Prediction of Bone Metastasis in Breast Cancer Based on Minimal Driver Gene Set in Gene Dependency Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sets and Pre-Processing
2.2. Construction of Gene Dependency Network
- (1)
- In order to reduce the false discovery rate, only the gene pairs involved in the protein–protein interaction network were used as candidates. Here, all the gene pairs in HIPPIE [22] were set as candidates for gene dependency pairs.
- (2)
- For each candidate gene dependency pair (e.g., gene A and gene B), we set the expression levels of gene A and gene B and the bone-metastasis risks of all the patients as a triple. The triple was sorted according to the expression level of gene B in ascending order. Then, the conditional mutual information was calculated by using Equation (1):where is the mutual information of gene A and the bone-metastasis risks of 35% cancer patients whose expression levels of gene B are high. is the mutual information of gene A and the bone-metastasis risks of 35% of cancer patients whose expression levels of gene B are low.
- (3)
- A permutation test was proposed to calculate the p-value of the conditional mutual information for each gene pair. First, we randomly permuted the expression levels of gene B. Secondly, a random CMI was calculated for gene A and gene B using the method described in step (2). Thirdly, the random permutation was repeated 1000 times and 1000 random CMIs were obtained, and the p-value of the CMI was calculated based on the 1000 random CMIs. Finally, all the significant gene dependency pairs (p-value < 0.05) were combined as the gene dependency network.
2.3. Structural Controllability of Networks
2.4. Selecting the Driver Gene Sets by Controllability
2.5. Construction of the Classifier
2.6. Tools and Package
3. Results
3.1. Basic Information of the Gene Dependency Network
3.2. Functional Annotation of the Driver Nodes
3.3. The Selected Features
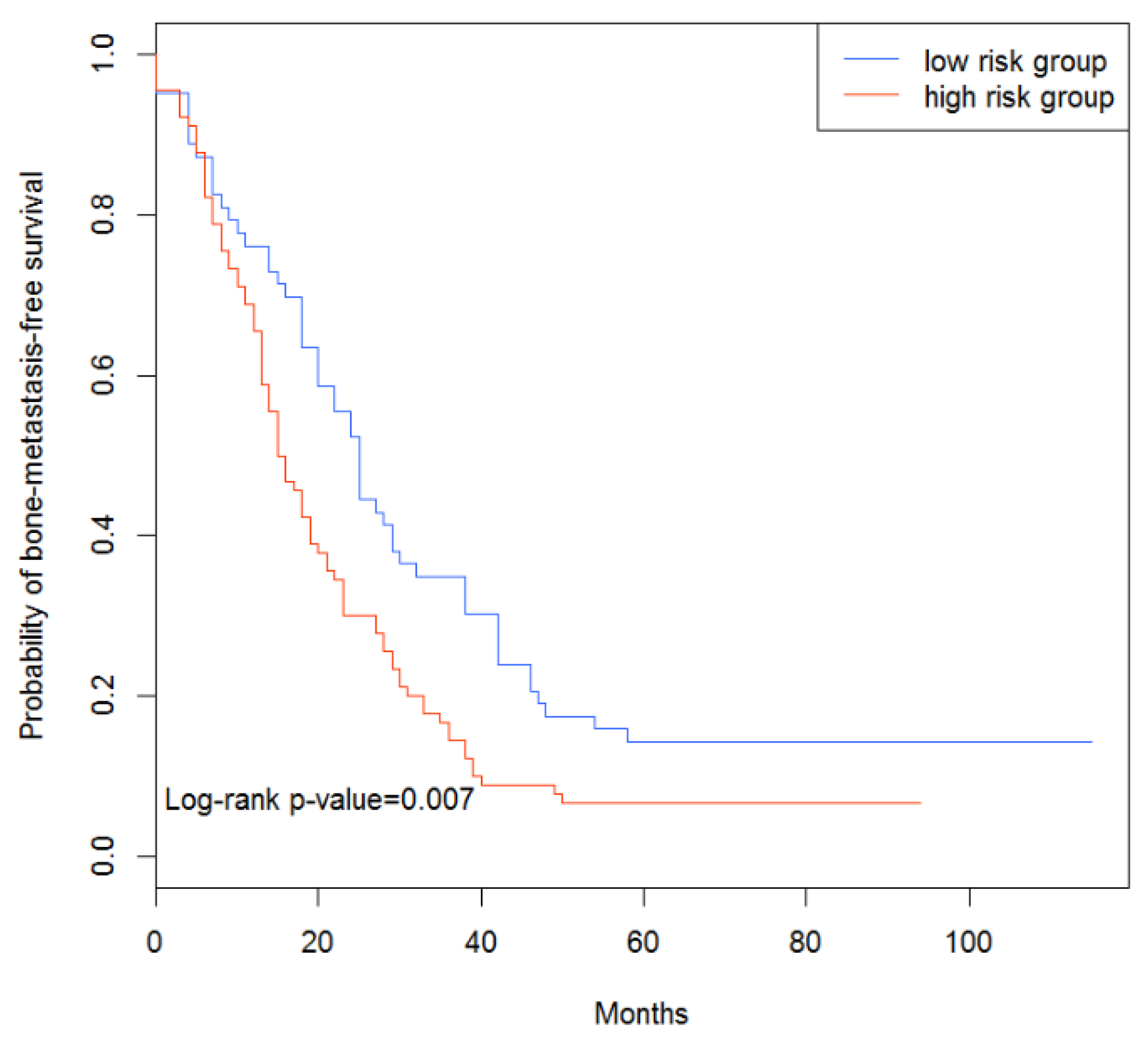
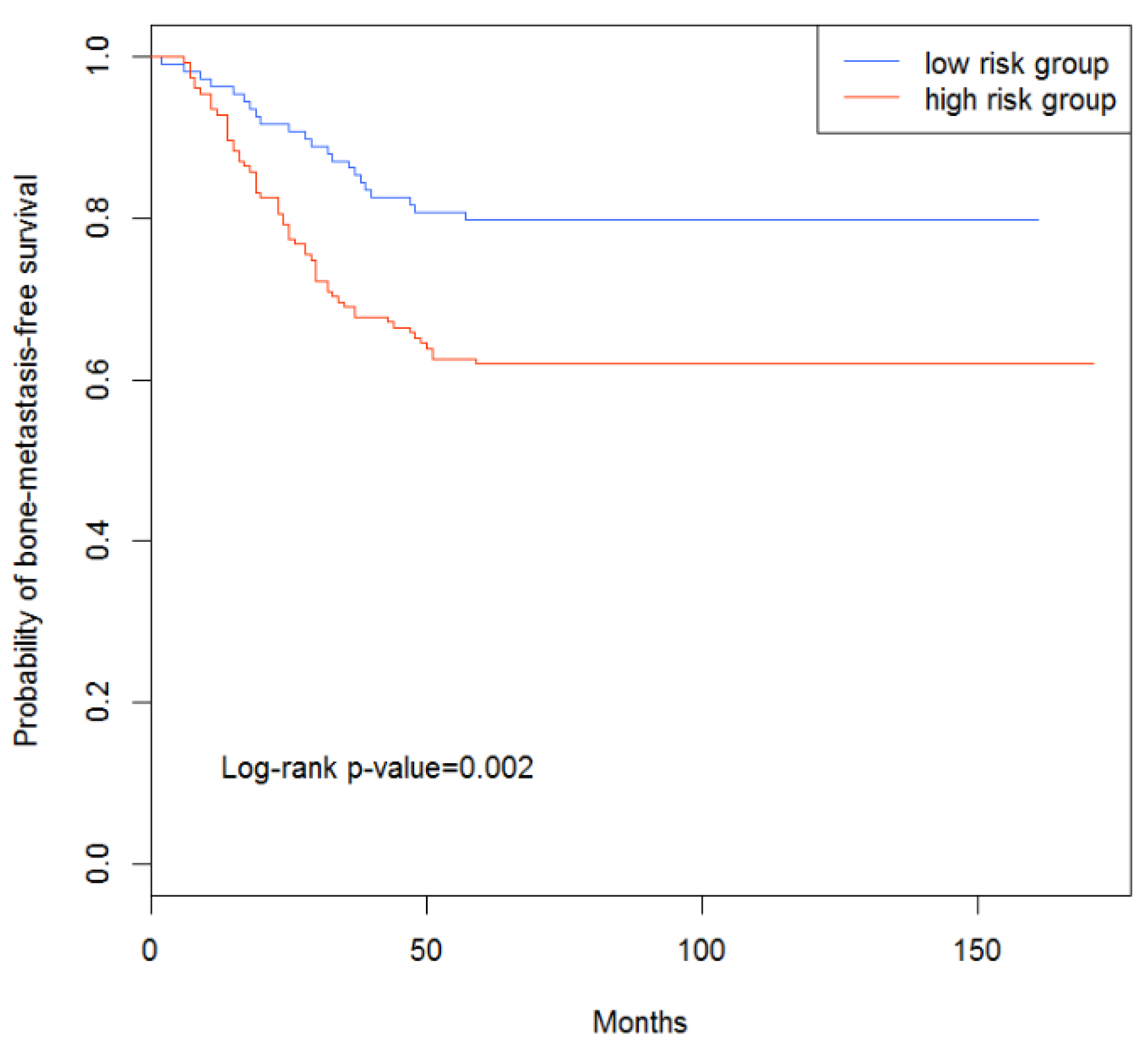
