Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep
Abstract
1. Introduction
2. Methods
2.1. Ethical Statement
2.2. Animals and Sample Collection
2.3. cDNA Library Preparation
2.4. Sequencing and Transcriptome Assembly
2.5. Prediction of the Differentially Expressed mRNAs and Long Non-Coding RNAs
2.6. Target Gene Prediction of the lncRNAs
2.7. GO and KEGG Enrichment Analysis
2.8. Quantitative Real-Time Polymerase Chain Reaction Validation
3. Results
3.1. Blood Biochemical Parameters
3.2. Read Mapping
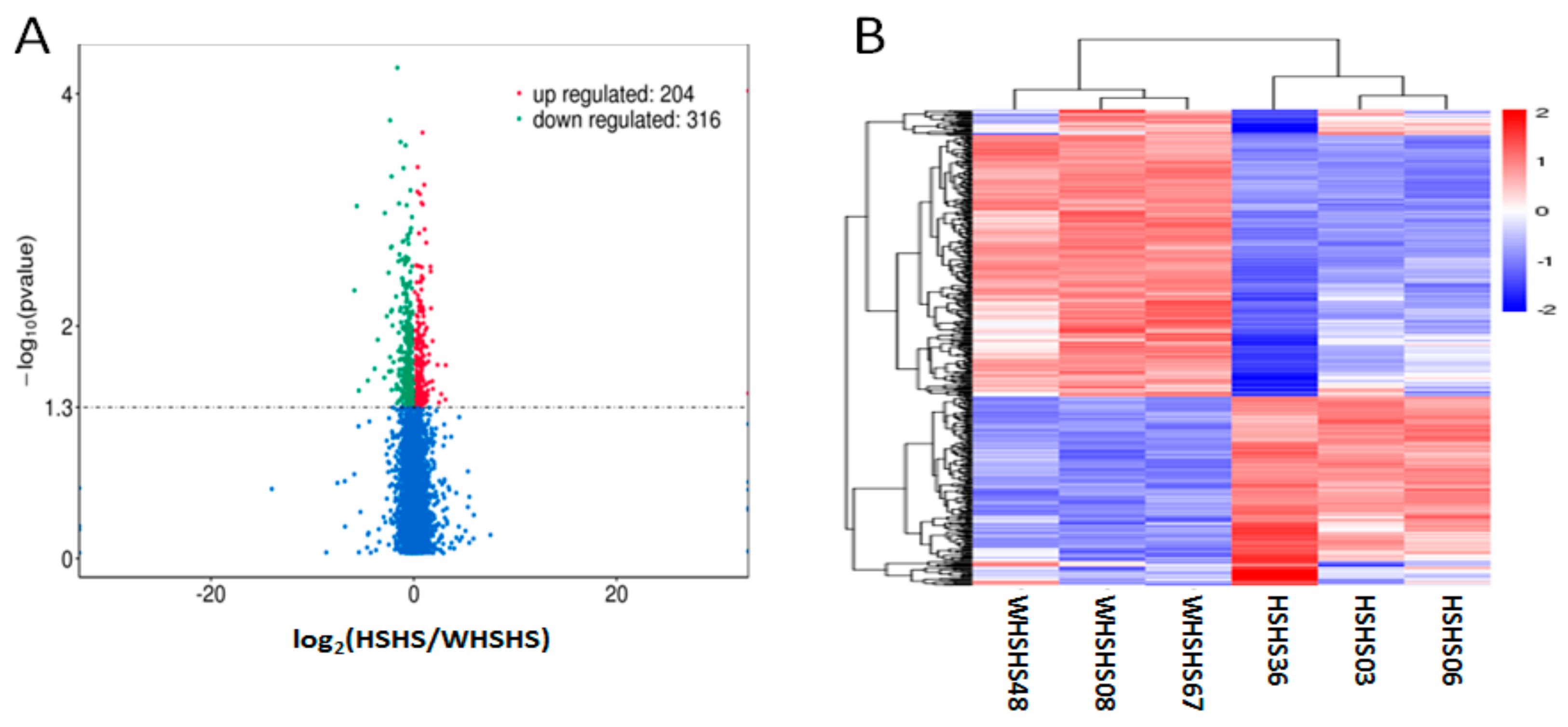
3.3. Enrichment Analysis of the Differentially Expressed mRNAs
3.4. Enrichment Analysis of the Differentially Expressed lncRNAs
3.5. Combined Analysis of the Differentially Expressed lncRNAs and Differentially Expressed mRNAs
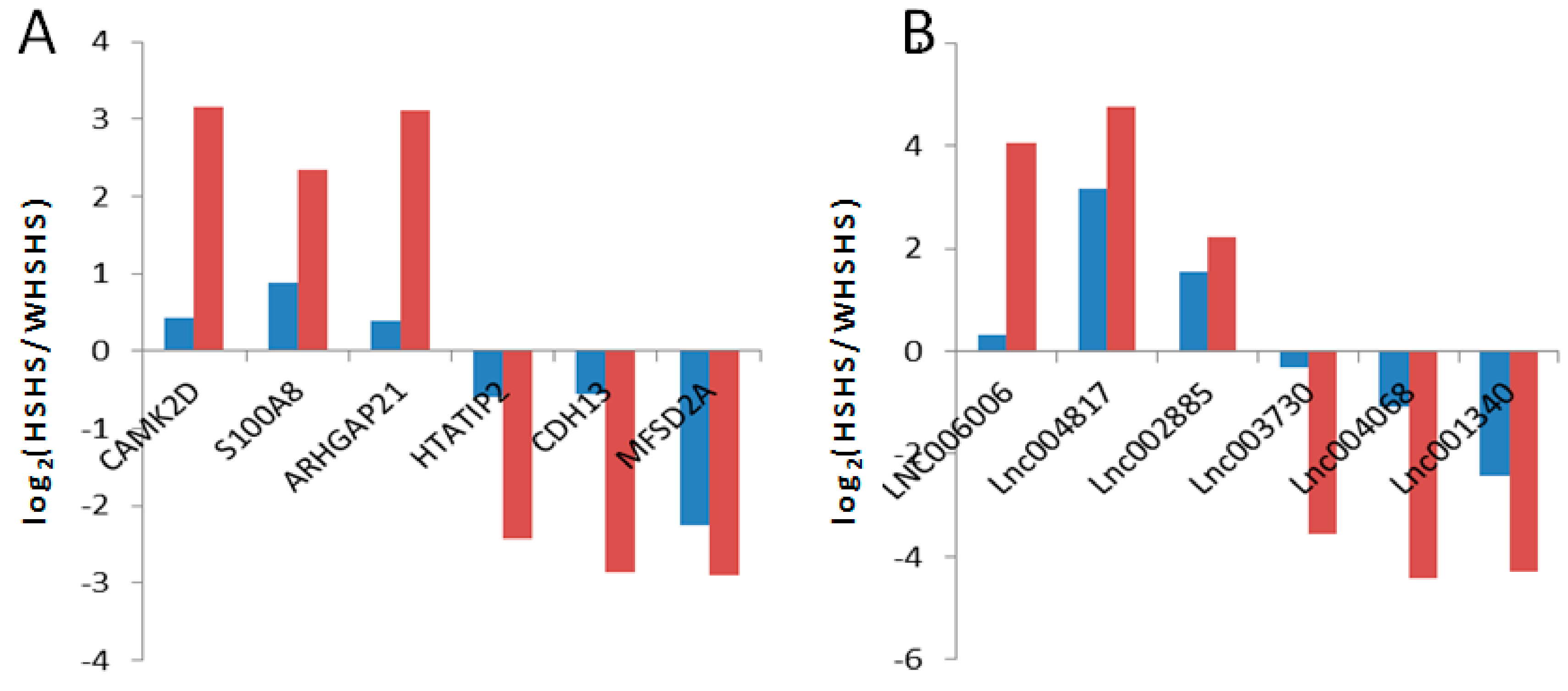
3.6. Validation of Differentially Expressed lncRNAs and mRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baumgard, L.H.; Rhoads, R.P., Jr. Effects of heat stress on postabsorptive metabolism and energetics. Annu. Rev. Anim. Biosci. 2013, 1, 311–337. [Google Scholar] [CrossRef]
- Fuquay, J.W. Heat stress as it affects animal production. J. Anim. Sci. 1981, 52, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Quinn, C.M.; Duran, R.M.; Audet, G.N.; Nisha, C.; Leon, L.R. Cardiovascular and thermoregulatory biomarkers of heat stroke severity in a conscious rat model. J. Appl. Physiol. 2014, 117, 971–978. [Google Scholar] [CrossRef]
- Daramola, J.O.; Abioja, M.O.; Onagbesan, O.M. Heat Stress Impact on Livestock Production. Springer: Berlin/Heidelberg, Germany, 2012; pp. 53–73. [Google Scholar]
- Shelton, M. Birth Weight and Mortality of Lambs Relation of Environmental Temperature During Gestation to. J. Anim. Sci. 1964, 23, 360–364. [Google Scholar] [CrossRef]
- Alexander, G.; Williams, D. Heat stress and development of the conceptus in domestic sheep. J. Agric. Sci. 1971, 76, 53–72. [Google Scholar] [CrossRef]
- MaraiI, F.M.; Bahgat, L.B.; Shalaby, T.H.; Abdel-Hafez, M.A. Fattening performance, some behavioural traits and physiological reactions of male lambs fed concentrates mixture alone with or without natural clay. Annals Arid Zone 2000, 39, 449–460. [Google Scholar]
- Marai, I.F.M.; El-Darawany, A.A.; Fadiel, A.; Abdel-Hafez, M.A.M. Physiological traits as affected by heat stress in sheep—A review. Small Rumin. Res. 2007, 71, 1–12. [Google Scholar] [CrossRef]
