Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. AON Design
2.3. Subjects
2.4. iPSC Differentiation into Photoreceptor Precursor Cells (PPCs)
2.5. RNA Analysis
2.6. qPCR
2.7. AON Classification and Common Properties
3. Results
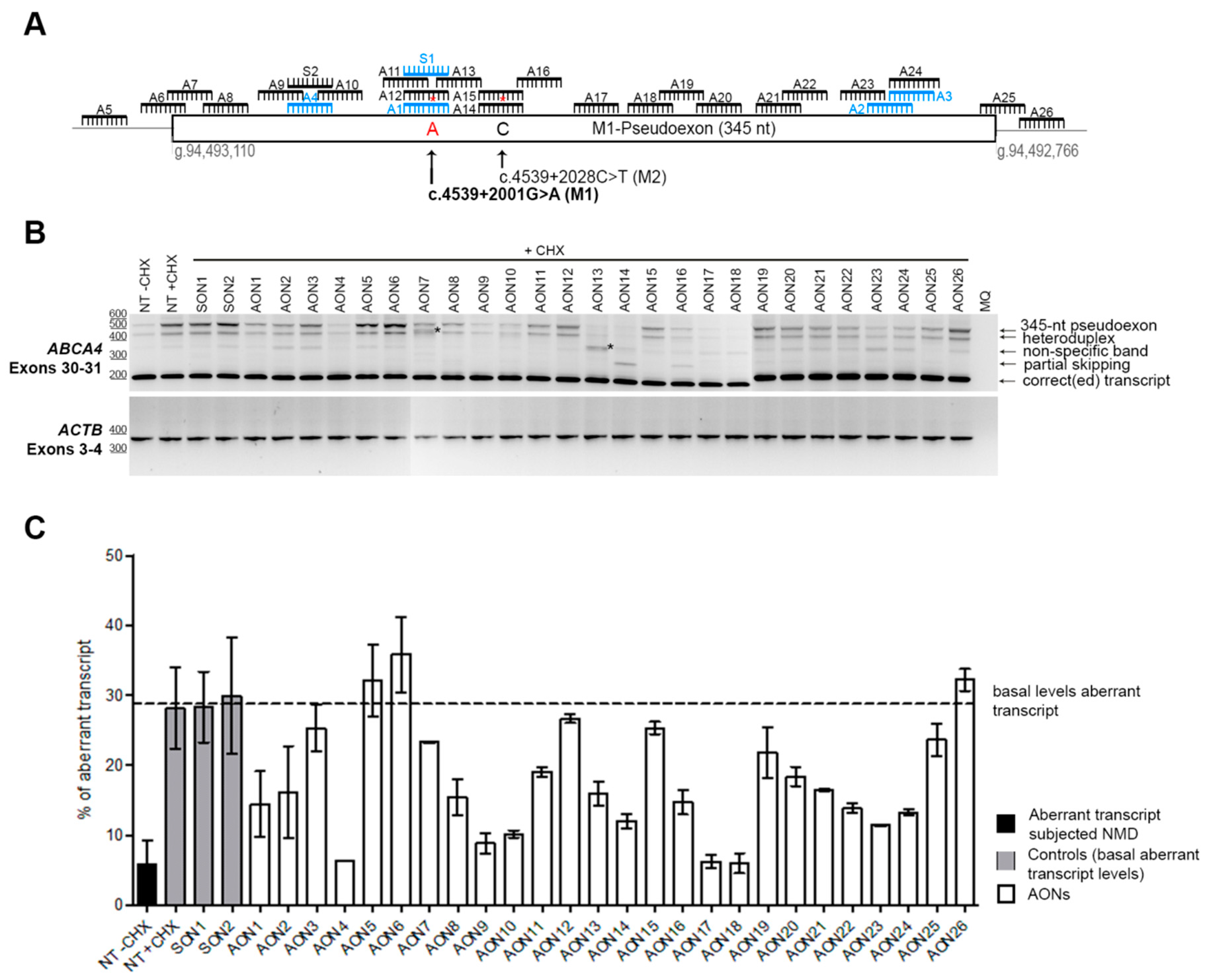
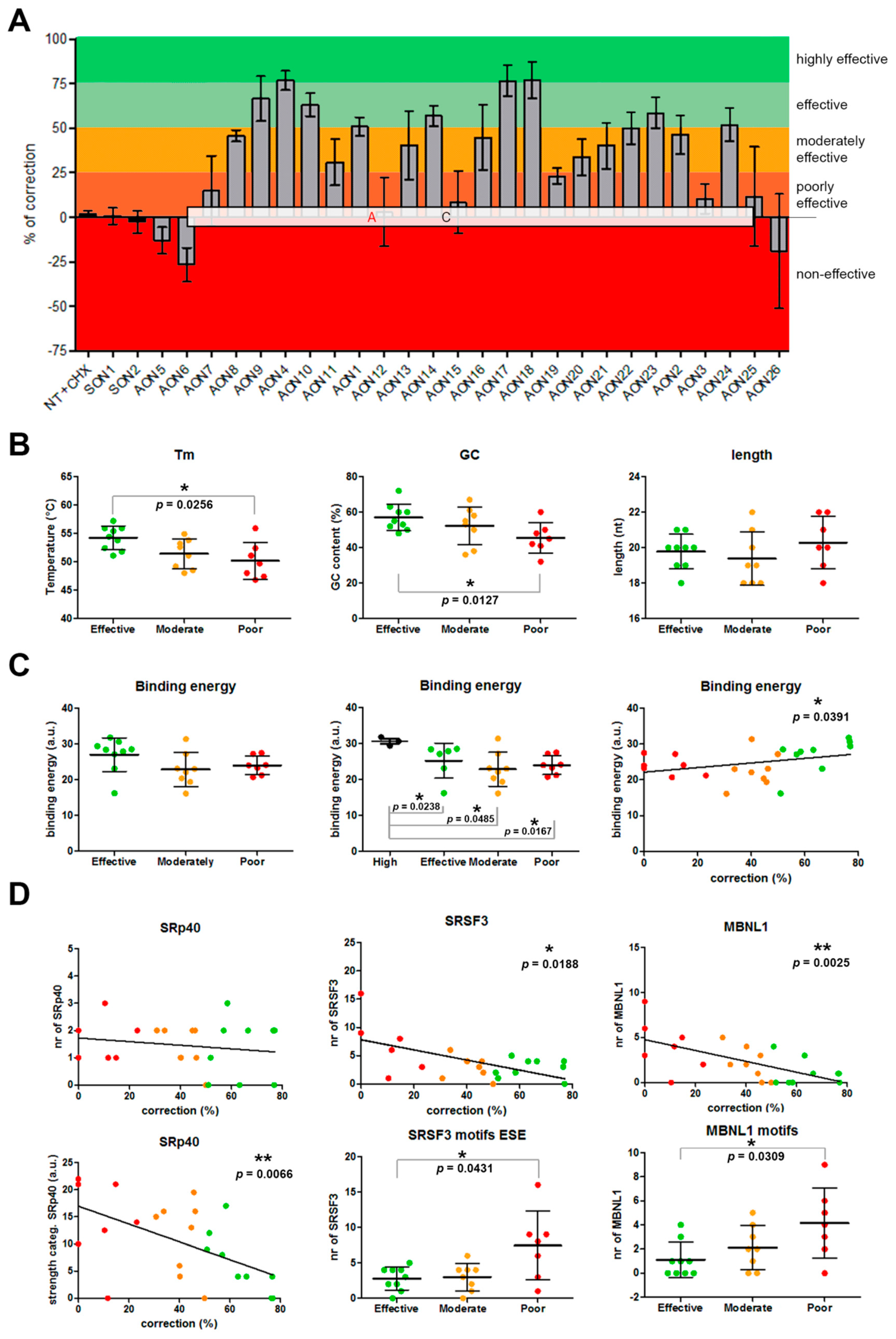
3.1. Screening and Selection of AONs
3.2. Analysis of the Properties of the AONs
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stargardt, K. Über familiäre, progressive Degeneration in der Maculagegend des Auges. Albrecht von Graefes Archiv für Ophthalmologie 1909, 71, 534–550. [Google Scholar] [CrossRef]
- Birnbach, C.D.; Jarvelainen, M.; Possin, D.E.; Milam, A.H. Histopathology and immunocytochemistry of the neurosensory retina in fundus flavimaculatus. Ophthalmology 1994, 101, 1211–1219. [Google Scholar] [CrossRef]
- Sahel, J.A.; Marazova, K.; Audo, I. Clinical characteristics and current therapies for inherited retinal degenerations. Cold Spring Harb. Perspect. Med. 2014, 5, a017111. [Google Scholar] [CrossRef] [PubMed]
- Zernant, J.; Xie, Y.A.; Ayuso, C.; Riveiro-Alvarez, R.; Lopez-Martinez, M.A.; Simonelli, F.; Testa, F.; Gorin, M.B.; Strom, S.P.; Bertelsen, M.; et al. Analysis of the ABCA4 genomic locus in Stargardt disease. Hum. Mol. Genet. 2014, 23, 6797–6806. [Google Scholar] [CrossRef] [PubMed]
- Vasiliou, V.; Vasiliou, K.; Nebert, D.W. Human ATP-binding cassette (ABC) transporter family. Hum. Genom. 2009, 3, 281–290. [Google Scholar] [CrossRef]
- Molday, R.S. Insights into the Molecular Properties of ABCA4 and Its Role in the Visual Cycle and Stargardt Disease. Prog. Mol. Biol. Transl. Sci. 2015, 134, 415–431. [Google Scholar] [PubMed]
- Molday, R.S.; Zhong, M.; Quazi, F. The role of the photoreceptor ABC transporter ABCA4 in lipid transport and Stargardt macular degeneration. Biochim. Biophys. Acta 2009, 1791, 573–583. [Google Scholar] [CrossRef]
- Quazi, F.; Lenevich, S.; Molday, R.S. ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat. Commun. 2012, 3, 925. [Google Scholar] [CrossRef]
