Abstract
Inositol polyphosphate 5-phosphatase (5PTase), a key enzyme that hydrolyzes the 5′ position of the inositol ring, has essential functions in growth, development, and stress responses in plants, yeasts, and animals. However, the evolutionary history and patterns of 5PTases have not been examined systematically. Here, we report a comprehensive molecular evolutionary analysis of the 5PTase gene family and define four groups. These four groups are different from former classifications, which were based on in vitro substrate specificity. Most orthologous groups appear to be conserved as single or low-copy genes in all lineages in Groups II–IV, whereas 5PTase genes in Group I underwent several duplication events in angiosperm, resulting in multiple gene copies. Whole-genome duplication (WGD) was the main mechanism for 5PTase duplications in angiosperm. Plant 5PTases have more members than that of animals, and most plant 5PTase genes appear to have evolved under strong purifying selection. The paralogs have diverged in substrate specificity and expression pattern, showing evidence of selection pressure. Meanwhile, the increase in 5PTases and divergences in sequence, expression, and substrate might have contributed to the divergent functions of 5PTase genes, allowing the angiosperms to successfully adapt to a great number of ecological niches.
1. Introduction
Phosphoinositides are phospholipids that are present ubiquitously in all eukaryotic cell membranes which regulate numerous cellular processes, including vesicular trafficking, cytoskeletal dynamics, proliferation, and survival [1,2,3,4]. The spatial and temporal localization of phosphoinositide signals is tightly regulated by phosphoinositide kinases and phosphatases, including the inositol polyphosphate 5-phosphatases (5PTases) (Figure 1). 5PTases, one of the key enzymes in the phosphatidylinositol (PI) pathway, are found in all kingdoms of life, and can dephosphorylate the second messenger molecules, inositol 1,4,5-trisphosphate (Ins(1,4,5)P3) and inositol 1,3,4,5-tetrakisphosphate (Ins(1,3,4,5)P4) [2]. The 5PTases contain a highly conserved catalytic 5-phosphatase domain with a length of approximately 350 amino acids. Previous studies have demonstrated the structure of the 5-phosphatase catalytic domain of Schizosaccharomyces pombe synaptojanin (SP synaptojanin), which is similar to those of Mg2+-dependent nucleases [5]. However, the catalytic mechanism of 5PTases remains unclear at present.
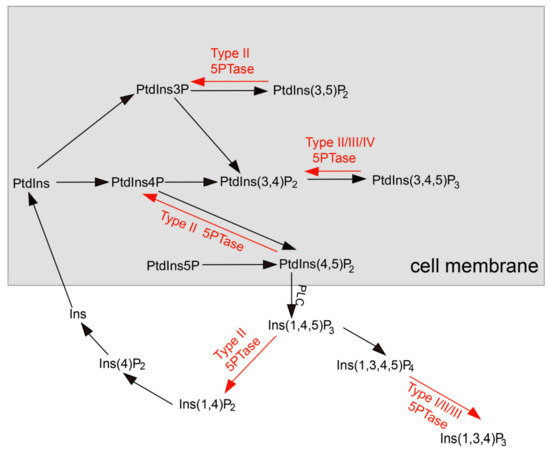
Figure 1.
The phosphoinositide metabolic pathway in plants. The free hydroxyl groups of the phosphatidylinositols (PtdIns) can be phosphorylated at positions 3, 4, and/or 5 by distinct kinases (black arrows) to generate various phosphoinositides. Phospholipase C (PLC) can convert substrate PtdIns(4,5)P2 into the second messenger Ins(1,4,5)P3. The action of inositol polyphosphate 5-phosphatases (red arrows) modulates the signaling functions of these molecules.
5Ptases regulate a range of cellular processes involved in growth, development, and stress responses in plants, yeasts, and mammals [6,7,8,9,10]. 5PTases exhibit different substrate specificities, and hydrolyze the 5′ position phosphate in the inositol ring of water-soluble inositol polyphosphates (Ins(1,4,5)P3, Ins(1,3,4,5)P4), and/or phosphoinositides (PtdIns(4,5)P2, PtdIns(3,4,5)P3, PtdIns(3,5)P2) (Figure 1) [2]. Based on their substrate specificity, mammalian 5PTases can be further divided into four types [2]. Type I 5PTases hydrolyze phosphate from Ins(1,4,5)P3 and Ins(1,3,4,5)P4. Type II 5PTases hydrolyze phosphate from both water-soluble inositol polyphosphates and phosphoinositides. Type III 5PTases hydrolyze phosphate from PI(3,4,5)P3 and Ins(1,3,4,5)P4, and type IV 5PTases specifically dephosphorylate PI(3,4,5)P3.
Eukaryotes have a family of diverse 5PTases, with four members in yeast, ten in mice, ten in humans, 15 in Arabidopsis, and 21 in rice [1,9,11,12,13,14,15,16]. All of the 5PTases are characterized by a highly conserved phosphatase domain of about 350 residues, and some plant 5PTases also contain several WD40 domains that function in protein–protein interactions [17,18,19]. The yeast 5PTase genes, INP51, INP52, INP53, and INP54, show different cell localization: INP52 localizes to plasma-membrane-derived endocytic vesicles, INP53 localizes to the Golgi, endosomal, or vesicular membranes, and INP54 localizes to the endoplasmic reticulum (ER) [20,21,22]. Loss of any single yeast 5PTase has little phenotypic effect, while the loss of any two 5PTases results in marked phenotypic changes, suggesting a degree of redundancy [23]. In mammals, 5PTases play important roles in the regulation of hematopoietic stem cell proliferation, synaptic vesicle recycling, insulin signaling, endocytosis, vesicular trafficking, and actin polymerization. Loss of 5PTase function in mammals results in a broad spectrum of diseases and disorders [7,24]. For example, mutations in OCRL1 results in the X-linked OculoCerebroRenal syndrome of Lowe and Type-2 Dent disease [25,26]. Loss of function of SHIP1 results in myeloproliferative syndrome [27,28]. SHIP2 is implicated in obesity, insulin resistance, and hypertension [29,30,31]. In addition, mutations in INPP5E can cause Lowe cerebrorenal syndromes [32]. These results reveal the functional differences in mammal 5PTases.
