RNA-seq Transcriptome Analysis in Ovarian Tissue of Pelibuey Breed to Explore the Regulation of Prolificacy
Abstract
1. Introduction
2. Materials and Methods
2.1. Synchronize Estrus and Sample Preparation
2.2. RNA Extraction
2.3. Library Preparation and Sequencing
2.4. Data Analysis and Differential Expression Analysis
2.5. Analysis of Gene Ontology Category and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway
2.6. Validation of RNA-Seq Data by Quantitative Real-Time Polymerase Chain Reaction
3. Results
3.1. Identification of Differentially Expressed Genes
3.2. Gene Ontology and KEGG Analysis
3.3. Validation of RNA-seq Data by qRT-PCR
4. Discussion
Gene Ontology and KEGG Analysis: Metabolic Functions in Folliculogenesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Feng, X.; Li, F.; Wang, F.; Zhang, G.; Pang, J.; Ren, C.; Zhang, T.; Yang, H.; Wang, Z.; Zhang, Y. Genome-wide differential expression profiling of mRNAs and lncRNAs associated with prolificacy in Hu sheep. Biosci. Rep. 2018, 38, BSR20171350. [Google Scholar] [CrossRef] [PubMed]
- El-halawany, N.; Kandil, O.M.; Shawky, A.A.; Al-tohamy, A.F.M.; El-sayd, Y.A.; Abdel-shafy, H.; Abou-fandoud, E.I.; Abdel-azeem, S.N.; El-rahim, A.H.A.; Abdoon, A.S.S.; et al. Investigating the e ffect of GDF9, BMP15, BMP6 and BMPR1B polymorphisms on Egyptian sheep fecundity and their transcripts expression in ovarian cells. Small Rumin. Res. 2018, 165, 34–40. [Google Scholar] [CrossRef]
- Galina, M.A.; Morales, R.; Silva, E.; Lpez, B. Reproductive performance of Pelibuey and Blackbelly sheep under tropical management systems in Mexico. Small Rumin. Res. 1996, 22, 31–37. [Google Scholar] [CrossRef]
- Ramírez, M.G.; Soto, G.; Poindron, M.; Álvarez, R.; Valencia, M.; González, D.; Terrazas, G. Maternal behaviour around birth and mother-young recognition in Pelibuey sheep. Vet. Mex. 2011, 42, 27–46. [Google Scholar]
- Abdoli, R.; Zamani, P.; Mirhoseini, S.Z.; Ghavi Hossein-Zadeh, N.; Nadri, S. A review on prolificacy genes in sheep. Reprod. Domest. Anim. 2016, 51, 631–637. [Google Scholar] [CrossRef]
- Mulsant, P.; Lecerf, F.; Fabre, S.; Schibler, L.; Monget, P.; Lanneluc, I.; Pisselet, C.; Riquet, J.; Monniaux, D.; Callebaut, I.; et al. Mutation in bone morphogenetic protein receptor-IB is associated with increased ovulation rate in Booroola Merino ewes. Proc. Natl. Acad. Sci. USA 2001, 98, 5104–5109. [Google Scholar] [CrossRef]
- Hanrahan, J.P.; Gregan, S.M.; Mulsant, P.; Mullen, M.; Davis, H.G.; Powell, R.; Galloway, M.S. Mutations in the Genes for Oocyte-Derived Growth Factors GDF9 and BMP15 Are Associated with Both Increased Ovulation Rate and Sterility in Cambridge and Belclare Sheep (Ovis aries). Biol. Reprod. 2004, 70, 900–909. [Google Scholar] [CrossRef]
- Vacca, G.M.; Dhaouadi, A.; Rekik, M.; Carcangiu, V.; Pazzola, M.; Dettori, M.L. Prolificacy genotypes at BMPR 1B, BMP15 and GDF9 genes in North African sheep breeds. Small Rumin. Res. 2010, 88, 67–71. [Google Scholar] [CrossRef]
- Eghbalsaied, S.; Rashidi, K.F.; Amini, H.R.; Farahi, M.; Davari, M.; Pirali, A.; Pourali, S.; Vatankhah, M.; Rostami, M.; Atashi, H. Variant GDF9 mRNA is likely not the main cause of larger litter size in Iranian Lori-Bakhtyari, Shal, Ghezel, and Afshari sheep breeds. Arch. Anim. Breed. 2017, 60, 119–129. [Google Scholar] [CrossRef]