3.4. Survival Analysis of Breast Cancer Patients
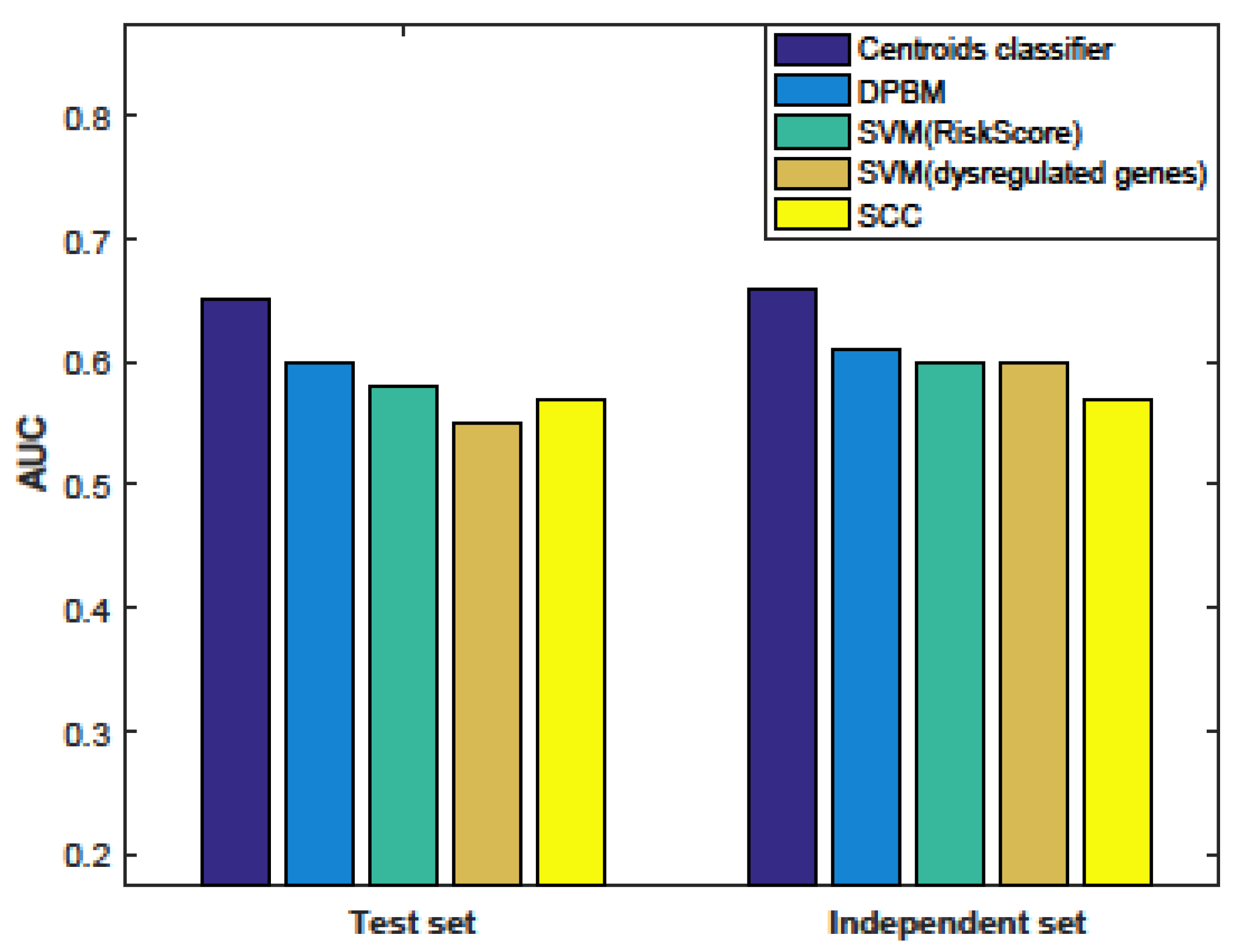
3.5. Comparison with Other Methods
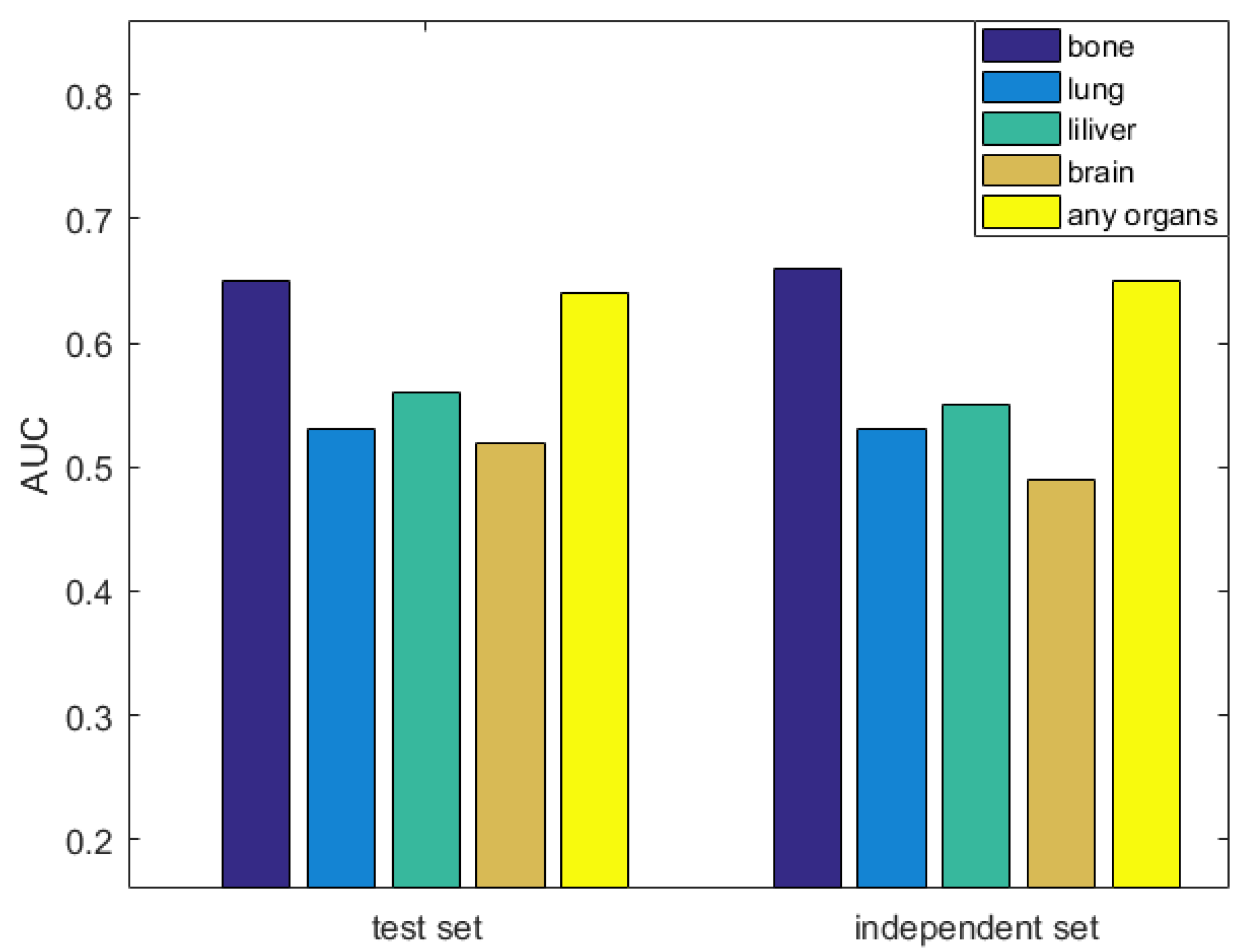
3.6. Evaluation the Significance of Our Method
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hedayat, A.; Parvin, H.; Abazar, R.; Seyyed Hamid, M.; Shokrallah, M.; Mojtaba, S.; Mohsen, B.; Ali, A. Relationships between Breast Cancer and Common Non-Communicable Disease Risk Factors: An Ecological Study. Asian Pac. J. Cancer Prev. APJCP 2013, 14, 5123–5125. [Google Scholar]
- Feig, S.A. 4–8 A Basal Epithelial Phenotype Is More Frequent in Interval Breast Cancers Compared with Screen Detected Tumors. Breast Dis. Year Book Q. 2006, 16, 337–338. [Google Scholar] [CrossRef]
- Thomas, L.; Amanda, J.; Akeila, B.; Nadia, R.; Soraya, S.; Berta Martin, A.; Angels, S.; Alain, B.; Jean-Marc, G.; Enrico, R. A six-gene signature predicting breast cancer lung metastasis. Cancer Res. 2008, 68, 6092. [Google Scholar]
- Müller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; Mcclanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Nogi, H.; Takeyama, H.; Uchida, K.; Agata, T.; Horiguchi-Yamada, J.; Yamada, H. Detection of MUC1 and keratin 19 mRNAs in the bone marrow by quantitative RT-PCR predicts the risk of distant metastasis in breast cancer patients. Breast Cancer 2003, 10, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Kingsley, L.A.; Fournier, P.G.; Chirgwin, J.M.; Guise, T.A. Molecular biology of bone metastasis. Mol. Cancer Ther. 2007, 6, 2609–2617. [Google Scholar] [CrossRef]
- Akhtari, M.; Mansuri, J.; Newman, K.A.; Guise, T.; Seth, P. Biology of breast cancer bone metastasis. Cancer Biol. Ther. 2008, 7, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Woelfle, U.; Cloos, J.; Sauter, G.; Riethdorf, L.; Jänicke, F.; Van, D.P.; Brakenhoff, R.; Pantel, K. Molecular signature associated with bone marrow micrometastasis in human breast cancer. Cancer Res. 2003, 63, 5679–5684. [Google Scholar]