Compared with the huge number of reports on mammal 5PTases, knowledge about the functions of plant 5PTases is rather limited. The Arabidopsis genome contains more 5PTases (At5PTases) than any yeasts or mammals [11]. Based on their protein structure, Arabidopsis 5PTases can be divided into two types, with 11 genes encoding type I 5PTases and four genes encoding type II 5PTases, which contain several WD40 domains [11]. Similar to mammal 5PTases, Arabidopsis 5PTases also display both overlapping and unique substrate preferences, suggesting the presence of different types of At5PTase with distinct biological functions. At5PTase1 and At5PTase2 both hydrolyze Ins(1,4,5)P3 and Ins(1,3,4,5)P4, and 5ptase1 and 5ptase2 single mutants show similar phenotypes, with faster seed germination and increased hypocotyl elongation when grown in the dark [33]. However, At5PTase1 and At5PTase2 are not functionally redundant, as the double mutant does not result in an additive effect [33]. The FRAGILE FIBER3 (RFA3), root-hair morphogenesis3 (MRH3; At5PTase5), cotyledon vascular pattern2 (CVP2; At5PTase6), and CVP2 LIKE 1 (CVL1; At5PTase4) genes are required for fiber cell development [34], root hair initiation [35], and cotyledon vascular patterning [36,37], respectively. Three type II At5PTase genes, At5PTase12, At5PTase13, and At5PTase14 are differentially expressed in Arabidopsis organs. In addition, the proteins display different substrate specificity along with At5PTase12 and At5PTase13, which display phosphatase activity toward Ins(1,4,5)P3, while At5PTase14 hydrolyzes PI(4,5)P2, PI(3,4,5)P3, and Ins(1,4,5)P3, with the highest substrate affinity being towards PI(4,5)P2 [18]. The At5PTase13 mutant results in defects in cotyledon vein development and root gravitropism [38,39]. In addition, mutations in At5PTase7 and its paralog At5PTase9 lead to a similar increased salt sensitivity phenotype with reduced production of reactive oxygen species (ROS) and decreased expression of stress-responsive genes in both mutants [40,41]. However, these two paralogs exhibit different substrate preferences, with At5PTase7 having a substrate preference for membrane-bound phosphoinositides (PtdIns(4,5)P2 and PtdIns(3,4,5)P3), whereas At5PTase9 dephosphorylates PtdIns(4,5)P2, PtdIns(3,5)P2, and PtdIns(3,4,5)P3) [40,41]. These results also indicate that At5PTase7 and At5PTase9 are non-redundant, similar to At5PTase1 and At5PTase2. Although the substrate preferences of many mammal and plant 5PTases have been reported, the comparative substrate specificity between mammal and plant 5PTase is not clear. Recently, maize BREVIS PLANT1 (BV1), an inositol polyphosphate 5-phosphatase with a WD40 domain, was reported to be involved in internode elongation [42]. Despite these important functions, information on the origins and evolution of 5PTases remains fundamentally unexplored.
The evolutionary study of plant and mammal 5PTase genes is very limited. Previous reports have focused on a single species or a few species only, and there has been no systematic studies of the 5PTase family in plants or mammals. Large-scale sequenced genome data provides deeper sampling to gain insights into the evolution of the 5PTase family during the history of angiosperms. In this study, an evolutionary analysis of the 5PTase genes in major plant lineages is presented with phylogenetic analyses. Phylogenetic analyses were performed to delineate the evolutionary history of the 5PTase family in major angiosperm lineages, and exon/intron structure analyses were performed to gain insight into the possible mechanisms of the structural diversity of the 5PTase gene family. The tissue specificity and inducibility of 5PTase gene expression in plants were characterized by examining publicly available microarray data. The results obtained here will broaden our understanding of the roles and evolution of 5PTases and provide a framework for further functional investigations of these genes in plants.
2. Materials and Methods
2.1. Data Sources and Homolog Searches
Several datasets (Ensembl Genome, JGI Genome, Phytozome) and multiple steps were used to search for 5PTase sequences. We selected representative species from animals, plants, and fungi, including 16 plants, five animals, and one fungus. Proteomic data from animals and fungi were downloaded from the ENSEMBL (release 91, http://www.ensembl.org) and JGI (http://genome.jgi.doe.gov/) databases, respectively. Plant sequences were obtained from Phytozome v12.1 (http://www.phytozome.net/). The sequences for Amborella trichopoda were retrieved from the Amborella Genome Database (http://www.Amborella.org/). The number of 5PTase genes in different organisms can be found in Figure 2.
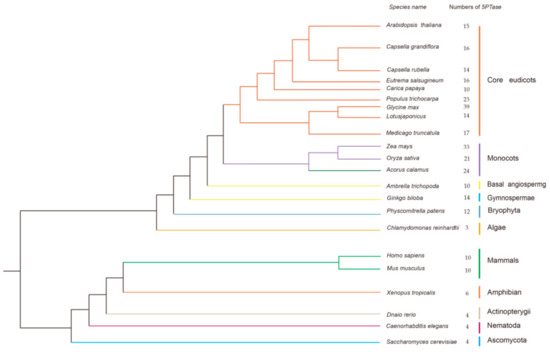
Figure 2.
Phylogenetic relationships of species that were used in this study. The species tree was constructed based on the phylogeny of conserved nuclear genes [56,57,58]. The total number of 5PTase proteins found in the genome of each species is indicated.
The hidden Markov model (HMM) program (version 3.0) [43] was employed with the Hidden Markov model to retrieve all eukaryotic 5PTase homologs. The Hidden Markov model profile of the 5PTase protein domain (PF03372 in Pfam database) was downloaded and used in local searches of the datasets [44]. These sequences were verified using the protein families (PFAM) database (http://pfam.xfam.org/search), the Conserved Domains Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), available from the National Center for Biotechnology Information, and the SMART (a Simple Modular Architecture Research Tool) database (http://smart.embl-heidelberg.de/), with a threshold e-value of less than 1e–10 [44,45,46].
2.2. Phylogenetic Analyses
Phylogenetic analyses were conducted using two methods: neighbor-joining (NJ), and maximum likelihood (ML). NJ trees were constructed using MEGA 5.0 with Poisson correction, pairwise deletion, and bootstrap (1000 replicates) [47]. Multiple sequence alignments were performed using ClustalW with default parameters in MEGA 5.0 (Table S1, Figure S2). PhyML 3.0 and RaxML v7.0.4 were used to construct ML trees, with the Jones, Taylor, and Thorton (JTT) model and gamma distribution option, and rooted using phosphoinositide-specific phospholipase C AT1G13680 and AT1G49740 as outgroups, as these two phospholipases belong to families other than the 5PTases [48,49].