- Demars, J.; Fabre, S.; Sarry, J.; Rossetti, R.; Gilbert, H.; Persani, L.; Tosser-Klopp, G.; Mulsant, P.; Nowak, Z.; Drobik, W.; et al. Genome-Wide Association Studies Identify Two Novel BMP15 Mutations Responsible for an Atypical Hyperprolificacy Phenotype in Sheep. PLoS Genet. 2013, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- López-Ramírez, R.B.; Magaña-Sevilla, H.F.; Zamora-Bustillos, R.; Ramón-Ugalde, J.P.; González-Mendoza, D. Analysis of the 3’ End Regions of the GDF9 and BMPR1B Genes in Blackbelly Sheep from Yucatán, Mexico. Cienc. E Investig. Agrar. 2014, 41, 23–24. [Google Scholar] [CrossRef]
- Nagdy, H.; Gh, K.; Mahmoud, M.; Kandiel, M.M.M.; Helmy, N.A.; Ibrahim, S.S.; Nawito, M.F.; Othman, O.E. International Journal of Veterinary Science and Medicine PCR-RFLP of bone morphogenetic protein 15 (BMP15/FecX) gene as a candidate for proli fi cacy in sheep. Int. J. Vet. Sci. Med. 2018, 6, S68–S72. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Shen, H.; Jia, B.; Zhang, Y.S.; Wang, X.H.; Zeng, X.C. Differential gene expression in ovaries of Qira black sheep and Hetian sheep using RNA-seq technique. PLoS ONE 2015, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Jia, R.; Ying, S.; Wang, Z.; Wang, F. Analysis of genes that influence sheep follicular development by different nutrition levels during the luteal phase using expression profiling. Anim. Genet. 2016, 47, 354–364. [Google Scholar] [CrossRef]
- Patel, R.K.; Mukesh, J. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PLoS ONE 2012, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Am. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Aoki-kinoshita, K.F.; Kanehisa, M. Gene Annotation and Pathway Mapping in KEGG. Methods Mol. Biol. 2007, 396, 71–91. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO : A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, w293–w297. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Uri-belapolsky, S.; Miller, I.; Shaish, A.; Levi, M.; Harats, D.; Ninio-Many, L.; Kamari, Y.; Shalgi, R. Interleukin 1-α deficiency increases the expression of follicle stimulating hormone receptors in granulosa cells. Mol. Reprod. Dev. 2017, 84, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Lakshminarayana, B.N.V.; Chakravarthi, V.P.; Brahmaiah, K.V.; Rao, V.H. Quantification of P450 aromatase gene expression in cultured and in vivo grown ovarian follicles in sheep. Small Rumin. Res. 2014, 117, 66–72. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, L.; Feng, G.; Xiang, W.; Zhang, K.; Chu, M.; Wang, P. MicroRNA Mediating Networks in Granulosa Cells Associated with Ovarian Follicular Development. Biomed. Res. Int. 2017, 2017, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Pokharel, K.; Peippo, J.; Andersson, G.; Li, M.; Kantanen, J. Transcriptome profiling of Finnsheep ovaries during out-of-season breeding period. Agric. Food Sci. 2015, 24, 1–9. [Google Scholar] [CrossRef]