- Zhou, X.; Liu, J. A computational model to predict bone metastasis in breast cancer by integrating the dysregulated pathways. BMC Cancer 2014, 14, 618. [Google Scholar] [CrossRef]
- Khalid, S.; Hanif, R.; Tareen, S.H.K.; Siddiqa, A.; Bibi, Z.; Ahmad, J. Formal modeling and analysis of ER-αassociated Biological Regulatory Network in breast cancer. PeerJ 2016, 4, e2542. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J. Inferring gene dependency network specific to phenotypic alteration based on gene expression data and clinical information of breast cancer. PLoS ONE 2014, 9, e92023. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gao, H.; Wang, J.; Wu, F. Control principles for complex biological networks. Brief. Bioinform. 2018. [Google Scholar] [CrossRef] [PubMed]
- Yang-Yu, L.; Jean-Jacques, S.; Albert-László, B. Controllability of complex networks. Nature 2011, 473, 167. [Google Scholar]
- Motakis, E.; Ivshina, A.V.; Kuznetsov, V.A. Data-driven approach to predict survival of cancer patients: Estimation of microarray genes’ prediction significance by Cox proportional hazard regression model. IEEE Eng. Med. Biol. Mag. 2009, 28, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Harrell, J.C.; Aleix, P.; Parker, J.S.; Cheng, F.; Xiaping, H.; Lisa, C.; Carey, A.; Matthew, E.; Perou, C.M. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res. Treat. 2012, 132, 523–535. [Google Scholar]
- Hayes, D.F. 2–20 Gene-Expression Profiles to Predict Distant Metastasis of Lymph-Node-Negative Primary Breast Cancer. Lancet 2006, 17, 154–155. [Google Scholar] [CrossRef]
- Minn, A.J.; Gupta, G.P.; Siegel, P.M.; Bos, P.D.; Weiping, S.; Giri, D.D.; Agnes, V.; Olshen, A.B.; Gerald, W.L.; Joan, M. Genes that mediate breast cancer metastasis to lung. Nature 2005, 436, 518–524. [Google Scholar] [CrossRef]
- Bos, P.D.; Zhang, X.H.F.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; van de Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef]
- Aziz, N.A.A.; Mokhtar, N.M.; Harun, R.; Mollah, M.M.H.; Rose, I.M.; Sagap, I.; Tamil, A.M.; Wan, Z.W.N.; Jamal, R. A 19-Gene expression signature as a predictor of survival in colorectal cancer. BMC Med. Genomics 2016, 9, 58. [Google Scholar]
