed highly in stem, leaf, and embryo; OsPYL3 mainly expressed in stem and leaf; OsPYL5 had a higher expression level in leaf; OsPYL7, OsPYL8 had a higher expression level in embryo. The gene expression levels of OsPYLs under ABA treatment were regulated differently, with the downregulation of OsPYL1, OsPYL2/9, and OsPYL3; the upregulation of OsPYL4; the stable expression of OsPYL5, OsPYL7/8, and OsPYL10 [37]. In Gossypium, 21, 20, 40, and 39 PYL genes were identified in the genomes of Gossypium arboretum, Gossypium raimondii, Gossypium hirsutum, and Gossypium barbadense, respectively. The transcription levels of many GhPYLs were inhibited by ABA treatment and induced by osmotic stress [38]. In this study, according to the results of promoter cis-elements analysis, all the BjuPYLs promoters contained diversities of cis-elements responsive to plant hormones (ABA, auxin, and SA), abiotic stresses (drought, cold, and salt stresses), and pathogen stresses, indicating that the BjuPYLs were regulated by hormone, abiotic stresses, and biotic stresses (Figure 4). The gene expression levels of BjuPYL4-3, BjuPYL4-4, BjuPYL5-4, BjuPYL6-2, BjuPYL6-3, BjuPYL8-1, BjuPYL8-2, BjuPYL8-4, and BjuPYL11-2 were very low in all tissues, indicating that these BjuPYLs genes had limited function in the regulation of plant growth and development (Figure 5). According to the results, we analyzed the expression levels of BjuPYLs using qPCR assay under various stresses. The results showed that BjuPYLs were induced by NaCl, low-temperature, and ABA, especially for BjuPYL5-5 and BjuPYL8-3, suggesting that BjuPYLs played roles in plant response to abiotic stress (Figure 6). P. brassicae is a crucial pathogen, which leads to the formation of clubroot. The induction of BjuPYL3, BjuPYL4-2, BjuPYL5-2, BjuPYL6-1, BjuPYL7-3, BjuPYL8s, and BjuPYL13 by P. brassicae indicated that they might be involved in plant response to P. brassicae (Figure 7). Interestingly, we also found that BjuPYL4-3, BjuPYL5-3, BjuPYL5-5, BjuPYL6-2, and BjuPYL8-2 were highly induced at D4 stage (the fast-growing stage), suggesting that these BjuPYLs might play roles in stem swelling of B. juncea var. tumida (Figure 8). In conclusion, our study identified 25 BjuPYLs in B. juncea var. tumida genome and analyzed their transcript levels under biotic stress, abiotic stresses, and different development stages, indicating that the BjuPYLs might potentially be utilized for improving the tolerance of B. juncea var. tumida to stresses and regulating stem swelling.
5. Conclusions
In this study, a total of 25 PYL homologous genes were identified in the B. juncea var. tumida genome. Based on the bioinformatics analysis, PYL homologous genes shared similar gene characteristics and high conservation. We also found that all the promoters of BjuPYLs contained hormone and stress-related cis-elements. Gene expression analysis showed that the BjuPYLs were induced by abiotic stress (NaCl, low-temperature, and ABA) and biotic stress (P. brassicae), and these ABA receptors also played roles in regulation stem swelling in B. juncea var. tumida. Our results indicated that BjuPYLs played a crucial role in plant response to stresses and organ development, and the study laid a foundation for further investigations of PYL family genes in B. juncea var. tumida.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/6/470/s1; Table S1: The primers used in this study, Figure S1: The phylogenic tree of BjuPYLs and AtPYLs, Figure S2: The alignment of PYLs proteins and protein domain prediction.
Author Contributions
C.L. and C.C. conceived the project and designed the experiments; Y.Z., Z.C., and R.S. performed the experiments and analyzed the data; C.L. and C.C. prepared the manuscript; C.C., C.L., and Z.C. revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Program Nos. 31701451), Chongqing technology innovation and application demonstration project (social livelihood) (Program No. cstc2018jscx-msybx0228), Chongqing Basic Research and Frontier Exploration Project (Program No. cstc2018jcyjAX0628), the Science and Technology Research Program of Chongqing Municipal Education Commission (Grant No. KJQN201801434), and the Yangtze Normal University Startup Foundation for Dr. Scientific Research (Program Nos. 2017KYQD151 and 2017KYQD152).
Conflicts of Interest
The authors declare that they have no competing interests for this research.