- Chen, A.Q.; Guang, Z.; Rong, Z.; Dong, Y.S.; Gang, Y.Z. Analysis of gene expression in granulosa cells of ovine antral growing follicles using suppressive subtractive hybridization. Anim. Reprod. Sci. J. 2009, 115, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Price, C.A. Mechanisms of fibroblast growth factor signaling in the ovarian follicle. J. Endocrinol. 2016, 228, R31–R43. [Google Scholar] [CrossRef] [PubMed]
- Portela, V.M.; Machado, M.; Buratini, J., Jr.; Zamberlam, G.; Amorim, R.L.; Goncalves, P.; Price, C.A. Expression and Function of Fibroblast Growth Factor 18 in the Ovarian Follicle in Cattle. Biol. Reprod. 2010, 83, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Imtaiyaz, H.M.; Shajee, B.; Waheed, A.; Ahmad, F.; Sly, W.S. Structure, function and applications of carbonic anhydrase isozymes. Bioorg. Med. Chem. 2013, 21, 1570–1582. [Google Scholar] [CrossRef]
- Hynninen, P.; Hämäläinen, J.M.; Pastorekova, S.; Pastorek, J.; Waheed, A.; Sly, W.S.; Tomas, E.; Kirkinen, P.; Parkkila, S. Transmembrane carbonic anhydrase isozymes IX and XII in the female mouse reproductive organs. Reprod. Biol. Endocrinol. 2004, 2, 1–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Friedley, N.J.; Rosen, S. Carbonic anhydrase activity in the mammalian ovary, fallopian tube, and uterus: histochemical and biochemical studies. Biol. Reprod. 1975, 12, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Nagao, Y.; Batanian, J.; Clemente, M.; Sly, W. Genomic Organization of the Human Gene (CA5) and Pseudogene for Mitochondrial Cabonic Anhydrase V and Their Localization to Chromosomes 16q and 16p. Genomics 1995, 28, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Van Karnebeek, C.D.; Sly, W.S.; Ross, C.J.; Salvarinova, R.; Yaplito-lee, J.; Santra, S.; Shyr, C.; Horvath, G.A.; Eydoux, P.; Lehman, A.M.; et al. Mitochondrial Carbonic Anhydrase VA Deficiency Resulting from CA5A Alterations Presents with Hyperammonemia in Early Childhood. Am. J. Hum. Genet. 2014, 94, 453–461. [Google Scholar] [CrossRef]
- Jorgez, C.J.; Klysik, M.; Jamin, S.P.; Behringer, R.R.; Matzuk, M.M. Granulosa Cell-Specific Inactivation of Follistatin Causes Female Fertility Defects. Mol. Endocrinol. 2004, 18, 953–967. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Q.; Pan, Y.; Gao, Z.; Liu, Y.; Chen, S. Effects of over-expressing resistin on glucose and lipid metabolism in mice. J. Zhejiang Univ. Sci. B 2008, 9, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Rak-Mardyła, A.; Durak, M.; Gregoraszczuk, E.L. Effects of resistin on porcine ovarian follicle steroidogenesis in prepubertal animals: An in vitro study. Reprod. Biol. Endocrinol. 2013, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dankert, J.F.; Pagan, J.K.; Starostina, N.G.; Kipreos, E.T.; Pagano, M. FEM1 proteins are ancient regulators of SLBP degradation. Cell Cycle. 2017, 16, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liao, S.; Zhuang, X.; Han, C. Mouse Fem1b interacts with and induces ubiquitin-mediated degradation of Ankrd37. Gene 2011, 485, 153–159. [Google Scholar] [CrossRef]