- Cornelis, S.S.; Bax, N.M.; Zernant, J.; Allikmets, R.; Fritsche, L.G.; den Dunnen, J.T.; Ajmal, M.; Hoyng, C.B.; Cremers, F.P. In Silico Functional Meta-Analysis of 5,962 ABCA4 Variants in 3,928 Retinal Dystrophy Cases. Hum. Mutat. 2017, 38, 400–408. [Google Scholar] [CrossRef]
- Maugeri, A.; van Driel, M.A.; van de Pol, D.J.; Klevering, B.J.; van Haren, F.J.; Tijmes, N.; Bergen, A.A.; Rohrschneider, K.; Blankenagel, A.; Pinckers, A.J.; et al. The 2588G→C mutation in the ABCR gene is a mild frequent founder mutation in the Western European population and allows the classification of ABCR mutations in patients with Stargardt disease. Am. J. Hum. Genet. 1999, 64, 1024–1035. [Google Scholar] [CrossRef]
- Sheffield, V.C.; Stone, E.M. Genomics and the eye. N. Engl. J. Med. 2011, 364, 1932–1942. [Google Scholar] [CrossRef] [PubMed]
- Zernant, J.; Collison, F.T.; Lee, W.; Fishman, G.A.; Noupuu, K.; Yuan, B.; Cai, C.; Lupski, J.R.; Yannuzzi, L.A.; Tsang, S.H.; et al. Genetic and clinical analysis of ABCA4-associated disease in African American patients. Hum. Mutat. 2014, 35, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.A.; Mullins, R.F.; Wagner, A.H.; Andorf, J.L.; Johnston, R.M.; Bakall, B.B.; Deluca, A.P.; Fishman, G.A.; Lam, B.L.; Weleber, R.G.; et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum. Mol. Genet. 2013, 22, 5136–5145. [Google Scholar] [CrossRef] [PubMed]
- Bauwens, M.; De Zaeytijd, J.; Weisschuh, N.; Kohl, S.; Meire, F.; Dahan, K.; Depasse, F.; De Jaegere, S.; De Ravel, T.; De Rademaeker, M.; et al. An augmented ABCA4 screen targeting noncoding regions reveals a deep intronic founder variant in Belgian Stargardt patients. Hum. Mutat. 2015, 36, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Bax, N.M.; Sangermano, R.; Roosing, S.; Thiadens, A.A.; Hoefsloot, L.H.; van den Born, L.I.; Phan, M.; Klevering, B.J.; Westeneng-van Haaften, C.; Braun, T.A.; et al. Heterozygous deep-intronic variants and deletions in ABCA4 in persons with retinal dystrophies and one exonic ABCA4 variant. Hum. Mutat. 2015, 36, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Sangermano, R.; Bax, N.M.; Bauwens, M.; van den Born, L.I.; De Baere, E.; Garanto, A.; Collin, R.W.; Goercharn-Ramlal, A.S.; den Engelsman-van Dijk, A.H.; Rohrschneider, K.; et al. Photoreceptor Progenitor mRNA Analysis Reveals Exon Skipping Resulting from the ABCA4 c.5461-10T→C Mutation in Stargardt Disease. Ophthalmology 2016, 123, 1375–1385. [Google Scholar] [CrossRef] [PubMed]