- Correa, M.P.C.; Cardoso, M.T.; Castanheira, M.; Landim, A.V.; Dallago, B.S.L.; Louvandini, H.; McManus, C. Heat tolerance in three genetic groups of lambs in central Brazil. Small Rumin. Res. 2012, 104, 70. [Google Scholar] [CrossRef]
- Mcmanus, C.; Dallago, B.S.L.; Lehugeur, C.; Ribeiro, L.A.; Hermuche, P.; Guimaraes, R.F.; de Carvalho, O.A.; Paiva, S.R. Patterns of heat tolerance in different sheep breeds in Brazil. Small Rumin. Res. 2016, 144, 290. [Google Scholar] [CrossRef]
- Srikanth, K.; Lee, E.; Kwan, A.; Lim, Y.; Lee, J.; Jang, G.; Chung, H. Transcriptome analysis and identification of significantly differentially expressed genes in Holstein calves subjected to severe thermal stress. Int. J. Biometeorol. 2017, 61, 1993–2008. [Google Scholar] [CrossRef]
- Cabrera, M.C.; Saadoun, A. An overview of the nutritional value of beef and lamb meat from South America. Meat Sci. 2014, 98, 435–444. [Google Scholar] [CrossRef]
- Hao, Y.; Feng, Y.J.; Yang, P.G.; Cui, Y.J.; Liu, J.R.; Yang, C.H.; Gu, X.H. Transcriptome analysis reveals that constant heat stress modifies the metabolism and structure of the porcine longissimus dorsi skeletal muscle. Mol. Genet. Genom. 2016, 291, 2101–2115. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.Y.; Jiang, R.S.; Xu, S.Y.; Zhang, Z.B.; Xu, G.Y.; Zheng, J.X.; Qu, L.J. Transcriptome responses to heat stress in hypothalamus of a meat-type chicken. J. Anim. Sci. Biotechnol. 2015, 6, 6. [Google Scholar] [CrossRef]
- Li, Y.J.; Huang, J.Q.; Liu, Z.; Zhou, Y.J.; Xia, B.P.; Wang, Y.J.; Kang, Y.J.; Wang, J.F. Transcriptome analysis provides insights into hepatic responses to moderate heat stress in the rainbow trout (Oncorhynchus mykiss). Gene 2017, 619, 1–9. [Google Scholar] [CrossRef]
- Cheng, A.Y.Y.; Leiter, L.A. PPAR-alpha: Therapeutic role in diabetes-related cardiovascular disease. Diabetes Obes. Metab. 2008, 10, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Y.; Zhang, W.X.; Tang, C.Q.; Tang, X.L.; Liu, L.; Liu, C. Vanin-1 Is a Key Activator for Hepatic Gluconeogenesis. Diabetes 2014, 63, 2073–2085. [Google Scholar] [CrossRef]
- van Diepen, J.A.; Jansen, P.A.; Ballak, D.B.; Hijmans, A.; Hooiveld, G.J.; Rommelaere, S.; Galland, F.; Naquet, P.; Rutjes, F.P.J.T.; Mensink, R.P.; et al. PPAR-alpha dependent regulation of vanin-1 mediates hepatic lipid metabolism. J. Hepatol. 2014, 61, 366–372. [Google Scholar] [CrossRef]
- Li, Y.Y.; Wang, X.G.; Yu, J.F.; Shao, F.; Zhang, Y.P.; Lu, X.Y.; Gu, Z.L. MiR-122 targets the vanin 1 gene to regulate its expression in chickens. Poultry Sci. 2016, 95, 1145–1150. [Google Scholar] [CrossRef] [PubMed]
