Retinal miRNA Functions in Health and Disease
Abstract
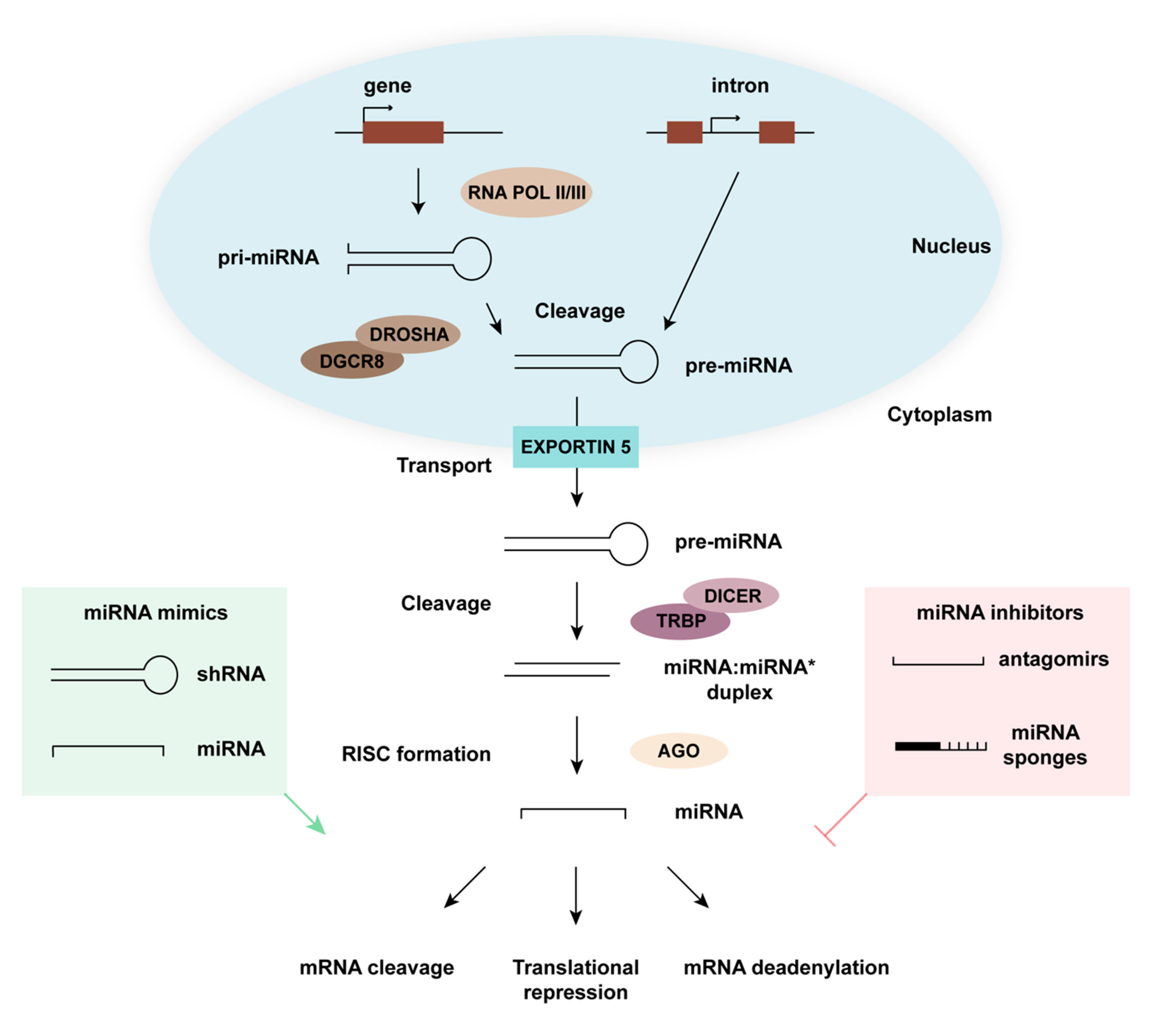
:1. miRNA Biogenesis and Function
2. Controlling Cellular miRNA Expression
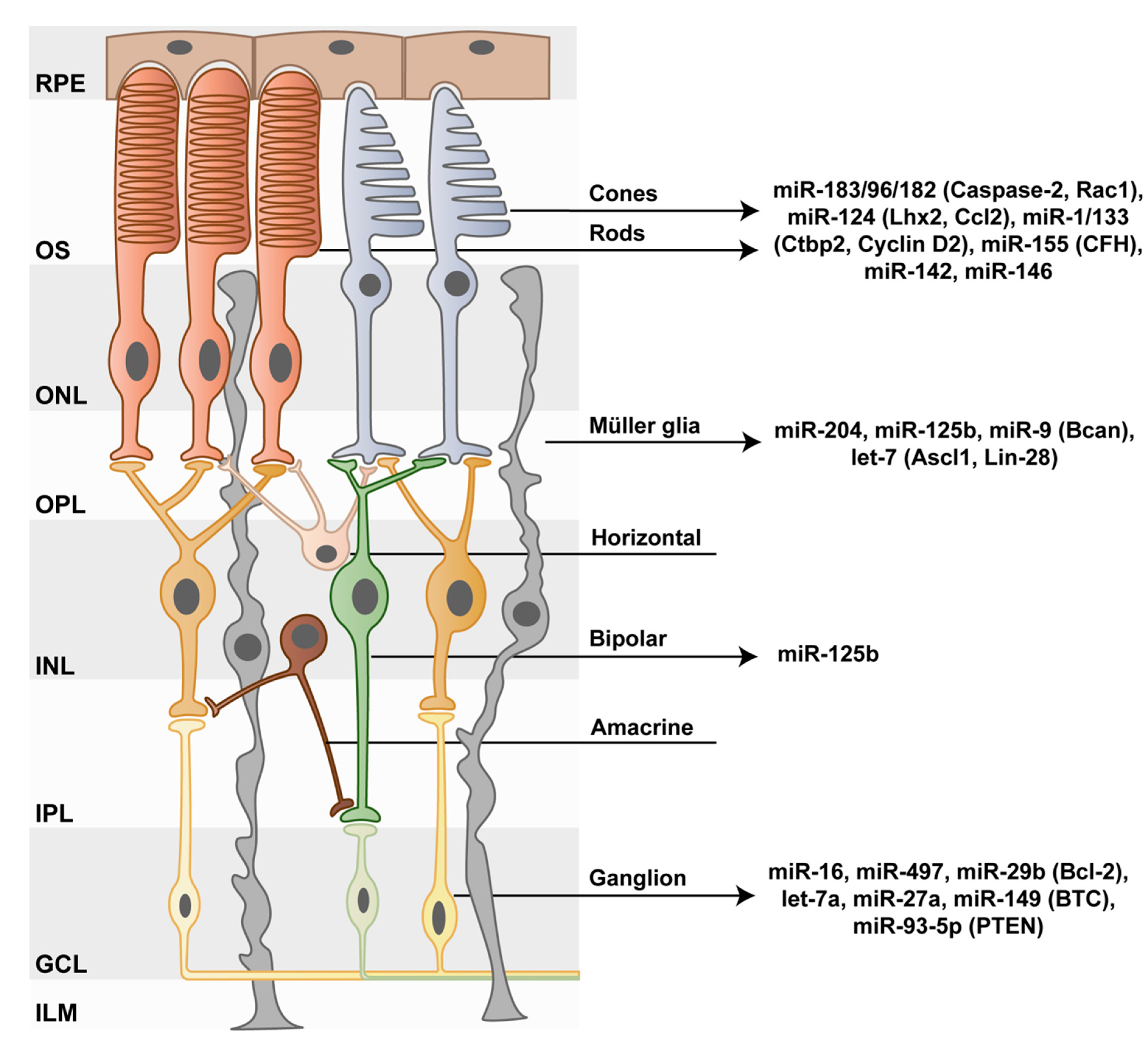
3. Photoreceptor–miRNAs as Cell Maintenance and Survival Regulators
4. The Impact of the miR-183/96/182 Cluster on Photoreceptors
5. miR-124 Protects Photoreceptors from Apoptosis
6. miRNA Functions in Inner Retinal Neurons
7. The role of miRNAs in Müller Glia Development and Function
8. miRNAs in Müller Glia De-Differentiation and Their Potential Regeneration Capacity
9. Global miRNA Alterations in Retinal Diseases
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Westholm, J.O.; Lai, E.C. Mirtrons: microRNA biogenesis via splicing. Biochimie 2011, 93, 1897–1904. [Google Scholar] [CrossRef] [Green Version]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Tan, C.L.; Plotkin, J.L.; Veno, M.T.; von Schimmelmann, M.; Feinberg, P.; Mann, S.; Handler, A.; Kjems, J.; Surmeier, D.J.; O’Carroll, D.; et al. MicroRNA-128 governs neuronal excitability and motor behavior in mice. Sci. (N.Y.) 2013, 342, 1254–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutsche, L.K.; Gysi, D.M.; Fallmann, J.; Lenk, K.; Petri, R.; Swiersy, A.; Klapper, S.D.; Pircs, K.; Khattak, S.; Stadler, P.F.; et al. Combined Experimental and System-Level Analyses Reveal the Complex Regulatory Network of miR-124 during Human Neurogenesis. Cell Syst. 2018, 7, 438–452 e438. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Ha, T.-Y. MicroRNAs in human diseases: From cancer to cardiovascular disease. Immune. Netw. 2011, 11, 135–154. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. Roles for microRNAs in conferring robustness to biological processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [Green Version]
- Sundermeier, T.R.; Palczewski, K. The impact of microRNA gene regulation on the survival and function of mature cell types in the eye. FASEB J. 2016, 30, 23–33. [Google Scholar] [CrossRef]
- Conte, I.; Hadfield, K.D.; Barbato, S.; Carrella, S.; Pizzo, M.; Bhat, R.S.; Carissimo, A.; Karali, M.; Porter, L.F.; Urquhart, J.; et al. MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc. Natl. Acad. Sci. USA 2015, 112, E3236–E3245. [Google Scholar] [CrossRef] [Green Version]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Paddison, P.J.; Caudy, A.A.; Bernstein, E.; Hannon, G.J.; Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002, 16, 948–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brummelkamp, T.R.; Bernards, R.; Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Sci. (N.Y.) 2002, 296, 550–553. [Google Scholar] [CrossRef] [PubMed]