2.3. Motif and Synteny Analyses
All 5PTase amino acid sequences were used to search against the PFAM, CDD, and SMART databases to uncover other known domains or motifs apart from the 5PTase domain. Multiple Em for Motif Elicitation (MEME) software (v4.9.0) was used to discover novel conserved motifs that might not be recorded in public databases [50]. Duplicate gene pairs were searched for any evidence of synteny using the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication) [51,52].
2.4. Expression Analysis
For the expression profile analysis of 5PTase genes in Arabidopsis and rice, ATH1 22k and Os 51k microarray data from the Genevestigator V3 database were used, and then the heat maps were constructed using the obtained gene expression datasets, respectively [53]. Transcriptome data of soybean 5PTase homologs in representative tissues were analyzed [54]. For heat, cold, and drought stress analysis, two-week-old soybean was exposed to 42 °C, 4 °C, and PEG400 for 1 h, respectively. Each treatment consisted of three biological replicates, and each biological replicate consisted of pools of six plants. Total RNA was isolated, and RNA sequencing (RNA-seq) was performed on the Illumina HiSeqTM2000 platform.
2.5. Calculation of Ka/Ks-Values
The ratio of non-synonymous to synonymous substitutions (Ka/Ks) was estimated to investigate the molecular evolution of 5PTase genes. The duplicated 5PTase coding sequences (CDS) were aligned using ClustalW (http://www.genome.jp/tools/clustalw/) to predict the divergence periods. Based on previous research, a Ka/Ks ratio of <1 indicates purifying selection, a ratio =1 indicates neutral evolution, and a ratio >1 indicates positive selection [55].
3. Results
3.1. Identification of 5PTase Genes in Plants, Animals, and Fungi
Using the Hidden Markov model (HMM) algorithm, the complete set of 5PTases genes were identified from a comprehensive dataset that contained selected plants, animals, and fungi. In total, 318 sequences were retrieved from 16 plants, five animals, and one fungus (Figure 2). 5PTase genes were determined based on whether the corresponding protein contained the 5PTase domain. The 5PTase proteins that were identified ranged in size from 169 to 1916 amino acids. Among the major lineages of green plants, 5PTase genes are present in algae, bryophyta, gymnosperms, and angiosperms (Figure 2). Further investigation revealed that the copy number of the 5PTase genes varied considerably among plants, ranging from three in the green algae Chlamydomonas reinhardtii to 12 in Physcomitrella patens (bryophyta), 21 in Oryza sativa (monocot), and 15 in Arabidopsis thaliana (eudicot), with the highest copy number being 39 in Glycine max (eudicot). Land plants contained higher copy numbers than algae, indicating that duplications of 5PTase genes likely occurred after land plants diverged from green algae. The 5PTase genes are also widespread among animals, from simple invertebrates, such as the Caenorhabditis elegans, to mammals, such as humans, with the gene copy number ranging from four to ten (Figure 2).
To standardize the gene names, we adopted a common nomenclature system from previous studies based on the names of Arabidopsis, human, and yeast genes [11,16]. For yeast, human, and Arabidopsis genes with known functions or previous research, the published gene names were retained and used as a reference. Rice genes that were found to be orthologous to Arabidopsis were named after the Arabidopsis genes. Finally, recent paralogs were distinguished using a lower-case letter after the number.
3.2. Phylogenetic Classification of 5PTase Genes into Four Groups
The overall sequence identity between different 5PTases was very low (approx. 10%). In order to explore the evolutionary relationships of eukaryotic 5PTase genes, we conducted phylogenetic analyses with amino acids containing the conserved 5PTase domain from representative species (Figure 3, Table S1, and Figure S2). Based on the phylogenetic analyses, the eukaryotic 5PTase genes can be divided into four major groups. Notably, these four groups are inconsistent with the four types of animal 5PTases reported by Majerus et al. [2]. Among these, Group I contains members from fungi, plants, and animals; Group II contains genes from plants and animals; whereas Group III only contains genes from plants (Figure 3).
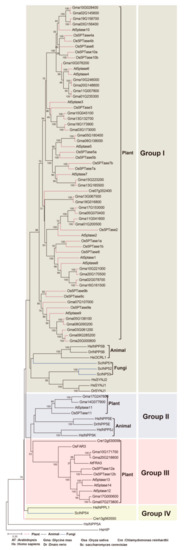
Figure 3.
Phylogeny of representative 5PTase genes from plants, animals, and fungi. Tree topology generated via maximum-likelihood (ML) using MAFFT (MSA tool that uses Fast Fourier Transforms) alignment is shown here. The protein sequences containing the conserved 5PTase domain were used for the alignment. At, Arabidopsis; Os, rice; Pp, Physcomitrella patens; Cre, Chlamydomonas reinhardtii; Hs, Homo sapiens; Dr, Danio rerio; Ce, Caenorhabditis elegans; Sc, Saccharomyces cerevisiae.
Group I contains genes from fungi, animals, and plants, indicating an early origin from the most recent common ancestor (MRCA) of the three kingdoms. More angiosperm 5PTase genes were identified in Group I than in fungi and animal 5PTases, indicating that this group of genes was expanded in angiosperms. Group II contains plant and animal 5PTase genes, indicating that they originated before the separation of plants and animals. Four Arabidopsis 5PTase genes (At5PTase12/13/14, and AtFAR3) were identified in Group III, all of which contained several WD40 domains in their N-terminus [11]. Group IV also contains genes from fungi, animals, and plants, but their homologs were lost in angiosperm (Figure 3). In addition, angiosperm in Groups I and III had higher gene copy numbers compared with fungi and animals, suggesting the expansion and functional divergence of 5PTase genes in angiosperms.