References
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; Hanhart, C.J.; Hilhorst, H.W.; Karssen, C.M. In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 1989, 90, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- Kim, T.H.; Bohmer, M.; Hu, H.; Nishimura, N.; Schroeder, J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010, 61, 561–591. [Google Scholar] [CrossRef]
- Kong, L.; Cheng, J.; Zhu, Y.; Ding, Y.; Meng, J.; Chen, Z.; Xie, Q.; Guo, Y.; Li, J.; Yang, S.; et al. Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat. Commun. 2015, 6, 8630. [Google Scholar] [CrossRef]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an abscisic acid signalling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernandez, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Lackman, P.; Gonzalez-Guzman, M.; Tilleman, S.; Carqueijeiro, I.; Pérez, A.C.; Moses, T.; Seo, M.; Kanno, Y.; Häkkinen, S.T.; Van-Montagu, M.C.; et al. Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci. USA 2011, 108, 5891–5896. [Google Scholar] [CrossRef] [PubMed]
- Bueso, E.; Rodriguez, L.; Lorenzo-Orts, L.; Gonzalez-Guzman, M.; Sayas, E.; Muñoz-Bertomeu, J.; Ibañez, C.; Serrano, R.; Rodriguez, P.L. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. Plant J. 2015, 80, 1057–1071. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, K.; Hwang, H.; Bhatnagar, N.; Kim, D.Y.; Yoon, I.S.; Byun, M.O.; Kim, S.T.; Jung, K.H.; Kim, B.G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014, 65, 453–464. [Google Scholar] [CrossRef]
- Di, F.; Jian, H.; Wang, T.; Chen, X.; Ding, Y.; Du, H.; Lu, K.; Li, J.; Liu, L. Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to abiotic stress in Brassica napus. Genes 2018, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y.; et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef]
- Shi, H.; Wang, L.L.; Sun, L.T.; Dong, L.L.; Liu, B.; Chen, L.P. Cell division and endoreduplication play important roles in stem swelling of tuber mustard (Brassica juncea Coss. var. tumida Tsen et Lee). Plant Biol. 2012, 14, 956–963. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Melcher, K.; Ng, L.M.; Zhou, X.E.; Soon, F.F.; Xu, Y.; Suino-Powell, K.M.; Park, S.Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009, 462, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Katsura, K.; Maruyama, K.; Narusaka, Y.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 2006, 60, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Nag, R.; Maity, M.K.; Dasgupta, M. Dual DNA binding property of ABA insensitive 3 like factors targeted to promoters responsive to ABA and auxin. Plant Mol. Biol. 2005, 59, 821–838. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Hagen, G.; Guilfoyle, T. Multiple auxin response modules in the soybean SAUR 15A promoter. Plant Sci. 1997, 126, 193–201. [Google Scholar] [CrossRef]
- Redman, J.; Whitcraft, J.; Johnson, C.; Arias, J. Abiotic and biotic stress differentially stimulate as-1 element activity in Arabidopsis. Plant Cell Rep. 2002, 21, 180–185. [Google Scholar]
- Iwasaki, T.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Identification of a cis -regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol. Gen. Genet. 1995, 247, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Xue, G.P. Characterisation of the DNA-binding profile of barley HvCBF1 using an enzymatic method for rapid, quantitative and high-throughput analysis of the DNA-binding activity. Nucleic Acids Res. 2002, 30, e77. [Google Scholar] [CrossRef]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H.; et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef]
- Brown, R.L.; Kazan, K.; McGrath, K.C.; Maclean, D.J.; Manners, J.M. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003, 132, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, S. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 2003, 15, 3033–3050. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.P. The DNA-binding activity of an AP2 transcriptional activator HvCBF2 involved in regulation of low-temperature responsive genes in barley is modulated by temperature. Plant J. 2003, 33, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.S.; Wilhelm, K.S.; Thomashow, M.F. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 1994, 24, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, Z.; Ren, Z.; Zhi, L.; Yao, B.; Su, C.; Liu, L.; Li, X. SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PLoS Genet. 2017, 13, e1006947. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Piao, Y.; Liu, Y.; Li, X.; Piao, Z. Genome-wide identification and expression analysis of chitinase gene family in Brassica rapa reveals its role in clubroot resistance. Plant Sci. 2018, 270, 257–267. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice 2015, 8, 28. [Google Scholar] [CrossRef]
- Zhang, G.; Lu, T.; Miao, W.; Sun, L.; Tian, M.; Wang, J.; Hao, F. Genome-wide identification of ABA receptor PYL family and expression analysis of PYLs in response to ABA and osmotic stress in Gossypium. PeerJ 2017, 5, e4126. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).