- Busskamp, V.; Krol, J.; Nelidova, D.; Daum, J.; Szikra, T.; Tsuda, B.; Juttner, J.; Farrow, K.; Scherf, B.G.; Alvarez, C.P.; et al. miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron 2014, 83, 586–600. [Google Scholar] [CrossRef]
- Michel, U.; Malik, I.; Ebert, S.; Bahr, M.; Kugler, S. Long-term in vivo and in vitro AAV-2-mediated RNA interference in rat retinal ganglion cells and cultured primary neurons. Biochem. Biophys. Res. Commun. 2005, 326, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Busskamp, V.; Markiewicz, I.; Stadler, M.B.; Ribi, S.; Richter, J.; Duebel, J.; Bicker, S.; Fehling, H.J.; Schubeler, D.; et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 2010, 141, 618–631. [Google Scholar] [CrossRef]
- Trapani, I.; Auricchio, A. Seeing the Light after 25 Years of Retinal Gene Therapy. Trends Mol. Med. 2018, 24, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Hudry, E.; Vandenberghe, L.H. Therapeutic AAV Gene Transfer to the Nervous System: A Clinical Reality. Neuron 2019, 101, 839–862. [Google Scholar] [CrossRef]
- Sundermeier, T.R.; Zhang, N.; Vinberg, F.; Mustafi, D.; Kohno, H.; Golczak, M.; Bai, X.; Maeda, A.; Kefalov, V.J.; Palczewski, K. DICER1 is essential for survival of postmitotic rod photoreceptor cells in mice. FASEB J. 2014, 28, 3780–3791. [Google Scholar] [CrossRef] [Green Version]
- Aldunate, E.Z.; Di Foggia, V.; Di Marco, F.; Hervas, L.A.; Ribeiro, J.C.; Holder, D.L.; Patel, A.; Jannini, T.B.; Thompson, D.A.; Martinez-Barbera, J.P.; et al. Conditional Dicer1 depletion using Chrnb4-Cre leads to cone cell death and impaired photopic vision. Sci. Rep. 2019, 9, 2314. [Google Scholar] [CrossRef]
- Lagos-Quintana, M. New microRNAs from mouse and human. RNA 2003, 9, 175–179. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Witmer, P.D.; Lumayag, S.; Kovacs, B.; Valle, D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 2007, 282, 25053–25066. [Google Scholar] [CrossRef]
- Weston, M.D.; Pierce, M.L.; Rocha-Sanchez, S.; Beisel, K.W.; Soukup, G.A. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006, 1111, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Aldrich, B.T.; Frakes, E.P.; Kasuya, J.; Hammond, D.L.; Kitamoto, T. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience 2009, 164, 711–723. [Google Scholar] [CrossRef] [Green Version]
- Jin, Z.B.; Hirokawa, G.; Gui, L.; Takahashi, R.; Osakada, F.; Hiura, Y.; Takahashi, M.; Yasuhara, O.; Iwai, N. Targeted deletion of miR-182, an abundant retinal microRNA. Mol. Vis. 2009, 15, 523–533. [Google Scholar]
- Dambal, S.; Shah, M.; Mihelich, B.; Nonn, L. The microRNA-183 cluster: The family that plays together stays together. Nucleic Acids Res. 2015, 43, 7173–7188. [Google Scholar] [CrossRef]