- Uemura, T.; Goto, T.; Kang, M.S.; Mizoguchi, N.; Hirai, S.; Lee, J.Y.; Nakano, Y.; Shono, J.; Hoshino, S.; Taketani, K.; et al. Diosgenin, the main aglycon of fenugreek, inhibits LXRalpha activity in HepG2 cells and decreases plasma and hepatic triglycerides in obese diabetic mice. J. Nutr. 2011, 141, 17–23. [Google Scholar] [CrossRef]
- Jakobsson, T.; Treuter, E.; Gustafsson, J.A.; Steffensen, K.R. Liver X receptor biology and pharmacology: new pathways, challenges and opportunities. Trends Pharmacol. Sci. 2012, 33, 394–404. [Google Scholar] [CrossRef]
- Zhang, B.; Shang, P.; Qiangba, Y.; Xu, A.; Wang, Z.; Zhang, H. The association of NR1H3 gene with lipid deposition in the pig. Lipids Health Dis. 2016, 15, 99. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, E.D.; Daige, C.L.; Petrowski, M.; Dedman, H.; Pattison, J.; Juliano, J.; Li, A.C.; Schulman, I.G. Non-redundant roles for LXRalpha and LXRbeta in atherosclerosis susceptibility in low density lipoprotein receptor knockout mice. J. Lipid Res. 2010, 51, 900–906. [Google Scholar] [PubMed]
- Sandoval, A.; Duran, P.; Gandini, M.A.; Andrade, A.; Almanza, A.; Kaja, S.; Felix, R. Regulation of L-type CaV1.3 channel activity and insulin secretion by the cGMP-PKG signaling pathway. Cell Calc. 2017, 66, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Miranda, J.; Williams, C.; Lairon, D. Dietary, physiological, genetic and pathological influences on postprandial lipid metabolism. Br. J. Nutr. 2007, 98, 458–473. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, L.A.; Olivier, M.; Hubacek, J.A.; Cohen, J.C.; Cox, D.R.; Fruchart, J.C.; Krauss, R.M.; Rubin, E.M. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 2001, 294, 169–173. [Google Scholar] [CrossRef] [PubMed]
- van der Vliet, H.N.; Sammels, M.G.; Leegwater, A.C.; Levels, J.H.; Reitsma, P.H.; Boers, W.; Chamuleau, R.A. Apolipoprotein A-V: a novel apolipoprotein associated with an early phase of liver regeneration. J. Biol. Chem. 2001, 276, 44512–44520. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.; Park, M.; Kong, H.; Segre, J. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protocol. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.T.; Bu, D.C.; Zhao, G.G.; Yu, K.T.; Zhang, C.H.; Liu, Y.N.; Chen, R.S.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Christison, G.I.; Johnson, H.D. Cortisol turnover in heat stressed cows. J. Anim. Sci. 1972, 53, 1005–1010. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Wang, L.Q.; Qiu, P.; Kostich, M.; Greene, J.; Hernandez, M. Domain-oriented functional analysis based on expression profiling. BMC Genom. 2002, 3, 32. [Google Scholar] [CrossRef]