- Zernant, J.; Lee, W.; Nagasaki, T.; Collison, F.T.; Fishman, G.A.; Bertelsen, M.; Rosenberg, T.; Gouras, P.; Tsang, S.H.; Allikmets, R. Extremely hypomorphic and severe deep intronic variants in the ABCA4 locus result in varying Stargardt disease phenotypes. Mol. Case Stud. 2018, 4, a002733. [Google Scholar] [CrossRef]
- Albert, S.; Garanto, A.; Sangermano, R.; Khan, M.; Bax, N.M.; Hoyng, C.B.; Zernant, J.; Lee, W.; Allikmets, R.; Collin, R.W.J.; et al. Identification and Rescue of Splice Defects Caused by Two Neighboring Deep-Intronic ABCA4 Mutations Underlying Stargardt Disease. Am. J. Hum. Genet. 2018, 102, 517–527. [Google Scholar] [CrossRef]
- Sangermano, R.; Khan, M.; Cornelis, S.S.; Richelle, V.; Albert, S.; Garanto, A.; Elmelik, D.; Qamar, R.; Lugtenberg, D.; van den Born, L.I.; et al. ABCA4 midigenes reveal the full splice spectrum of all reported noncanonical splice site variants in Stargardt disease. Genome Res. 2018, 28, 100–110. [Google Scholar] [CrossRef]
- Sangermano, R.; Garanto, A.; Khan, M.; Runhart, E.H.; Bauwens, M.; Bax, N.M.; van den Born, L.I.; Khan, M.I.; Cornelis, S.S.; Verheij, J.B.G.M.; et al. Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet. Med. 2019. [Google Scholar] [CrossRef]
- Shen, X.; Corey, D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018, 46, 1584–1600. [Google Scholar] [CrossRef] [PubMed]
- Collin, R.W.; den Hollander, A.I.; van der Velde-Visser, S.D.; Bennicelli, J.; Bennett, J.; Cremers, F.P. Antisense Oligonucleotide (AON)-based Therapy for Leber Congenital Amaurosis Caused by a Frequent Mutation in CEP290. Mol. Ther. Nucleic Acids 2012, 1, e14. [Google Scholar] [CrossRef] [PubMed]
- Gerard, X.; Perrault, I.; Hanein, S.; Silva, E.; Bigot, K.; Defoort-Delhemmes, S.; Rio, M.; Munnich, A.; Scherman, D.; Kaplan, J.; et al. AON-mediated Exon Skipping Restores Ciliation in Fibroblasts Harboring the Common Leber Congenital Amaurosis CEP290 Mutation. Mol. Ther. Nucleic Acids 2012, 1, e29. [Google Scholar] [CrossRef] [PubMed]