- Zhu, Q.; Sun, W.; Okano, K.; Chen, Y.; Zhang, N.; Maeda, T.; Palczewski, K. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J. Biol. Chem. 2011, 286, 31749–31760. [Google Scholar] [CrossRef] [PubMed]
- Lumayag, S.; Haldin, C.E.; Corbett, N.J.; Wahlin, K.J.; Cowan, C.; Turturro, S.; Larsen, P.E.; Kovacs, B.; Witmer, P.D.; Valle, D.; et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc. Natl. Acad. Sci. USA 2013, 110, E507–E516. [Google Scholar] [CrossRef] [Green Version]
- Krol, J.; Krol, I.; Alvarez, C.P.; Fiscella, M.; Hierlemann, A.; Roska, B.; Filipowicz, W. A network comprising short and long noncoding RNAs and RNA helicase controls mouse retina architecture. Nat. Commun. 2015, 6, 7305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davari, M.; Soheili, Z.S.; Samiei, S.; Sharifi, Z.; Pirmardan, E.R. Overexpression of miR-183/-96/-182 triggers neuronal cell fate in Human Retinal Pigment Epithelial (hRPE) cells in culture. Biochem. Biophys. Res. Commun. 2017, 483, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Palfi, A.; Hokamp, K.; Hauck, S.M.; Vencken, S.; Millington-Ward, S.; Chadderton, N.; Carrigan, M.; Kortvely, E.; Greene, C.M.; Kenna, P.F.; et al. microRNA regulatory circuits in a mouse model of inherited retinal degeneration. Sci. Rep. 2016, 6, 31431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol.: Cb. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414. [Google Scholar] [CrossRef]
- Karali, M.; Peluso, I.; Marigo, V.; Banfi, S. Identification and characterization of microRNAs expressed in the mouse eye. Invest. Ophthalmol. Vis. Sci. 2007, 48, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Sanuki, R.; Onishi, A.; Koike, C.; Muramatsu, R.; Watanabe, S.; Muranishi, Y.; Irie, S.; Uneo, S.; Koyasu, T.; Matsui, R.; et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 2011, 14, 1125–1134. [Google Scholar] [CrossRef]
- Chu-Tan, J.A.; Rutar, M.; Saxena, K.; Aggio-Bruce, R.; Essex, R.W.; Valter, K.; Jiao, H.; Fernando, N.; Wooff, Y.; Madigan, M.C.; et al. MicroRNA-124 Dysregulation is Associated With Retinal Inflammation and Photoreceptor Death in the Degenerating Retina. Invest. Ophthalmol. Vis. Sci. 2018, 59, 4094–4105. [Google Scholar] [CrossRef]
- Rutar, M.; Natoli, R.; Valter, K.; Provis, J.M. Early focal expression of the chemokine Ccl2 by Muller cells during exposure to damage-inducing bright continuous light. Invest. Ophthalmol. Vis. Sci. 2011, 52, 2379–2388. [Google Scholar] [CrossRef]