3.3. Domain Features of 5PTase Proteins
To better comprehend the evolutionary relationship of 5PTase genes, we analyzed the protein domain features of plant (Arabidopsis, rice, and soybean) and human 5PTases (Figure 4). The most closely related 5PTases display common motifs in plants and humans, respectively (Figure 4A,B). Four Arabidopsis 5PTase proteins (At5PTase12/13/14, FRA3), two rice 5PTase proteins (Os5PTase12a/12b), and four soybean 5PTase proteins (Gma10G171700, Gma20G218600, Gma17G000600, Gma07G273800) have multiple WD40 domains in their N-termini, indicating that these 5PTases might function by interacting with other proteins (Figure 4A). Interestingly, human 5PTases exhibited various combinations of different conserved motifs, including pleckstrin homology (PH), SH2, RRM, SAC1, SAM, and RhoGTPase-activating protein (RhoGAP), which determine protein subcellular localization and/or protein–protein interactions. These different motifs determine unique cellular and subcellular distributions and specialized functions of human 5PTases. For example, Synaptojanin 1 (SYNJ1) is a major neuronal 5-phosphatase [55,59], whereas OCRL1 is a major 5-phosphatase in fibroblasts [60,61].

Figure 4.
Phylogenetic relationships, exon/intron structure, and domain organization of 5PTase genes in Arabidopsis, rice, soybean (A), and humans (B). The motif architecture is demonstrated as colored boxes. IPPC: inositol polyphosphate phosphatase catalytic domain; WD40: WD40/beta-transducin repeats domain; PPR_3: pentatricopeptide repeat domain; DYW: DYW_deaminase domain. The yellow boxes represent exons, the black lines represent introns, and the gray boxes refer to untranslated regions (UTRs).
3.4. Structural Analysis of 5PTase Family Genes
Intron loss and gain have relative importance in eukaryotic evolution [62]. The exon/intron organizations of different 5PTase genes were examined for available genome sequences. Most plant members exhibit a similar exon/intron organization in terms of exon length and intron number, whereas there was a considerably more diverse number and length of introns in human 5PTase genes (Figure 4). For instance, 5PTase genes in plants have 6–12 introns; 6% (27/75) of the genes have nine introns, 20% (15/75) have ten, 2.6% (2/75) have 11, and 2.6% (2/75) have 12 introns (Figure 4A). Whereas human 5PTase genes have 9–31 introns and are strikingly distinct in intron arrangement and number between different genes (Figure 4B). The great differences in exon/intron organization between plant and human 5PTases suggest that intron gain or loss may play an important role in the structural evolution of the 5PTase family.
3.5. Identification of Multiple Duplication Events in Land Plant 5PTases
To further investigate the evolution and duplication events of the 5PTase gene family, we conducted phylogenetic analyses in plants (Figure 5). 5PTase genes were divided into two subgroups, with the corresponding proteins of most members in Subgroup II containing several WD40 domains (Figure 4). Within Subgroup I, there were three independent duplications in angiosperm and eudicots, suggesting that the duplication events likely occurred in the ancestor of a corresponding lineage (Figure 5). The duplication of eudicot At5PTase10/6 occurred in the common ancestor of core eudicots, and the two duplications of At5PTase3/5/7 occurred before the divergence of eudicots and monocots. Within Subgroup II, there was one independent duplication in core eudicots, suggesting that the duplication events likely occurred in the ancestor of core eudicots (Figure 5). In addition, 39 5PTase genes were identified in Glycine max, along with 16 soybean leaf pairs (Figure 5), demonstrating that more gene duplications occurred in soybean 5PTase.
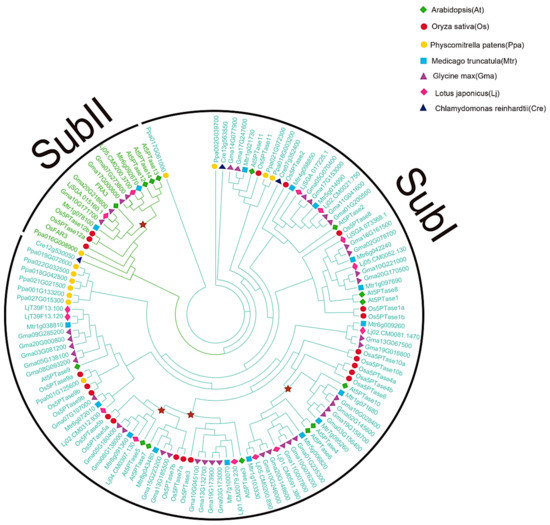
Figure 5.
Phylogenetic analysis of plant 5PTase genes. The tree topology of 5PTase protein sequences was constructed by Maximum Likelihood (ML) using the Jones, Taylor, and Thorton model among Arabidospis, rice, moss, Medicago truncatula, soybean, Lotus japonicas, and Chalamydomonas reinhardtii. The 5PTase protein sequences were aligned using MAFFT (MSA tool that uses Fast Fourier Transforms). The red stars represent major duplication events.
WGDs are common in angiosperms, so in order to explore whether plant 5PTase genes were caused by genome duplication, we searched for possible synteny in genomic regions containing the 5PTase genes. Two and seven pairs of Arabidopsis and rice 5PTases, respectively, were found in syntenic genomic regions, indicating that these multiple gene copies are the result of whole genome or segmental duplications (Figure 6, Table 1). To investigate the molecular evolution of 5PTase genes, the ratio of non-synonymous to synonymous substitutions (Ka/Ks) was estimated (Table 1), which revealed that the Ka/Ks ratios varied from 0.06 to 1.87 among three differential species (Table 1). Most of the 5PTase duplicates displayed lower Ka/Ks ratios (less than 1), indicating that these duplicates experienced strong purifying selection. The Ka/Ks ratio of Os5PTase9b/9c was 1.01, suggesting that these two 5PTase genes were constrained by neutral evolution in rice, whereas the Ka/Ks ratio of Os5PTase1a/1b was 1.87, indicating that its evolution was driven by positive selection, thus allowing it to neo-functionalize. Soybean contains multiple gene duplicates and Ka/Ks ratios that range from 0.07 to 0.39, with none being greater than 1 (Table 1).
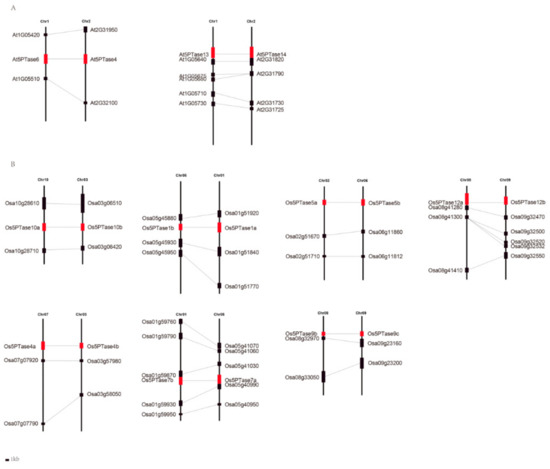
Figure 6.