- Garanto, A.; Chung, D.C.; Duijkers, L.; Corral-Serrano, J.C.; Messchaert, M.; Xiao, R.; Bennett, J.; Vandenberghe, L.H.; Collin, R.W. In vitro and in vivo rescue of aberrant splicing in CEP290-associated LCA by antisense oligonucleotide delivery. Hum. Mol. Genet. 2016, 25, 2552–2563. [Google Scholar] [PubMed]
- Parfitt, D.A.; Lane, A.; Ramsden, C.M.; Carr, A.J.; Munro, P.M.; Jovanovic, K.; Schwarz, N.; Kanuga, N.; Muthiah, M.N.; Hull, S.; et al. Identification and Correction of Mechanisms Underlying Inherited Blindness in Human iPSC-Derived Optic Cups. Cell Stem Cell 2016, 18, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Duijkers, L.; van den Born, L.I.; Neidhardt, J.; Bax, N.M.; Pierrache, L.H.M.; Klevering, B.J.; Collin, R.W.J.; Garanto, A. Antisense Oligonucleotide-Based Splicing Correction in Individuals with Leber Congenital Amaurosis due to Compound Heterozygosity for the c.2991+1655A>G Mutation in CEP290. Int. J. Mol. Sci. 2018, 19, 753. [Google Scholar] [CrossRef] [PubMed]
- Dulla, K.; Aguila, M.; Lane, A.; Jovanovic, K.; Parfitt, D.A.; Schulkens, I.; Chan, H.L.; Schmidt, I.; Beumer, W.; Vorthoren, L.; et al. Splice-Modulating Oligonucleotide QR-110 Restores CEP290 mRNA and Function in Human c.2991+1655A>G LCA10 Models. Mol. Ther. Nucleic Acids 2018, 12, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Slijkerman, R.W.; Vache, C.; Dona, M.; Garcia-Garcia, G.; Claustres, M.; Hetterschijt, L.; Peters, T.A.; Hartel, B.P.; Pennings, R.J.; Millan, J.M.; et al. Antisense Oligonucleotide-based Splice Correction for USH2A-associated Retinal Degeneration Caused by a Frequent Deep-intronic Mutation. Mol. Ther. Nucleic Acids 2016, 5, e381. [Google Scholar] [CrossRef]
- Garanto, A.; van der Velde-Visser, S.D.; Cremers, F.P.M.; Collin, R.W.J. Antisense Oligonucleotide-Based Splice Correction of a Deep-Intronic Mutation in CHM Underlying Choroideremia. Adv. Exp. Med. Biol. 2018, 1074, 83–89. [Google Scholar]