- Newman, A.M.; Gallo, N.B.; Hancox, L.S.; Miller, N.J.; Radeke, C.M.; Maloney, M.A.; Cooper, J.B.; Hageman, G.S.; Anderson, D.H.; Johnson, L.V.; et al. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 2012, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Otani, A.; Oishi, A.; Kojima, H.; Makiyama, Y.; Nakagawa, S.; Yoshimura, N. Knockout of ccr2 alleviates photoreceptor cell death in a model of retinitis pigmentosa. Exp. Eye Res. 2012, 104, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutar, M.; Natoli, R.; Provis, J.M. Small interfering RNA-mediated suppression of Ccl2 in Muller cells attenuates microglial recruitment and photoreceptor death following retinal degeneration. J. Neuroinflammation 2012, 9, 221. [Google Scholar] [CrossRef] [PubMed]
- Sennlaub, F.; Auvynet, C.; Calippe, B.; Lavalette, S.; Poupel, L.; Hu, S.J.; Dominguez, E.; Camelo, S.; Levy, O.; Guyon, E.; et al. CCR2(+) monocytes infiltrate atrophic lesions in age-related macular disease and mediate photoreceptor degeneration in experimental subretinal inflammation in Cx3cr1 deficient mice. Embo. Mol. Med. 2013, 5, 1775–1793. [Google Scholar] [CrossRef]
- Fu, Y.; Hou, B.; Weng, C.; Liu, W.; Dai, J.; Zhao, C.; Yin, Z.Q. Functional ectopic neuritogenesis by retinal rod bipolar cells is regulated by miR-125b-5p during retinal remodeling in RCS rats. Sci. Rep. 2017, 7, 1011. [Google Scholar] [CrossRef]
- Jayaram, H.; Cepurna, W.O.; Johnson, E.C.; Morrison, J.C. MicroRNA Expression in the Glaucomatous Retina. Invest. Ophthalmol. Vis. Sci. 2015, 56, 7971–7982. [Google Scholar] [CrossRef] [Green Version]
- Nie, X.G.; Fan, D.S.; Huang, Y.X.; He, Y.Y.; Dong, B.L.; Gao, F. Downregulation of microRNA-149 in retinal ganglion cells suppresses apoptosis through activation of the PI3K/Akt signaling pathway in mice with glaucoma. Am. J. Physiol. Cell Physiol. 2018, 315, C839–C849. [Google Scholar] [CrossRef]
- Li, R.; Jin, Y.; Li, Q.; Sun, X.; Zhu, H.; Cui, H. MiR-93-5p targeting PTEN regulates the NMDA-induced autophagy of retinal ganglion cells via AKT/mTOR pathway in glaucoma. Biomed. Pharm. 2018, 100, 1–7. [Google Scholar] [CrossRef]
- Li, H.J.; Sun, Z.L.; Pan, Y.B.; Sun, Y.Y.; Xu, M.H.; Feng, D.F. Inhibition of miRNA-21 promotes retinal ganglion cell survival and visual function by modulating Muller cell gliosis after optic nerve crush. Exp. Cell Res. 2019, 375, 10–19. [Google Scholar] [CrossRef]
- Zhang, Q.L.; Wang, W.; Li, J.; Tian, S.Y.; Zhang, T.Z. Decreased miR-187 induces retinal ganglion cell apoptosis through upregulating SMAD7 in glaucoma. Biomed. Pharm. 2015, 75, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, N.; Luo, X. Intraocular miR-211 exacerbates pressure-induced cell death in retinal ganglion cells via direct repression of FRS2 signaling. Biochem. Biophys. Res. Commun. 2018, 503, 2984–2992. [Google Scholar] [CrossRef]