Collinearity analysis of duplicated 5PTase gene pairs. At, Arabidopsis thaliana; Os, Oryza sativa. The syntenic paralogous genes are connected by lines.

Table 1.
Divergence between 5PTase gene pairs in plants.
3.6. Expression and Functional Divergence of 5PTase
To investigate differences in the expression of 5PTase genes, we analyzed the expression profiles of At5PTases and Os5PTases during plant development, based on the microarray data reported in the Genevestigator (Figure 7). Overall, all of the At5PTase and Os5PTase genes are expressed during the vegetative and reproductive development stages, and display strong organ specificity. In Arabidopsis, most Group I 5PTases are mainly expressed in vegetative organs: At5PTase3, At5PTase5, and At5PTase9 are expressed the highest in the root; At5PTase2, At5PTase4, and At5PTase7 are mainly expressed in the leaf; whereas most of the Group III 5PTases are mainly expressed in the reproductive organs (Figure 7A). In addition, differential expression patterns also exist among 5PTases in the same group. For example, At5PTase8, one 5PTase gene in Group I, is mainly expressed in old flowers but not in the vegetative organs. In rice, most Os5PTases are highly expressed in the reproductive organs, and other Os5PTases are mainly expressed in the leaves and shoots. Consistent with At5PTase expression, rice 5PTases also display both similar and differential expression patterns among the same groups (Figure 7B). Apparently, Os5PTase6, Os5PTase4b, and Os5PTase10b are mainly expressed in early inflorescence; Os5PTase11, Os5PTase7a, Os5PTase5a, Os5PTase5b, and Os5PTase12b are only highly expressed in anthers; whereas, Os5PTase1b, Os5PTase3, Os5PTase9b, Os5PTase8, Os5PTase4a, and Os5PTase10a are only highly expressed in pistil. All of these results indicate both the conservation and diversification of expression among 5PTase genes in Arabidopsis and rice.
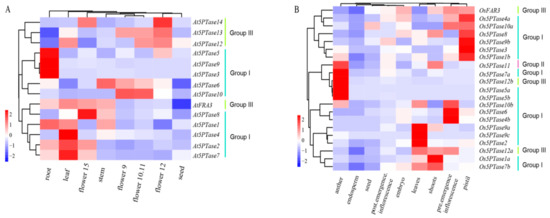
Figure 7.
Developmental expression patterns of 5PTase family genes in Arabidopsis and rice. Expression profiles of (A) At5PTases and (B) Os5PTase in different developmental stages obtained from microarray data reported in the Genevestigator. Results are shown as heat maps in blue/white/red (low to high) that reflect the percentage of expression.
The Ka/Ks ratios were less than 1 in soybean 5PTase paralogs, suggesting that soybean 5PTase genes have undergone strong negative selection pressure (Table 1). To gain insight into the potential developmental roles of Gm5PTase genes, the expression profiles of Gm5PTase genes were analyzed in 28 developmental stages/tissues, based on previous transcriptome data [54]. Overall, many genes displayed a distinct tissue-specific expression pattern, suggesting specific roles in particular stages of development (Figure S3). For example, seven Gm5PTase genes (Gma03G081200, Gma20G000800, Gma09G285200, Gma08G093200, Gma19G173900, Gma05G070400, and Gma13G185500) are mainly expressed in the root, and seven Gm5PTase genes (Gma17G153000, Gma10G045100, Gma03G173000, Gma07G107000, Gma13G132700, Gma05G180400, and Gma08G138000) are mainly expressed in flowers. These tissue-specific expression patterns are consistent with the expression patterns of At5PTases and Os5PTases (Figure 7).
To further investigate the functional roles of Gm5PTase genes, we analyzed the transcriptomic datasets of soybean 5PTases under different abiotic stresses, such as drought, cold, and high temperature (Li, Wang, and Yuan, unpublished data) (Figure S3). Many Gm5PTase genes showed specific induction under the cold, heat, and drought. For example, seven, nine, and seven Gm5PTase genes were induced under cold, drought, and heat stress, respectively (Figure S4), indicating that soybean 5PTase genes might play important roles in stress responses.
4. Discussion
4.1. 5PTases Genes Have Been Conserved during Speciation
Multiple 5PTase genes were found in each of the plant, animal, and fungal genomes analyzed, including dicots, monocots, bryophytes, vertebrates, and invertebrates (Figure 2). This indicates that 5PTase genes were present prior to the diversification of plants, animals, and fungi (Figure 3). In land plants, more 5PTase genes were identified than in animals, indicating that the number of 5PTase genes in plants underwent expansion. Based on the phylogenetic analyses, the 5PTases diverged earlier during specification (Figure 3). In addition, the Ka/Ks ratio revealed that most plant 5PTase protein families underwent a process of purifying selection (Table 1), suggesting that the 5PTase protein families tended to be stable during the long evolutionary process.
Multiple gene copies were found for the plant 5PTases, and many of them were localized in syntenic genomic regions, indicating that they resulted from whole genome duplications (WGDs) or segmental duplications, and in our study, 17 Gm5PTase paralogous pairs were identified in the soybean genome. In a previous study, the soybean genome was shown to have experienced two WGD events; one occurred 59 million years ago (mya) and the second occurred 13 mya [63,64]. The divergence period of Gm5PTase gene pairs ranged from 6.41 to 15.68 mya, indicating that these Gm5PTase pairs were formed by the most recent WGD, which also suggests that the WGD duplication was the main mechanism for the 5PTase gene family expansion during the evolution of soybean. It has been previously reported that regulatory genes (i.e., protein kinases and transcription factors) and signaling genes are more likely to be retained after duplication events compared to the genome-wide average [65,66,67]. During cell growth, development, and stress responses, 5PTases are the key enzyme of the phosphatidylinositol (PI) pathway. They serve as molecular “hubs” during PI signaling and have, therefore, been conserved throughout evolution.