- Bonifert, T.; Gonzalez Menendez, I.; Battke, F.; Theurer, Y.; Synofzik, M.; Schols, L.; Wissinger, B. Antisense Oligonucleotide Mediated Splice Correction of a Deep Intronic Mutation in OPA1. Mol. Ther. Nucleic Acids 2016, 5, e390. [Google Scholar] [CrossRef]
- Bauwens, M.; Garanto, A.; Sangermano, R.; Naessens, S.; Weisschuh, N.; De Zaeytijd, J.; Khan, M.; Sadler, F.; Balikova, I.; Van Cauwenbergh, C.; et al. ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: Novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet. Med. Off. J. Am. Coll. Med. Genet. 2019. [Google Scholar] [CrossRef] [PubMed]
- Cideciyan, A.V.; Jacobson, S.G.; Drack, A.V.; Ho, A.C.; Charng, J.; Garafalo, A.V.; Roman, A.J.; Sumaroka, A.; Han, I.C.; Hochstedler, M.D.; et al. Effect of an intravitreal antisense oligonucleotide on vision in Leber congenital amaurosis due to a photoreceptor cilium defect. Nat. Med. 2019, 25, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Homolova, K.; Zavadakova, P.; Doktor, T.K.; Schroeder, L.D.; Kozich, V.; Andresen, B.S. The deep intronic c.903+469T>C mutation in the MTRR gene creates an SF2/ASF binding exonic splicing enhancer, which leads to pseudoexon activation and causes the cblE type of homocystinuria. Hum. Mutat. 2010, 31, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Rincon, A.; Aguado, C.; Desviat, L.R.; Sanchez-Alcudia, R.; Ugarte, M.; Perez, B. Propionic and methylmalonic acidemia: Antisense therapeutics for intronic variations causing aberrantly spliced messenger RNA. Am. J. Hum. Genet. 2007, 81, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Aartsma-Rus, A. Overview on AON design. Methods Mol. Biol. 2012, 867, 117–129. [Google Scholar] [PubMed]
- Collin, R.W.; Garanto, A. Applications of antisense oligonucleotides for the treatment of inherited retinal diseases. Curr. Opin. Ophthalmol. 2017, 28, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Flamier, A.; Barabino, A.; Gilbert, B. Differentiation of Human Embryonic Stem Cells into Cone Photoreceptors. Bio-Protoc. 2016, 6, e1870. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Slijkerman, R.; Kremer, H.; van Wijk, E. Antisense Oligonucleotide Design and Evaluation of Splice-Modulating Properties Using Cell-Based Assays. Methods Mol. Biol. 2018, 1828, 519–530. [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acid Res. 2013, 31, 3406–3415. [Google Scholar] [CrossRef]