- Kong, N.; Lu, X.; Li, B. Downregulation of microRNA-100 protects apoptosis and promotes neuronal growth in retinal ganglion cells. BMC Mol. Biol. 2014, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Yu, Y.; Zhou, Q.; Li, C.; Yang, L.; Pei, C.G. Inhibition of miR-134 Protects Against Hydrogen Peroxide-Induced Apoptosis in Retinal Ganglion Cells. J. Mol. Neurosci. 2015, 56, 461–471. [Google Scholar] [CrossRef]
- Damiani, D.; Alexander, J.J.; O’Rourke, J.R.; McManus, M.; Jadhav, A.P.; Cepko, C.L.; Hauswirth, W.W.; Harfe, B.D.; Strettoi, E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J. Neurosci. 2008, 28, 4878–4887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinter, R.; Hindges, R. Perturbations of microRNA function in mouse dicer mutants produce retinal defects and lead to aberrant axon pathfinding at the optic chiasm. Plos ONE 2010, 5, e10021. [Google Scholar] [CrossRef]
- Iida, A.; Shinoe, T.; Baba, Y.; Mano, H.; Watanabe, S. Dicer plays essential roles for retinal development by regulation of survival and differentiation. Invest. Ophthalmol. Vis. Sci 2011, 52, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Reh, T.A.; Hindges, R. MicroRNAs in Retinal Development. Annu. Rev. Vis. Sci. 2018, 4, 25–44. [Google Scholar] [CrossRef]
- Georgi, S.A.; Reh, T.A. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J. Neurosci. 2010, 30, 4048–4061. [Google Scholar] [CrossRef] [Green Version]
- La Torre, A.; Georgi, S.; Reh, T.A. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, E2362–E2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X.; Ahmad, I. let-7 microRNA regulates neurogliogenesis in the mammalian retina through Hmga2. Dev. Biol. 2016, 410, 70–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karali, M.; Manfredi, A.; Puppo, A.; Marrocco, E.; Gargiulo, A.; Allocca, M.; Corte, M.D.; Rossi, S.; Giunti, M.; Bacci, M.L.; et al. MicroRNA-restricted transgene expression in the retina. Plos ONE 2011, 6, e22166. [Google Scholar] [CrossRef]
- Hackler, L., Jr.; Wan, J.; Swaroop, A.; Qian, J.; Zack, D.J. MicroRNA profile of the developing mouse retina. Invest. Ophthalmol. Vis. Sci. 2010, 51, 1823–1831. [Google Scholar] [CrossRef]
- Jeon, C.J.; Strettoi, E.; Masland, R.H. The major cell populations of the mouse retina. J. Neurosci. 1998, 18, 8936–8946. [Google Scholar] [CrossRef]
- Quintero, H.; Gomez-Montalvo, A.I.; Lamas, M. MicroRNA changes through Muller glia dedifferentiation and early/late rod photoreceptor differentiation. Neuroscience 2016, 316, 109–121. [Google Scholar] [CrossRef]
- Wohl, S.G.; Reh, T.A. The microRNA expression profile of mouse Muller glia in vivo and in vitro. Sci. Rep. 2016, 6, 35423. [Google Scholar] [CrossRef]
- Wohl, S.G.; Jorstad, N.L.; Levine, E.M.; Reh, T.A. Muller glial microRNAs are required for the maintenance of glial homeostasis and retinal architecture. Nat. Commun. 2017, 8, 1603. [Google Scholar] [CrossRef]