- Tomich, P.L.; Helgeson, V.S.J.P.-O. Five years later: A cross-sectional comparison of breast cancer survivors with healthy women. Psycho-Oncol. J. Psychol. Soc. Behav. Dimens. Cancer 2010, 11, 154–169. [Google Scholar] [CrossRef]
- Snyder, C.F.; Frick, K.D.; Peairs, K.S.; Kantsiper, M.E.; Herbert, R.J.; Blackford, A.L.; Wolff, A.C.; Earle, C.C. Comparing care for breast cancer survivors to non-cancer controls: A five-year longitudinal study. J. Gen. Intern. Med. 2009, 24, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.H.; Fontaine, J.; Vinayagam, A.; Porras, P.; Wanker, E.E.; Andradenavarro, M.A. HIPPIE: Integrating protein interaction networks with experiment based quality scores. PLoS ONE 2012, 7, e31826. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.; Pearson, J. Structural controllability of multiinput linear systems. IEEE Trans. Autom. Control 1976, 21, 203–212. [Google Scholar] [CrossRef]
- Galil, Z. Efficient algorithms for finding maximal matching in graphs. In Colloquium on Trees in Algebra and Programming; Springer: Berlin/Heidelberg, Germany, 1983; Volume 159, pp. 90–113. [Google Scholar] [CrossRef]
- Bedo, J.; Sanderson, C.; Kowalczyk, A. An efficient alternative to SVM based recursive feature elimination with applications in natural language processing and bioinformatics. In Proceedings of the Australian Joint Conference on Artificial Intelligence: Advances in Artificial Intelligence, Hobart, Australia, 4–8 December 2006. [Google Scholar]
- Abraham, G.; Kowalczyk, A.; Loi, S.; Haviv, I.; Zobel, J. Prediction of breast cancer prognosis using gene set statistics provides signature stability and biological context. BMC Bioinform. 2010, 11, 277. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.G.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Jinhua, X.; Yinghua, C.; Olopade, O.I. MYC and Breast Cancer. Genes Cancer 2010, 1, 629–640. [Google Scholar]