- Garanto, A.; Riera, M.; Pomares, E.; Permanyer, J.; de Castro-Miro, M.; Sava, F.; Abril, J.F.; Marfany, G.; Gonzalez-Duarte, R. High transcriptional complexity of the retinitis pigmentosa CERKL gene in human and mouse. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5202–5214. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.; Cieply, B.; Carstens, R.; Ramamurthy, V.; Stoilov, P. The Musashi 1 Controls the Splicing of Photoreceptor-Specific Exons in the Vertebrate Retina. PLoS Genet. 2016, 12, e1006256. [Google Scholar] [CrossRef] [PubMed]
| AON# | Sequence (5′to 3′) | L | Tm | GC | FE-A | FE-D | BE | Remarks |
|---|---|---|---|---|---|---|---|---|
| AON1 | ACAGGAGUCCUCAGCAUUG | 19 | 51.1 | 53 | −0.1 | −12.4 | 16.2 | Specific for c.4539+2001G>A-pseudoexon Previously reported in Reference [18] |
| AON2 | UUUUGUCCAGGGACCAAGG | 19 | 51.1 | 53 | −1.6 | −15.6 | 23.1 | Previously reported in Reference [18] |
| AON3 | CUGUUACAUUUUGUCCAGG | 19 | 46.8 | 42 | −0.9 | −7.3 | 20.7 | Previously reported in Reference [18] |
| AON4 | GGGGCACAGAGGACUGAGA | 19 | 55.4 | 63 | −0.8 | −5.9 | 30.6 | Previously reported in Reference [18] |
| AON5 | GAGAGAAAAUAUUGCUUGAGAA | 22 | 47.4 | 32 | 1.7 | −5.0 | 27.5 | |
| AON6 | GCAGAUGAGCUGUGAUUCAA | 20 | 49.7 | 45 | −2.5 | −8.8 | 24.0 | |
| AON7 | UAUGAUGCAGCAGAUGAGCUG | 21 | 52.4 | 48 | −3.9 | −12.2 | 24.1 | |
| AON8 | UGGGAUCCCUAUGAUGCAGC | 20 | 53.8 | 55 | −1.1 | −17.4 | 19.4 | |
| AON9 | AGAGGACUGAGACAAGUUCC | 20 | 51.8 | 50 | −4.2 | −10.0 | 23.1 | |
| AON10 | GCUUCCUCUUGGGGCACAGA | 20 | 55.9 | 60 | −5.1 | −12.0 | 28.4 | |
| AON11 | CCUCAGCAUUGACAGCAA | 18 | 48 | 50 | −0.6 | −3.2 | 16.1 | |
| AON12 | ACAGGAGCCCUCAGCAUUG | 19 | 53.2 | 58 | −0.4 | −9.3 | 11.1 | One mismatch in c.4539+2001G>A-pseudoexon |
| AON13 | UGGAGGCAGCCACAGGAG | 18 | 54.9 | 67 | −1.3 | −11.8 | 31.4 | |