- Jones, B.W.; Watt, C.B.; Frederick, J.M.; Baehr, W.; Chen, C.K.; Levine, E.M.; Milam, A.H.; Lavail, M.M.; Marc, R.E. Retinal remodeling triggered by photoreceptor degenerations. J. Comp. Neurol. 2003, 464, 1–16. [Google Scholar] [CrossRef]
- Chung, S.H.; Shen, W.; Jayawardana, K.; Wang, P.; Yang, J.; Shackel, N.; Gillies, M.C. Differential gene expression profiling after conditional Muller-cell ablation in a novel transgenic model. Invest. Ophthalmol. Vis. Sci. 2013, 54, 2142–2152. [Google Scholar] [CrossRef]
- Chung, S.H.; Gillies, M.; Sugiyama, Y.; Zhu, L.; Lee, S.R.; Shen, W. Profiling of microRNAs involved in retinal degeneration caused by selective Muller cell ablation. Plos ONE 2015, 10, e0118949. [Google Scholar] [CrossRef] [PubMed]
- Byrne, L.C.; Khalid, F.; Lee, T.; Zin, E.A.; Greenberg, K.P.; Visel, M.; Schaffer, D.V.; Flannery, J.G. AAV-mediated, optogenetic ablation of Muller Glia leads to structural and functional changes in the mouse retina. Plos ONE 2013, 8, e76075. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Lee, S.R.; Araujo, J.; Chung, S.H.; Zhu, L.; Gillies, M.C. Effect of glucocorticoids on neuronal and vascular pathology in a transgenic model of selective Muller cell ablation. Glia 2014, 62, 1110–1124. [Google Scholar] [CrossRef]
- Ramachandran, R.; Fausett, B.V.; Goldman, D. Ascl1a regulates Muller glia dedifferentiation and retinal regeneration through a Lin-28-dependent, let-7 microRNA signalling pathway. Nat. Cell Biol. 2010, 12, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Goldman, D. Muller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 2014, 15, 431–442. [Google Scholar] [CrossRef]
- Yao, K.; Qiu, S.; Tian, L.; Snider, W.D.; Flannery, J.G.; Schaffer, D.V.; Chen, B. Wnt Regulates Proliferation and Neurogenic Potential of Muller Glial Cells via a Lin28/let-7 miRNA-Dependent Pathway in Adult Mammalian Retinas. Cell Rep. 2016, 17, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Gupta, S.; Chaudhary, M.; Khursheed, M.A.; Mitra, S.; Kurup, A.J.; Ramachandran, R. let-7 MicroRNA-Mediated Regulation of Shh Signaling and the Gene Regulatory Network Is Essential for Retina Regeneration. Cell Rep. 2018, 23, 1409–1423. [Google Scholar] [CrossRef] [PubMed]
- Jorstad, N.L.; Wilken, M.S.; Grimes, W.N.; Wohl, S.G.; VandenBosch, L.S.; Yoshimatsu, T.; Wong, R.O.; Rieke, F.; Reh, T.A. Stimulation of functional neuronal regeneration from Muller glia in adult mice. Nature 2017, 548, 103–107. [Google Scholar] [CrossRef]
- Papagiannakopoulos, T.; Kosik, K.S. MicroRNA-124: Micromanager of neurogenesis. Cell Stem Cell 2009, 4, 375–376. [Google Scholar] [CrossRef]