- Brown, A.M. Wnt signaling in breast cancer: Have we come full circle? Breast Cancer Res. 2001, 3, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.; Cole, M. Turning the tables: Myc activates Wnt in breast cancer. Cell Cycle 2007, 6, 2625–2627. [Google Scholar] [CrossRef]
- Anne-Lise, B.R.D. TP53 and breast cancer. Hum. Mutat. 2003, 21, 292–300. [Google Scholar]
- Chen, C.P.; Sun, Z.L.; Lu, X.; Wu, W.X.; Guo, W.L.; Lu, J.J.; Han, C.; Huang, J.Q.; Fang, Y. miR-340 suppresses cell migration and invasion by targeting MYO10 in breast cancer. Oncol. Rep. 2015, 35, 709. [Google Scholar] [CrossRef]
- Antti, A.; Riina, K.; Elina, M.; Pegah, R.; Gunilla, H.G.S.; Harri, S.; Miller, B.W.; Morton, J.P.; Elmar, B.; Pekka, T. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J. Clin. Investig. 2014, 124, 1069. [Google Scholar]
- Mary-Claire, K.; Marks, J.H.; Mandell, J.B. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 2003, 302, 643–646. [Google Scholar]
- Kleer, C.G.; Qi, C.; Sooryanarayana, V.; Ronglai, S.; Ichiro, O.; Tomlins, S.A.; Debashis, G.; Sewalt, R.G.A.B.; Otte, A.P.; Hayes, D.F. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.E.; Li, X.; Toy, K.; DuPrie, M.; Ventura, A.C.; Banerjee, M.; Ljungman, M.; Merajver, S.D.; Kleer, C.G. Downregulation of EZH2 decreases growth of estrogen receptor-negative invasive breast carcinoma and requires BRCA1. Oncogene 2009, 28, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Bangxing, H.; Haiyan, L.; Mingjun, Z.; Jingda, X.; Yong, L.; Yuhuan, Z.; Jianfei, Q.; Chang, J.T.; Jing, Y.; Qing, Y. p38 MAPK inhibits breast cancer metastasis through regulation of stromal expansion. Int. J. Cancer 2015, 136, 34–43. [Google Scholar]
- Yao, Y.; Fang, Z.P.; Chen, H.; Yue, L.; Min, D.L.; Tang, L.N.; Yu, W.X.; Kung, H.F.; Lin, M.C.; Shen, Z. HGFK1 inhibits bone metastasis in breast cancer through the TAK1/p38 MAPK signaling pathway. Cancer Gene Ther. 2012, 19, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Guan, J.L. Focal adhesion kinase: A prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett. 2010, 289, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Bagi, C.M.; Roberts, G.W.; Andresen, C.J. Dual focal adhesion kinase/Pyk2 inhibitor has positive effects on bone tumors: Implications for bone metastases. Cancer 2010, 112, 2313–2321. [Google Scholar] [CrossRef]