4.2. Functional Divergence in the Evolution of 5PTase Genes
Gene duplication provides the raw material for functional innovation [68]. 5PTase proteins are present in nearly all domains of life and display an ancient origin before the divergence of fungi, plants, and animals (Figure 3). This long evolutionary history has allowed a great deal of sequence divergence and resulted in low sequence identities between different subfamilies. Yet, the key functional domain of 5PTases remained constant in most members, with likely conservation of the original 5-phosphatases activity being the possible result of strong selection.
Previous studies have shown that functional divergence could have occurred by changes in gene expression patterns or protein subcellular localizations [69,70,71]. Intron loss or gain is important in generating structural diversity and complexity, which further promotes variability [71,72]. Members of the 5PTase pairs showed diversity in their transcription profile. As shown in Figure 7, AT5PTases and Os5PTases have shown diversity in different developmental stages: sub-cellular localization in the nucleus, chloroplast, cytoplasm, plasma membrane, and substrate specificity to inositol polyphosphates and phosphoinositides (Figure 8). In addition, the numbers of introns and the intron arrangement are strikingly distinct between plants and animals, which also contributes to the functional divergence and diversity of 5PTase family proteins during the evolutionary process in plants and animals. Therefore, the functional divergence of 5PTase has apparently arisen from protein characteristics (including sub-cellular localization, and substrate specificity), gene structural diversity (exon/intron structure), and expression patterns. These results indicate the functional divergence of 5PTases in plants and animals during their long-term evolutionary history.
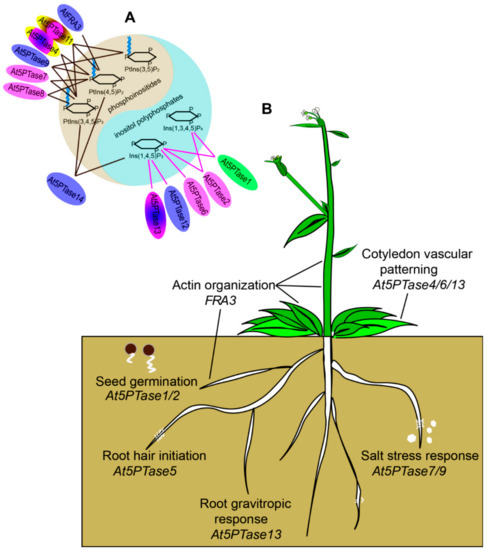
Figure 8.
Functionally characterized 5PTase genes from Arabidopsis. (A) Substrate specificity and protein localization of At5PTases. FRA3, 5PTase9, 5PTase12, and 5PTase14 were localized in the nucleus; 5PTase2, 5PTase6, 5PTase7, and 5PTase8 were localized in the cytoplasm; 5PTase1 was localized in chloroplasts; 5PTase13 was localized in the nucleus and cytoplasm; 5PTase11 was localized in the cytoplasm and plasma membrane; and 5PTase4 was localized in the nucleus, cytoplasm, and plasma membrane. Different localizations were displayed with different colors. 5PTase 1, 5PTase 2, 5PTase 6, 5PTase 12, and 5PTase 13 belong to Type I 5PTases and purple lines are used to connect them with the 5-phosphatase substrates. 5PTase 4, 5PTase 7, 5PTase 8, 5PTase 9, 5PTase 11, 5PTase14, and FRA3 belong to Type II 5PTases and black lines are used to connect them with 5-phosphatase substrates [2]. (B) 5PTases are crucial for multiple processes of plant growth, including seed germination [35], vascular patterning [36,37], root hair initiation [35], and salt stress [40,41].
5. Conclusions
Highly retained proportions of 5PTases were observed in plants and animals using phylogeny analysis. 5PTases in plants and animals play a complicated role in many growth and metabolic processes. Different 5PTases show complex and cross-linked substrate specificity (Figure 8). However, the function of each plant 5PTase and its molecular mechanisms are still unclear, and the interactions among 5PTases are still barely understood. Further studies will be carried out to explore the functions of 5PTase genes in plants.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/5/393/s1, Table S1. The amino acids of 5PTase genes, Figure S2. The sequence alignment of 5PTases, Figure S3. Heatmap showing the expression of soybean 5PTases in different tissues, Figure S4. Expression of soybean 5PTase genes under drought, cold, and heat treatment.
Author Contributions
Z.Z., H.Y. and L.C. designed the research. Z.Z. wrote the manuscript. Y.L., Z.L., S.K., and Y.Z. performed the identification of 5PTases, protein structure, evolution analysis, and expression analysis. C.Z. and W.Z. participated in manuscript preparation and revision. All authors read and approved the final manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors are financially supported by the Natural Science Foundation of Henan Provincial Science and Technology (No. 182300410063), Key Scientific Research Projects of Henan Higher Education Institutions (No. 18A180031), the National Natural Science Funds of China (No.U1604112), the Funding Scheme for Young Core Teachers of Henan Province (2017), and Nanhu Scholars Program for Young Scholars of Xinyang Normal University.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Erneux, C.; Govaerts, C.; Communi, D.; Pesesse, X. The diversity and possible functions of the inositol polyphosphate 5-phosphatases. Biochim. Biophys. Acta 1998, 1436, 185–199. [Google Scholar] [CrossRef]
- Majerus, P.W.; Kisseleva, M.V.; Norris, F.A. The role of phosphatases in inositol signaling reactions. J. Biol. Chem. 1999, 274, 10669–10672. [Google Scholar] [CrossRef] [PubMed]
- Munnik, T.; Testerink, C. Plant phospholipid signaling: “in a nutshell”. J. Lipid Res. 2009, 50, S260–S265. [Google Scholar] [CrossRef]
- Kerr, W.G.; Colucci, F. Inositol phospholipid signaling and the biology of natural killer cells. J. Innate Immun. 2011, 3, 249–257. [Google Scholar] [CrossRef]
- Tsujishita, Y.; Guo, S.; Stolz, L.E.; York, J.D.; Hurley, J.H. Specificity Determinants in Phosphoinositide Dephosphorylation: Crystal Structure of an Archetypal Inositol Polyphosphate 5-Phosphatase. Cell 2001, 105, 379–389. [Google Scholar] [CrossRef]