- Cheng, L.C.; Pastrana, E.; Tavazoie, M.; Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009, 12, 399–408. [Google Scholar] [CrossRef] [Green Version]
- Maiorano, N.A.; Mallamaci, A. Promotion of embryonic cortico-cerebral neuronogenesis by miR-124. Neural Dev. 2009, 4, 40. [Google Scholar] [CrossRef]
- Masserdotti, G.; Gillotin, S.; Sutor, B.; Drechsel, D.; Irmler, M.; Jorgensen, H.F.; Sass, S.; Theis, F.J.; Beckers, J.; Berninger, B.; et al. Transcriptional Mechanisms of Proneural Factors and REST in Regulating Neuronal Reprogramming of Astrocytes. Cell Stem Cell 2015, 17, 74–88. [Google Scholar] [CrossRef] [Green Version]
- Abrajano, J.J.; Qureshi, I.A.; Gokhan, S.; Zheng, D.; Bergman, A.; Mehler, M.F. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. Plos ONE 2009, 4, e7936. [Google Scholar] [CrossRef]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, J.W.; Lee, S.K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007, 21, 744–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conaco, C.; Otto, S.; Han, J.J.; Mandel, G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. USA 2006, 103, 2422–2427. [Google Scholar] [CrossRef] [Green Version]
- Nesti, E.; Corson, G.M.; McCleskey, M.; Oyer, J.A.; Mandel, G. C-terminal domain small phosphatase 1 and MAP kinase reciprocally control REST stability and neuronal differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, E3929–E3936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, M.; Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L.; Gill, G.N. Small CTD phosphatases function in silencing neuronal gene expression. Sci. (N.Y.) 2005, 307, 596–600. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, H.B.; Li, X.; Zhou, Y.; Xia, X.B.; Song, W.T. MiR-124 Promotes the Growth of Retinal Ganglion Cells Derived from Muller Cells. Cell Physiol. Biochem. 2018, 45, 973–983. [Google Scholar] [CrossRef]
- Ji, H.P.; Xiong, Y.; Song, W.T.; Zhang, E.D.; Gao, Z.L.; Yao, F.; Su, T.; Zhou, R.R.; Xia, X.B. MicroRNA-28 potentially regulates the photoreceptor lineage commitment of Muller glia-derived progenitors. Sci. Rep. 2017, 7, 11374. [Google Scholar] [CrossRef]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Loscher, C.J.; Hokamp, K.; Wilson, J.H.; Li, T.; Humphries, P.; Farrar, G.J.; Palfi, A. A common microRNA signature in mouse models of retinal degeneration. Exp. Eye Res. 2008, 87, 529–534. [Google Scholar] [CrossRef]
- Anasagasti, A.; Ezquerra-Inchausti, M.; Barandika, O.; Munoz-Culla, M.; Caffarel, M.M.; Otaegui, D.; Lopez de Munain, A.; Ruiz-Ederra, J. Expression Profiling Analysis Reveals Key MicroRNA-mRNA Interactions in Early Retinal Degeneration in Retinitis Pigmentosa. Invest. Ophthalmol. Vis. Sci. 2018, 59, 2381–2392. [Google Scholar] [CrossRef]