- Bussard, K.M.; Venzon, D.J.; Mastro, A.M. Osteoblasts are a major source of inflammatory cytokines in the tumor microenvironment of bone metastatic breast cancer. J. Cell. Biochem. 2010, 111, 1138–1148. [Google Scholar] [CrossRef]
- Kudo-Saito, C. FSTL1 promotes bone metastasis by causing immune dysfunction. OncoImmunology 2013, 2, e26528. [Google Scholar] [CrossRef]
- Johnson, R.W.; Merkel, A.R.; Page, J.M.; Ruppender, N.S.; Guelcher, S.A.; Sterling, J.A.J.C.; Metastasis, E. Wnt signaling induces gene expression of factors associated with bone destruction in lung and breast cancer. Clin. Exp. Metastasis 2014, 31, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Qiao, F.; Wei, P.; Chi, Y.; Wang, W.; Ni, S.; Wang, Q.; Chen, T.; Sheng, W.; Du, X. DIXDC1 activates the Wnt signaling pathway and promotes gastric cancer cell invasion and metastasis. Mol. Carcinog. 2016, 55, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Sucharita, B.; Pai, S.K.; Shigeru, H.; Sadahiro, H.; Yukio, T.; Ken, S.; David, P.; Therese, C.; Misako, W.; Gross, S.C. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene 2004, 23, 5675–5681. [Google Scholar]
- Cosphiadi, I.; Atmakusumah, T.D.; Siregar, N.C.; Abdulmuthalib; Harahap, A.; Mansyur, M.J.C.B.C. Bone Metastasis in Advanced Breast Cancer: Analysis of Gene Expression Microarray. Clin. Breast Cancer 2018, 18, e1117–e1122. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Zhang, Y.; Yi, Q.; Wu, Y.; Wan, R.; Tang, L. CRIM1, a newfound cancer-related player, regulates the adhesion and migration of lung cancer cells. Growth Factors 2015, 33, 384. [Google Scholar] [CrossRef]
- Naoki, H.; Tsuyoshi, Y.; Ryoichi, Y.; Yoshihisa, N.; Hiroshi, I. RanBP10 acts as a novel coactivator for the androgen receptor. Biochem. Biophys. Res. Commun. 2008, 368, 121–125. [Google Scholar]