- Stevenson, J.M.; Perera, I.Y.; Heilmann, I.I.; Persson, S.; Boss, W.F. Inositol signaling and plant growth. Trends Plant Sci. 2000, 5, 252–258. [Google Scholar] [CrossRef]
- Astle, M.V.; Seaton, G.; Davies, E.M.; Fedele, C.G.; Rahman, P.; Arsala, L.; Mitchell, C.A. Regulation of phosphoinositide signaling by the inositol polyphosphate 5-phosphatases. IUBMB Life 2006, 58, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Chen, X.; Li, G. Involvement of phospholipid signaling in plant growth and hormone effects. Curr. Opin. Plant Biol. 2007, 10, 483–489. [Google Scholar] [CrossRef]
- Xue, H.W.; Chen, X.; Mei, Y. Function and regulation of phospholipid signalling in plants. Biochem. J. 2009, 421, 145–156. [Google Scholar] [CrossRef]
- Pirruccello, M.; De Camilli, P. Inositol 5-phosphatases: Insights from the Lowe syndrome protein OCRL. Trends Biochem. Sci. 2012, 37, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Berdy, S.E.; Kudla, J.; Gruissem, W.; Gillaspy, G.E. Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating inositol trisphosphate signaling. Plant Physiol. 2001, 126, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Majerus, P.W. Inositols do it all. Genes Dev. 1996, 10, 1051–1053. [Google Scholar] [CrossRef] [PubMed]
- Stolz, L.E.; Huynh, C.V.; Thorner, J.; York, J.D. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics 1998, 148, 1715–1729. [Google Scholar]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef]
- Ai, J.; Maturu, A.; Johnson, W.; Wang, Y.; Marsh, C.B.; Tridandapani, S. The inositol phosphatase SHIP-2 down-regulates FcgammaR-mediated phagocytosis in murine macrophages independently of SHIP-1. Blood 2006, 107, 813–820. [Google Scholar] [CrossRef]
- Majerus, P.W.; York, J.D. Phosphoinositide phosphatases and disease. J. Lipid Res. 2009, 50, S249–S254. [Google Scholar] [CrossRef]
- Zhong, R.; Ye, Z.H. The SAC domain-containing protein gene family in Arabidopsis. Plant Physiol. 2003, 132, 544–555. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Ye, Z.H. Molecular and biochemical characterization of three WD-repeat-domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 1720–1728. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Lee, S.H.; Kim, H.; Jin, J.B.; Kim, D.H.; Hwang, I. A WD40 repeat protein, Arabidopsis Sec13 homolog 1, may play a role in vacuolar trafficking by controlling the membrane association of AtDRP2A. Mol. Cells 2006, 22, 210–219. [Google Scholar]
- Wiradjaja, F.; Ooms, L.M.; Whisstock, J.C.; McColl, B.; Helfenbaum, L.; Sambrook, J.F.; Gething, M.J.; Mitchell, C.A. The yeast inositol polyphosphate 5-phosphatase Inp54p localizes to the endoplasmic reticulum via a C-terminal hydrophobic anchoring tail: Regulation of secretion from the endoplasmic reticulum. J. Biol. Chem. 2001, 276, 7643–7653. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, C.; Wicky, S.; Schwarz, H.; Singer-Kruger, B. Sjl2p is specifically involved in early steps of endocytosis intimately linked to actin dynamics via the Ark1p/Prk1p kinases. Febs Lett. 2006, 580, 633–641. [Google Scholar] [CrossRef]
- Ha, S.A.; Bunch, J.T.; Hama, H.; DeWald, D.B.; Nothwehr, S.F. A novel mechanism for localizing membrane proteins to yeast trans-Golgi network requires function of synaptojanin-like protein. Mol. Biol. Cell 2001, 12, 3175–3190. [Google Scholar] [CrossRef]
- Roth, M.G. Phosphoinositides in constitutive membrane traffic. Physiol. Rev. 2004, 84, 699–730. [Google Scholar] [CrossRef]
- Ooms, L.M.; Horan, K.A.; Rahman, P.; Seaton, G.; Gurung, R.; Kethesparan, D.S.; Mitchell, C.A. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 2009, 419, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Attree, O.; Olivos, I.M.; Okabe, I.; Bailey, L.C.; Nelson, D.L.; Lewis, R.A.; McInnes, R.R.; Nussbaum, R.L. The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature 1992, 358, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Hoopes, R.R., Jr.; Shrimpton, A.E.; Knohl, S.J.; Hueber, P.; Hoppe, B.; Matyus, J.; Simckes, A.; Tasic, V.; Toenshoff, B.; Suchy, S.F.; et al. Dent Disease with mutations in OCRL1. Am. J. Hum. Genet. 2005, 76, 260–267. [Google Scholar] [CrossRef]
- Helgason, C.D.; Damen, J.E.; Rosten, P.; Grewal, R.; Sorensen, P.; Chappel, S.M.; Borowski, A.; Jirik, F.; Krystal, G.; Humphries, R.K. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998, 12, 1610–1620. [Google Scholar] [CrossRef] [PubMed]
- Paraiso, K.H.; Ghansah, T.; Costello, A.; Engelman, R.W.; Kerr, W.G. Induced SHIP deficiency expands myeloid regulatory cells and abrogates graft-versus-host disease. J. Immunol. 2007, 178, 2893–2900. [Google Scholar] [CrossRef] [PubMed]
- Suwa, A.; Kurama, T.; Shimokawa, T. SHIP2 and its involvement in various diseases. Expert Opin. Ther. Targets 2010, 14, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Sleeman, M.W.; Wortley, K.E.; Lai, K.M.; Gowen, L.C.; Kintner, J.; Kline, W.O.; Garcia, K.; Stitt, T.N.; Yancopoulos, G.D.; Wiegand, S.J.; et al. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat. Med. 2005, 11, 199–205. [Google Scholar] [CrossRef]
- Clement, S.; Krause, U.; Desmedt, F.; Tanti, J.F.; Behrends, J.; Pesesse, X.; Sasaki, T.; Penninger, J.; Doherty, M.; Malaisse, W.; et al. The lipid phosphatase SHIP2 controls insulin sensitivity. Nature 2001, 409, 92–97. [Google Scholar] [CrossRef]
- Bielas, S.L.; Silhavy, J.L.; Brancati, F.; Kisseleva, M.V.; Al-Gazali, L.; Sztriha, L.; Bayoumi, R.A.; Zaki, M.S.; Abdel-Aleem, A.; Rosti, R.O.; et al. Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat. Genet. 2009, 41, 1032–1036. [Google Scholar] [CrossRef]
- Gunesekera, B.; Torabinejad, J.; Robinson, J.; Gillaspy, G.E. Inositol polyphosphate 5-phosphatases 1 and 2 are required for regulating seedling growth. Plant Physiol. 2007, 143, 1408–1417. [Google Scholar] [CrossRef]