- Xu, W.B.; Chen, L.; He, P.; Yang, J.; Xu, C.D.; Wang, B.; Wang, Z.J.; Yang, H.Y.; Xie, M.H.; Yang, S.M.; et al. Analysis of Cf-12 Tomato Transcriptome Profile in Response to Cladosporium fulvum Infection with Hisat, StringTie and Ballgown. Int. J. Agric. Biol. 2018, 20, 2590. [Google Scholar]
- Han, D.X.; Sun, X.L.; Fu, Y.; Wang, C.J.; Liu, J.B.; Jiang, H.; Gao, Y.; Chen, C.Z.; Yuan, B.; Zhang, J.B. Identification of long non-coding RNAs in the immature and mature rat anterior pituitary. Sci. Rep. 2017, 7, 17780. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Jastrebski, S.F.; Lamont, S.J.; Schmidt, C.J. Chicken hepatic response to chronic heat stress using integrated transcriptome and metabolome analysis. PLoS ONE 2017, 12, e0181900. [Google Scholar] [CrossRef]
- Hocking, P.M.; Maxwell, M.H.; Mitchell, M.A. Haematology and blood composition at two ambient temperatures in genetically fat and lean adult broiler breeder females fed ad libitum or restricted throughout life. Br. Poult. Sci. 1994, 35, 799–807. [Google Scholar] [CrossRef]
- Zeng, T.; Zhang, L.P.; Li, J.J.; Wang, D.Q.; Tian, Y.; Lu, L.Z. De novo assembly and characterization of Muscovy duck liver transcriptome and analysis of differentially regulated genes in response to heat stress. Cell Stress Chaperones. 2015, 20, 483–493. [Google Scholar] [CrossRef]
- Bhusari, S.; Hearne, L.B.; Spiers, D.E.; Lamberson, W.R.; Antoniou, E. Transcriptional profiling of mouse liver in response to chronic heat stress. J. Therm. Biol. 2008, 33, 157–167. [Google Scholar] [CrossRef]
- Streffer, C. Aspects of metabolic change after hyperthermia. Recent Results Cancer Res. 1988, 107, 7–16. [Google Scholar] [PubMed]
- Baumgard, L.H.; Wheelock, J.B.; Sanders, S.R.; Moore, C.E.; Green, H.B.; Waldron, M.R.; Rhoads, R.P. Postabsorptive carbohydrate adaptations to heat stress and monensin supplementation in lactating Holstein cows. J. Dairy Sci. 2011, 94, 5620–5633. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, J.B.; Rhoads, R.P.; Vanbaale, M.J.; Sanders, S.R.; Baumgard, L.H. Effects of heat stress on energetic metabolism in lactating Holstein cows. J. Dairy Sci. 2010, 93, 644–655. [Google Scholar] [CrossRef]
- Pearce, S.C.; Upah, N.C.; Harris, A.; Gabler, N.K.; Ross, J.W.; Rhoads, R.P.; Baumgard, L.H. Effects of heat stress on energetic metabolism in growing pigs. FASEB J. 2011, 25, 1052.5. [Google Scholar]