- Yee, L.D.; Sabourin, C.L.; Liu, L.; Li, H.M.; Smith, P.J.; Seewaldt, V.; Kniss, D.A. Peroxisome proliferator-activated receptor γ activation in human breast cancer. Int. J. Oncol. 1999, 15, 967. [Google Scholar] [CrossRef]
- Zhang, X.H.F.; Wang, Q.; Gerald, W.; Hudis, C.A.; Norton, L.; Smid, M.; Foekens, J.A.; Massagué, J. Latent Bone Metastasis in Breast Cancer Tied to Src-Dependent Survival Signals. Cancer Cell 2009, 16, 67–78. [Google Scholar] [CrossRef]
| KEGG Gene Set Name | p-Value | FDR q-Value |
|---|---|---|
| MAPK signaling pathway | 7.31 × 10−19 | 1.36 × 10−16 |
| Neuroactive ligand-receptor interaction | 1.12 × 10−15 | 1.04 × 10−13 |
| Pathways in cancer | 2.82 × 10−13 | 1.75 × 10−11 |
| Focal adhesion | 4.05 × 10−11 | 1.88 × 10−9 |
| Cytokine-cytokine receptor interaction | 8.22 × 10−11 | 3.06 × 10−9 |
| Regulation of actin cytoskeleton | 2.72 × 10−10 | 8.43 × 10−9 |
| SNARE (SNAP Receptor) interactions in vesicular transport | 1.62 × 10−9 | 4.29 × 10−8 |
| Complement and coagulation cascades | 1.26 × 10−8 | 2.92 × 10−7 |
| Purine metabolism | 3.08 × 10−8 | 6.36 × 10−7 |
| Spliceosome | 5.07 × 10−8 | 9.42 × 10−7 |
| Gene Id | Gene Name | Frequency | p-Value |
|---|---|---|---|
| 85458 | DIXDC1 | 500 | 1.68192 × 10−6 |
| 29068 | ZBTB44 | 500 | 3.29096 × 10−6 |
| 51232 | CRIM1 | 500 | 1.46030e × 10−5 |
| 9986 | RCE1 | 500 | 2.12898 × 10−5 |
| 56888 | KCMF1 | 500 | 4.00030 × 10−5 |
| 1456 | CSNK1G3 | 500 | 4.58237 × 10−5 |
| 6256 | RXRA | 500 | 7.63144 × 10−5 |
| 55343 | SLC35C1 | 500 | 0.00012 |
| 55520 | ELAC1 | 500 | 0.00012 |
| 55081 | IFT57 | 500 | 0.00012 |
| 57610 | RANBP10 | 500 | 0.00018 |
| 5877 | RABIF | 500 | 0.00018 |
| 25839 | COG4 | 500 | 0.00019 |
| 23261 | CAMTA1 | 500 | 0.00020 |
| 3009 | HIST1H1B | 500 | 0.00020 |
| 3092 | HIP1 | 500 | 0.00026 |
| 246243 | RNASEH1 | 500 | 0.00030 |
| 3104 | ZBTB48 | 500 | 0.00031 |
| 10342 | TFG | 500 | 0.00032 |
| 6282 | S100A11 | 500 | 0.00033 |
| 10462 | CLEC10A | 500 | 0.00033 |
| 51199 | NIN | 500 | 0.00041 |