- Chang, B.; Hawes, N.L.; Hurd, R.E.; Davisson, M.T.; Nusinowitz, S.; Heckenlively, J.R. Retinal degeneration mutants in the mouse. Vis. Res. 2002, 42, 517–525. [Google Scholar] [CrossRef] [Green Version]
- Genini, S.; Guziewicz, K.E.; Beltran, W.A.; Aguirre, G.D. Altered miRNA expression in canine retinas during normal development and in models of retinal degeneration. BMC Genom. 2014, 15, 172. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: microRNAs can up-regulate translation. Sci. (N.Y.) 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Papermaster, D.; Cepko, C.L. A unique pattern of photoreceptor degeneration in cyclin D1 mutant mice. Proc. Natl. Acad. Sci. USA 1998, 95, 9938–9943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kutty, R.K.; Nagineni, C.N.; Samuel, W.; Vijayasarathy, C.; Hooks, J.J.; Redmond, T.M. Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochem. Biophys. Res. Commun. 2010, 402, 390–395. [Google Scholar] [CrossRef] [Green Version]
- Lukiw, W.J.; Surjyadipta, B.; Dua, P.; Alexandrov, P.N. Common micro RNAs (miRNAs) target complement factor H (CFH) regulation in Alzheimer’s disease (AD) and in age-related macular degeneration (AMD). Int. J. Biochem. Mol. Biol. 2012, 3, 105–116. [Google Scholar]
- Karali, M.; Persico, M.; Mutarelli, M.; Carissimo, A.; Pizzo, M.; Singh Marwah, V.; Ambrosio, C.; Pinelli, M.; Carrella, D.; Ferrari, S.; et al. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res 2016, 44, 1525–1540. [Google Scholar] [CrossRef] [Green Version]
- Ryan, D.G.; Oliveira-Fernandes, M.; Lavker, R.M. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol. Vis. 2006, 12, 1175–1184. [Google Scholar]
- Karali, M.; Peluso, I.; Gennarino, V.A.; Bilio, M.; Verde, R.; Lago, G.; Dolle, P.; Banfi, S. miRNeye: A microRNA expression atlas of the mouse eye. BMC Genom. 2010, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011, 12, 846–860. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuzic, M.; Rojo Arias, J.E.; Wohl, S.G.; Busskamp, V. Retinal miRNA Functions in Health and Disease. Genes 2019, 10, 377. https://doi.org/10.3390/genes10050377
Zuzic M, Rojo Arias JE, Wohl SG, Busskamp V. Retinal miRNA Functions in Health and Disease. Genes. 2019; 10(5):377. https://doi.org/10.3390/genes10050377
Chicago/Turabian StyleZuzic, Marta, Jesus Eduardo Rojo Arias, Stefanie Gabriele Wohl, and Volker Busskamp. 2019. "Retinal miRNA Functions in Health and Disease" Genes 10, no. 5: 377. https://doi.org/10.3390/genes10050377
APA StyleZuzic, M., Rojo Arias, J. E., Wohl, S. G., & Busskamp, V. (2019). Retinal miRNA Functions in Health and Disease. Genes, 10(5), 377. https://doi.org/10.3390/genes10050377