- Sano, H.; Takahashi, K.; Ambo, K.; Tsuda, T. Turnover and oxidation rates of blood glucose and heat production in sheep exposed to heat. J. Dairy Sci. 1983, 66, 856–861. [Google Scholar] [CrossRef]
- Sanders, S.R.; Cole, L.C.; Flann, K.L.; Baumgard, L.H.; Rhoads, R.P. Effects of acute heat stress on skeletal muscle gene expression associated with energy metabolism in rats. FASEB J. 2009, 23, 598.7. [Google Scholar]
- Ronchi, B.; Bernabucci, U.; Lacetera, N.; Supplizi, A.V.; Nardone, A. Distinct and common effects of heat stress and restricted feeding on metabolic status of Holstein heifers. Zootecnica E Nutrizione Animale 1999. [Google Scholar]
- Rommelaere, S.; Millet, V.; Gensollen, T.; Bourges, C.; Eeckhoute, J.; Hennuyer, N.; Bauge, E.; Chasson, L.; Cacciatore, I.; Staels, B.; et al. PPARalpha regulates the production of serum Vanin-1 by liver. FEBS Lett. 2013, 587, 3742–3748. [Google Scholar] [CrossRef] [PubMed]
- Duval, C.; Fruchart, J.C.; Staels, B. PPAR alpha, fibrates, lipid metabolism and inflammation. Arch. Mal. Coeur. Vaiss. 2004, 97, 665–672. [Google Scholar]
- Junjie, L.; Runzi, S. The Influence of heat stress on semen quality and serum biochemical indexon beef breeding bull. J. Hebei Agric. Univ. 2002, 25, 71. [Google Scholar]
- Wang, S.; Edens, F.W. Heat conditioning induces heat shock proteins in broiler chickens and turkey poults. Poult Sci. 1998, 77, 1636–1645. [Google Scholar] [CrossRef]
- Yahav, S.; Shamay, A.; Horev, G.; Bar-Ilan, D.; Genina, O.; Friedman-Einat, M. Effect of acquisition of improved thermotolerance on the induction of heat shock proteins in broiler chickens. Poultry Sci. 1997, 76, 1428–1434. [Google Scholar] [CrossRef]
- Koluman, N.; Daskiran, I. Effects of ventilation of the sheep house on heat stress, growth and thyroid hormones of lambs. Trop. Anim. Health Prod. 2011, 43, 1123–1127. [Google Scholar] [CrossRef]
- Todini, L.; Malfatti, A.; Valbonesi, A.; Trabalza-Marinucci, M.; Debenedetti, A. Plasma total T3 and T4 concentrations in goats at different physiological stages, as affected by the energy intake. Small Rumin. Res. 2007, 68, 285–290. [Google Scholar] [CrossRef]
- Attebery, J.T.; Johnson, H.D. Effects of environmental temperature, controlled feeding and fasting on rumen motility. J. Anim. Sci. 1969, 29, 734–737. [Google Scholar] [CrossRef]