| AON14 | GAUGCUGGAGGGUUUUGAGUG | 21 | 54.4 | 52 | −1.7 | −12.6 | 27.1 | One mismatch in c.4539+2028C>T-pseudoexon Perfect match in c.4539+2001G>A-pseudoexon |
| AON15 | GAUGCUGGAGAGUUUUGAGUG | 21 | 52.4 | 48 | −1.7 | −14.2 | 20.2 | Specific for c.4539+2028C>T-pseudoexon One mismatch in c.4539+2001G>A-pseudoexon |
| AON16 | GCCUUGACGUCCUGAUGCU | 19 | 53.2 | 58 | 1.4 | −10.3 | 20.4 | |
| AON17 | GCCAAGAGCUCAGGGUACAG | 20 | 55.9 | 60 | −0.9 | −19.9 | 31.8 | |
| AON18 | CUUGGCCUCCCCUCCCUC | 18 | 57.2 | 72 | 1.4 | −8.3 | 29.4 | |
| AON19 | AACACCAUGUAGGUAGGC | 18 | 48 | 50 | −1.6 | −6.8 | 21.2 | |
| AON20 | GUUUAGGAAAUGAAACACCAUG | 22 | 49.2 | 36 | −0.7 | −4.5 | 23.0 | |
| AON21 | GACCGCGUGGAAGUAAGG | 18 | 52.6 | 61 | −0.3 | −14.9 | 22.1 | |
| AON22 | AUAAGUUUCUAAGCUGGACAG | 21 | 48.5 | 38 | −0.4 | −8.1 | 27.2 | |
| AON23 | GGACCAAGGACCAACACUAC | 20 | 53.8 | 55 | −0.6 | −9.7 | 27.9 | |
| AON24 | GGCUGUUACAUUUUGUCCAGG | 21 | 52.4 | 48 | −1.0 | −7.5 | 28.5 | |
| AON25 | GGCAGGAACUGGCUUGCCUU | 20 | 55.9 | 60 | −8.6 | −20.2 | 27.2 | |
| AON26 | AGAAGUGAAAGAAAAUGGCAGG | 22 | 51.1 | 41 | 1.9 | −3.0 | 23.3 | |
| SON1 | CAAUGCUGAGGACUCCUGU | 19 | 51.1 | 53 | −0.7 | −11 | 6.0 | Sense sequence of AON1 Previously reported in Reference [18] |
| SON2 | UCUCAGUCCUCUGUGCCCC | 19 | 55.4 | 63 | −0.9 | −5.6 | 3.4 | Sense sequence of AON4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garanto, A.; Duijkers, L.; Tomkiewicz, T.Z.; Collin, R.W.J. Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease. Genes 2019, 10, 452. https://doi.org/10.3390/genes10060452
Garanto A, Duijkers L, Tomkiewicz TZ, Collin RWJ. Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease. Genes. 2019; 10(6):452. https://doi.org/10.3390/genes10060452
Chicago/Turabian StyleGaranto, Alejandro, Lonneke Duijkers, Tomasz Z. Tomkiewicz, and Rob W. J. Collin. 2019. "Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease" Genes 10, no. 6: 452. https://doi.org/10.3390/genes10060452
APA StyleGaranto, A., Duijkers, L., Tomkiewicz, T. Z., & Collin, R. W. J. (2019). Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease. Genes, 10(6), 452. https://doi.org/10.3390/genes10060452