| 10531 | PITRM1 | 500 | 0.00048 |
| 9856 | KIAA0319 | 500 | 0.00049 |
| 11167 | FSTL1 | 500 | 0.00049 |
| 3993 | LLGL2 | 500 | 0.00052 |
| 56729 | RETN | 500 | 0.00054 |
| 51514 | DTL | 500 | 0.00054 |
| 9202 | ZMYM4 | 500 | 0.00058 |
| 51302 | CYP39A1 | 500 | 0.00065 |
| 9971 | NR1H4 | 500 | 0.00067 |
| 79083 | MLPH | 500 | 0.00073 |
| 65082 | VPS33A | 500 | 0.00075 |
| 10179 | RBM7 | 500 | 0.00078 |
| 55794 | DDX28 | 500 | 0.00082 |
| 57405 | SPC25 | 500 | 0.00089 |
| 51659 | GINS2 | 500 | 0.00089 |
| 1852 | DUSP9 | 500 | 0.00092 |
| 57017 | COQ9 | 500 | 0.00096 |
| 10397 | NDRG1 | 500 | 0.00098 |
| 9911 | TMCC2 | 500 | 0.00128 |
| 55095 | SAMD4B | 500 | 0.00137 |
| 23649 | POLA2 | 500 | 0.00143 |
| 10615 | SPAG5 | 500 | 0.00143 |
| 7134 | TNNC1 | 500 | 0.00145 |
| 7083 | TK1 | 500 | 0.00146 |
| 9442 | MED27 | 500 | 0.00151 |
| 8449 | DHX16 | 500 | 0.00171 |
| 8817 | FGF18 | 500 | 0.00176 |
| 483 | ATP1B3 | 500 | 0.00179 |
| 2175 | FANCA | 500 | 0.00190 |
| Training Data Set | Test Data Set | Independent Data Set | |
|---|---|---|---|
| Centroids classifier | 0.22 | 0.15 | 0.20 |
| ER status | 0.18 | 0.16 | 0.07 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.-N.; Zhong, R.; Zhou, X.-H. Prediction of Bone Metastasis in Breast Cancer Based on Minimal Driver Gene Set in Gene Dependency Network. Genes 2019, 10, 466. https://doi.org/10.3390/genes10060466
Li J-N, Zhong R, Zhou X-H. Prediction of Bone Metastasis in Breast Cancer Based on Minimal Driver Gene Set in Gene Dependency Network. Genes. 2019; 10(6):466. https://doi.org/10.3390/genes10060466
Chicago/Turabian StyleLi, Jia-Nuo, Rui Zhong, and Xiong-Hui Zhou. 2019. "Prediction of Bone Metastasis in Breast Cancer Based on Minimal Driver Gene Set in Gene Dependency Network" Genes 10, no. 6: 466. https://doi.org/10.3390/genes10060466
APA StyleLi, J.-N., Zhong, R., & Zhou, X.-H. (2019). Prediction of Bone Metastasis in Breast Cancer Based on Minimal Driver Gene Set in Gene Dependency Network. Genes, 10(6), 466. https://doi.org/10.3390/genes10060466