- Pantoja, M.H.A.; Esteves, S.N.; Jacinto, M.A.C.; Pezzopane, J.R.M.; Paz, C.C.P.; Silva, J.; Lourenco Junior, J.B.; Brandao, F.Z.; Moura, A.B.B.; Romanello, N.; et al. Thermoregulation of male sheep of indigenous or exotic breeds in a tropical environment. J. Thermal Biol. 2017, 69, 302–310. [Google Scholar] [CrossRef]
| Biochemical Indexes | HSHS | WHSHS | p-Value |
|---|---|---|---|
| CK (U/L) | 126.13 ± 38.87 | 146.27 ± 31.80 | 0.132 |
| TP (g/L) | 71.30 ± 4.33 | 67.59 ± 3.40 | 0.014 |
| GLU (mmol/L) | 2.23 ± 0.58 | 2.32 ± 0.33 | 0.591 |
| BUN (mmol/L) | 7.82 ± 1.01 | 7.30 ± 0.70 | 0.115 |
| LDH (U/L) | 666.27 ± 185.10 | 441.00 ± 101.74 | 0.000 |
| Ca (mmol/L) | 2.68 ± 0.13 | 2.67 ± 0.05 | 0.883 |
| K+ (mmol/L) | 4.29 ± 0.17 | 4.21 ± 0.14 | 0.213 |
| Na+ (mmol/L) | 141.53 ± 2.38 | 145.13 ± 1.95 | 0.000 |
| SOD (U/mL) | 102.18 ± 4.68 | 98.48 ± 4.82 | 0.042 |
| T3 (ng/mL) | 13.10 ± 0.37 | 13.03 ± 0.39 | 0.585 |
| T4 (ng/mL) | 553.17 ± 9.35 | 503.56 ± 10.54 | 0.000 |
| Cor (ng/mL) | 16.88 ± 9.97 | 10.16 ± 6.24 | 0.035 |
| HSP70 (ng/mL) | 15.27 ± 0.81 | 15.91 ± 0.82 | 0.038 |
| Sample | HSHS03 | HSHS06 | HSHS36 | WHSHS08 | WHSHS48 | WHSHS67 |
|---|---|---|---|---|---|---|
| Raw Reads | 101,911,032 | 95,707,656 | 103,944,564 | 99,986,482 | 9,219,4420 | 104,239,158 |
| Clean Reads | 95,803,190 | 91,642,522 | 95,317,472 | 94,234,468 | 84,488,554 | 95,701,894 |
| Q30 (%) | 92.48 | 91.71 | 92.05 | 92.31 | 91.67 | 91.70 |
| GC Content (%) | 47.58 | 46.81 | 47.15 | 48.94 | 49.38 | 48.20 |
| Total Mapped | 86,231,172
(90.01%) | 80,485,006
(87.82%) | 84,145,628
(88.28%) | 85,369,917
(90.59%) | 72,563,302
(85.89%) | 83,970,873
(87.74%) |
| Gene Ontology (GO) Term (Biological Process) | Observed * | p-Value |
|---|---|---|
| Oxoacid metabolic process | 45 | 1.40 × 10−5 |
| Organic acid metabolic process | 45 | 1.40 × 10−5 |
| Carboxylic acid metabolic process | 42 | 1.40 × 10−5 |
| Regulation of fatty acid biosynthetic process | 8 | 4.72 × 10−4 |
| Positive regulation of protein deacetylation | 6 | 1.88 × 10−3 |
| Alpha-amino acid metabolic process | 17 | 2.60 × 10−3 |
| Cellular amino acid metabolic process | 23 | 4.37 × 10−3 |
| Small molecule metabolic process | 74 | 4.65 × 10−3 |
| Protein deacetylation | 10 | 7.33 × 10−3 |
| Regulation of protein deacetylation | 7 | 7.33 × 10−3 |
| Protein deacylation | 10 | 8.09 × 10−3 |
| Negative regulation of glucocorticoid receptor signaling pathway | 4 | 9.81 × 10−3 |
| Cellular metabolic process | 251 | 1.29 × 10−2 |
| Small molecule biosynthetic process | 22 | 1.51 × 10−2 |
| Regulation of glucocorticoid receptor signaling pathway | 4 | 1.86 × 10−2 |