- Zhong, R.; Burk, D.H.; Morrison, W.H., 3rd; Ye, Z.H. FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. Plant Cell 2004, 16, 3242–3259. [Google Scholar] [CrossRef]
- Jones, M.A.; Raymond, M.J.; Smirnoff, N. Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. Cell Mol. Biol. 2006, 45, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Carland, F.M.; Nelson, T. Cotyledon vascular pattern2-mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell 2004, 16, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Carland, F.; Nelson, T. CVP2- and CVL1-mediated phosphoinositide signaling as a regulator of the ARF GAP SFC/VAN3 in establishment of foliar vein patterns. Plant J. Cell Mol. Biol. 2009, 59, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.H.; Wang, Y.; Mueller-Roeber, B.; Brearley, C.A.; Xu, Z.H.; Xue, H.W. At5PTase13 modulates cotyledon vein development through regulating auxin homeostasis. Plant Physiol. 2005, 139, 1677–1691. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lin, W.H.; Chen, X.; Xue, H.W. The role of Arabidopsis 5PTase13 in root gravitropism through modulation of vesicle trafficking. Cell Res. 2009, 19, 1191–1204. [Google Scholar] [CrossRef]
- Golani, Y.; Kaye, Y.; Gilhar, O.; Ercetin, M.; Gillaspy, G.; Levine, A. Inositol polyphosphate phosphatidylinositol 5-phosphatase9 (At5ptase9) controls plant salt tolerance by regulating endocytosis. Mol. Plant 2013, 6, 1781–1794. [Google Scholar] [CrossRef]
- Kaye, Y.; Golani, Y.; Singer, Y.; Leshem, Y.; Cohen, G.; Ercetin, M.; Gillaspy, G.; Levine, A. Inositol polyphosphate 5-phosphatase7 regulates the production of reactive oxygen species and salt tolerance in Arabidopsis. Plant Physiol. 2011, 157, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Avila, L.M.; Cerrudo, D.; Swanton, C.; Lukens, L. Brevis plant1, a putative inositol polyphosphate 5-phosphatase, is required for internode elongation in maize. J. Exp. Bot. 2016, 67, 1577–1588. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Finn, R.D.; Miller, B.L.; Clements, J.; Bateman, A. iPfam: A database of protein family and domain interactions found in the Protein Data Bank. Nucleic Acids Res. 2014, 42, D364–D373. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef]
- Catchen, J.M.; Conery, J.S.; Postlethwait, J.H. Automated identification of conserved synteny after whole-genome duplication. Genome Res. 2009, 19, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.A.; Peng, D.L.; et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef]
- McPherson, P.S.; Garcia, E.P.; Slepnev, V.I.; David, C.; Zhang, X.; Grabs, D.; Sossin, W.S.; Bauerfeind, R.; Nemoto, Y.; De Camilli, P. A presynaptic inositol-5-phosphatase. Nature 1996, 379, 353–357. [Google Scholar] [CrossRef]
- Zeng, L.; Zhang, N.; Zhang, Q.; Endress, P.K.; Huang, J.; Ma, H. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic. New Phytol. 2017, 214, 1338–1354. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Zhang, Q.; Sun, R.; Kong, H.; Zhang, N.; Ma, H. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 2014, 5, 4956. [Google Scholar] [CrossRef]
- Huang, C.H.; Sun, R.; Hu, Y.; Zeng, L.; Ma, H. Resolution of Brassicaceae Phylogeny Using Nuclear Genes Uncovers Nested Radiations and Supports Convergent Morphological Evolution. Mol. Biol. Evol. 2015, 33, 394–412. [Google Scholar] [CrossRef] [PubMed]
- Voronov, S.V.; Frere, S.G.; Giovedi, S.; Pollina, E.A.; Borel, C.; Zhang, H.; Schmidt, C.; Akeson, E.C.; Wenk, M.R.; Cimasoni, L.; et al. Synaptojanin 1-linked phosphoinositide dyshomeostasis and cognitive deficits in mouse models of Down’s syndrome. Proc. Natl. Acad. Sci. USA 2008, 105, 9415–9420. [Google Scholar] [CrossRef]
- Suchy, S.F.; Olivos-Glander, I.M.; Nussabaum, R.L. Lowe syndrome, a deficiency of phosphatidylinositol 4,5-bisphosphate 5-phosphatase in the Golgi apparatus. Hum. Mol. Genet. 1995, 4, 2245–2250. [Google Scholar] [CrossRef]
- Zhang, X.; Hartz, P.A.; Philip, E.; Racusen, L.C.; Majerus, P.W. Cell lines from kidney proximal tubules of a patient with Lowe syndrome lack OCRL inositol polyphosphate 5-phosphatase and accumulate phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 1998, 273, 1574–1582. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. Rates of intron loss and gain: Implications for early eukaryotic evolution. Proc. Natl. Acad. Sci. USA 2005, 102, 5773–5778. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Cannon, S.B.; McKain, M.R.; Harkess, A.; Nelson, M.N.; Dash, S.; Deyholos, M.K.; Peng, Y.; Joyce, B.; Stewart, C.N., Jr.; Rolf, M.; et al. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol. Biol. Evol. 2015, 32, 193–210. [Google Scholar] [CrossRef]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef]
- Lei, L.; Zhou, S.L.; Ma, H.; Zhang, L.S. Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 2012, 12, 51. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Ganko, E.W.; Meyers, B.C.; Vision, T.J. Divergence in expression between duplicated genes in Arabidopsis. Mol. Biol. Evol. 2007, 24, 2298–2309. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Yang, Z.L.; Yang, X.; Liu, Y.J.; Wang, X.R.; Zeng, Q.Y. Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell 2009, 21, 3749–3766. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.L.; Liu, H.J.; Wang, X.R.; Zeng, Q.Y. Molecular evolution and expression divergence of the Populus polygalacturonase supergene family shed light on the evolution of increasingly complex organs in plants. New Phytol. 2013, 197, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, B.; Yu, L.; Feng, D.; Wang, H.; Wang, J. Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol. Biol. 2009, 9, 90. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).