| Organic acid biosynthetic process | 17 | 1.90 × 10−2 |
| Carboxylic acid biosynthetic process | 17 | 1.90 × 10−2 |
| Regulation of fatty acid metabolic process | 8 | 2.03 × 10−2 |
| Single-organism biosynthetic process | 22 | 2.03 × 10−2 |
| Organonitrogen compound metabolic process | 60 | 2.56 × 10−2 |
| Monocarboxylic acid biosynthetic process | 12 | 3.34 × 10−2 |
| Positive regulation of fatty acid biosynthetic process | 4 | 3.34 × 10−2 |
| Nitrogen compound metabolic process | 163 | 3.83 × 10−2 |
| Fatty acid biosynthetic process | 10 | 3.87 × 10−2 |
| Heterochromatin organization | 4 | 3.87 × 10−2 |
| Regulation of lipid biosynthetic process | 10 | 4.11 × 10−2 |
| Negative regulation of intracellular steroid hormone receptor signaling pathway | 6 | 4.35 × 10−2 |
| Histone modification | 22 | 4.35 × 10−2 |
| Biosynthetic process | 145 | 4.35 × 10−2 |
| Positive regulation of cellular biosynthetic process | 56 | 4.96 × 10−2 |
| KEGG Pathway | Number of Genes | p-Value |
|---|---|---|
| One carbon pool by folate | 5 | 6.99 × 10−4 |
| Carbon metabolism | 11 | 1.17 × 10−3 |
| Biosynthesis of amino acids | 8 | 3.92 × 10−3 |
| Spliceosome | 11 | 1.15 × 10−2 |
| Protein processing in endoplasmic reticulum | 12 | 1.34 × 10−2 |
| Glyoxylate and dicarboxylate metabolism | 4 | 1.43 × 10−2 |
| Type I diabetes mellitus | 6 | 1.45 × 10−2 |
| Metabolic pathways | 54 | 1.69 × 10−2 |
| Phagosome | 11 | 2.33 × 10−2 |
| Circadian rhythm | 4 | 2.37 × 10−2 |
| Graft-versus-host disease | 5 | 2.45 × 10−2 |
| Citrate cycle (TCA cycle) | 4 | 2.60 × 10−2 |
| PPAR signaling pathway | 6 | 2.91 × 10−2 |
| Allograft rejection | 5 | 3.43 × 10−2 |
| Vitamin digestion and absorption | 3 | 4.00 × 10−2 |
| Cysteine and methionine metabolism | 4 | 4.52 × 10−2 |
| Herpes simplex infection | 12 | 4.53 × 10−2 |
| Transcript ID | Gene ID | FPKM (HSHS/WHSHS) | log2 (HSHS/WHSHS) | p-Value |
|---|---|---|---|---|
| LNC_005888 | XLOC_171029 | 67.33/30.65 | 1.135395178 | 1.80 × 10−3 |
| LNC_004811 | XLOC_139887 | 1.43/2.70 | −0.914641699 | 8.78 × 10−3 |
| LNC_003332 | XLOC_095440 | 7.80/12.88 | −0.723050426 | 1.12 × 10−2 |
| LNC_001340 | XLOC_039194 | 0.21/4.06 | −4.299912802 | 1.14 × 10−2 |
| LNC_001712 | XLOC_048154 | 2.81/0.94 | 1.580309792 | 1.43 × 10−2 |
| LNC_003730 | XLOC_106375 | 0.24/2.81 | −3.559291973 | 1.63 × 10−2 |
| LNC_000765 | XLOC_023257 | 0.85/2.74 | −1.690587291 | 1.70 × 10−2 |
| LNC_000193 | XLOC_005239 | 10.93/0.00 | – | 1.73 × 10−2 |
| LNC_003439 | XLOC_098618 | 43.58/11.56 | 1.915191489 | 2.23 × 10−2 |
| LNC_001699 | XLOC_047869 | 6.44/2.94 | 1.130446672 | 3.06 × 10−2 |
| LNC_006006 | XLOC_173090 | 1.12/11.56 | 4.062706206 | 3.27 × 10−2 |
| LNC_004068 | XLOC_115897 | 0.18/2.94 | −4.411271344 | 3.30 × 10−2 |
| LNC_004817 | XLOC_140078 | 1.92/0.07 | 4.754006517 | 3.51 × 10−2 |
| LNC_002885 | XLOC_083427 | 1.47/3.89 | 2.215701391 | 3.64 × 10−2 |
| LNC_004624 | XLOC_133494 | 5.93/2.28 | 1.377877824 | 3.84 × 10−2 |
| LNC_001782 | XLOC_050277 | 1.04/3.95 | −1.923392878 | 3.92 × 10−2 |
| LNC_005777 | XLOC_169074 | 5.09/5.98 | −0.231523482 | 4.02 × 10−2 |
| LNC_003630 | XLOC_103678 | 3.66/2.05 | 0.833707374 | 4.28 × 10−2 |
| LNC_004460 | XLOC_127505 | 1.58/6.24 | −1.978854313 | 4.74 × 10−2 |
| LNC_004835 | XLOC_140490 | 4.44/1.61 | 1.465355203 | 4.98 × 10−2 |
| GO Term: Biological Process | Observed * | p-Value |
|---|---|---|
| GO:0072593~reactive oxygen species metabolic process | 4 | 1.07 × 10−9 |
| GO:0051239~regulation of multicellular organismal process | 4 | 9.47 × 10−9 |
| GO:0045927~positive regulation of growth | 4 | 9.47 × 10−9 |
| GO:0010897~negative regulation of triglyceride catabolic process | 4 | 3.67 × 10−8 |
| GO:0090209~negative regulation of triglyceride metabolic process | 4 | 1.74 × 10−7 |
| GO:2000377~regulation of reactive oxygen species metabolic process | 4 | 1.74 × 10−7 |
| GO:0010896~regulation of triglyceride catabolic process | 4 | 2.83 × 10−7 |
| GO:0048583~regulation of response to stimulus | 4 | 2.83 × 10−7 |
| GO:0060192~negative regulation of lipase activity | 4 | 4.30 × 10−7 |
| GO:0016043~cellular component organization | 8 | 7.78 × 10−3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Kong, L.; Deng, M.; Lian, Z.; Han, Y.; Sun, B.; Guo, Y.; Liu, G.; Liu, D. Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep. Genes 2019, 10, 395. https://doi.org/10.3390/genes10050395
Li Y, Kong L, Deng M, Lian Z, Han Y, Sun B, Guo Y, Liu G, Liu D. Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep. Genes. 2019; 10(5):395. https://doi.org/10.3390/genes10050395
Chicago/Turabian StyleLi, Yaokun, Lingxuan Kong, Ming Deng, Zhiquan Lian, Yinru Han, Baoli Sun, Yongqing Guo, Guangbin Liu, and Dewu Liu. 2019. "Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep" Genes 10, no. 5: 395. https://doi.org/10.3390/genes10050395
APA StyleLi, Y., Kong, L., Deng, M., Lian, Z., Han, Y., Sun, B., Guo, Y., Liu, G., & Liu, D. (2019). Heat Stress-Responsive Transcriptome Analysis in the Liver Tissue of Hu Sheep. Genes, 10(5), 395. https://doi.org/10.3390/genes10050395