- Goodarzi, M.O.; Maher, J.F.; Cui, J.; Guo, X.; Taylor, K.D.; Azziz, R. FEM1A and FEM1B: Novel candidate genes for polycystic ovary syndrome. Hum. Reprod. 2008, 23, 2842–2849. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.J.; Lima, P.D.A.; Salehi, R.; Lee, D.R.; Tsang, B.K. Regulation of androgen receptor signaling by ubiquitination during folliculogenesis and its possible dysregulation in polycystic ovarian syndrome. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Light, A.; Hammes, S.R. LH-Induced Steroidogenesis in the Mouse Ovary, but Not Testis, Requires Matrix Metalloproteinase 2- and 9-Mediated Cleavage of Upregulated EGF Receptor Ligands. Biol. Reprod. 2015, 93, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huet, C.; Monget, P.; Pisselet, C.; Hennequet, C.; Locatelli, A.; Monniaux, D. Chronology of events accompanying follicular atresia in hypophysectomized ewes.Changes in levels of steroidogenic enzymes, connexin 43, insulin-like growth factor II/mannose 6 phosphate receptor, extracellular matrix components, and matrix metalloprotein. Biol. Reprod. 1998, 58, 175–185. [Google Scholar] [CrossRef]
- Choudhary, D.; Jansson, I.; Schenkman, J.B.; Sarfarazi, M.; Stoilov, I. Comparative expression profiling of 40 mouse cytochrome P450 genes in embryonic and adult tissues. Arch. Biochem. Biophys. 2003, 414, 91–100. [Google Scholar] [CrossRef]
- Taylor, H.S.; Vanden Heuvel, G.B.; Igarashi, P. A Conserved HOX Axis in the Mouse and Human Female Reproductive System: Late Establishment and Persistent Adult Expression of the Hoxa Cluster Genes. Biol. Reprod. 1997, 57, 1338–1345. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.Y.; Ladanyi, A.; Lengyel, E.; Naora, H. Expression of the Homeobox Gene HOXA9 in Ovarian Cancer Induces Peritoneal Macrophages to Acquire an M2 Tumor-Promoting Phenotype. Am. J. Pathol. 2014, 184, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Martín-Romero, F.J.; Ortíz-de-Galistea, J.R.; Lara-Laranjeira, J.; Domínguez-Arroyo, J.A.; González-Carrera, E.; Álvarez, I.S. Store-Operated Calcium Entry in Human Oocytes and Sensitivity to Oxidative Stress. Biol. Reprod. 2008, 315, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Arur, S. Conserved Insulin Signaling in the Regulation of Oocyte Growth, Development, and Maturation. Mol. Reprod. Dev. 2017, 84, 444–459. [Google Scholar] [CrossRef]
- Hicks, J.E.; Petters, R.M.; Sommer, J.R.; Farin, C.E. Nr4a1 mRNA regulation of FSH-induced meiotic resumption in bovine cumulus-oocyte complexes cultured in vitro. J. Bras. Reprod. Assist. 2014, 18, 69–75. [Google Scholar] [CrossRef]
- Schmitt, A.; Nebreda, A.R. Signalling pathways in oocyte meiotic maturation. J. Cell Sci. 2002, 115, 2457–2459. [Google Scholar]
- Yu, F.Q.; Han, C.S.; Yang, W.; Jin, X.; Hu, Z.Y.; Liu, Y.X. Activation of the p38 MAPK pathway by follicle-stimulating hormone regulates steroidogenesis in granulosa cells differentially. J. Endocrinol. 2005, 186, 85–96. [Google Scholar] [CrossRef]

| Name | Size (pb) | Primer Sequence | Tm (C°) | Efficiency |
|---|---|---|---|---|
| CA5A | 154 | F: 5′-CCCCTTGGAAAACCACTACA-3′ | 55.3 | 100.0 |
| R: 5′-TGGTATTTCACAGCGTTCCA-3′ | ||||
| FEM1B | 159 | F: 5′-AAGGTTTCACCTTGCTGCAT-3′ | 52.0 | 106.1 |
| R: 5’-CATTGTCCACAGCATTCACC-3′ | ||||
| β-actin | 146 | F: 5′-TGCGGCATTCACGAAACTAC-3′ | 55.9 | 100 |
| R: 5′-AGGGCAGTGATCTCTTTCTG-3′ |
| File Name | Group M | Group U |
|---|---|---|
| Total number of reads | 64,998,983 | 61,941,256 |
| Average length of readings (n) | 75.4 | 75.4 |
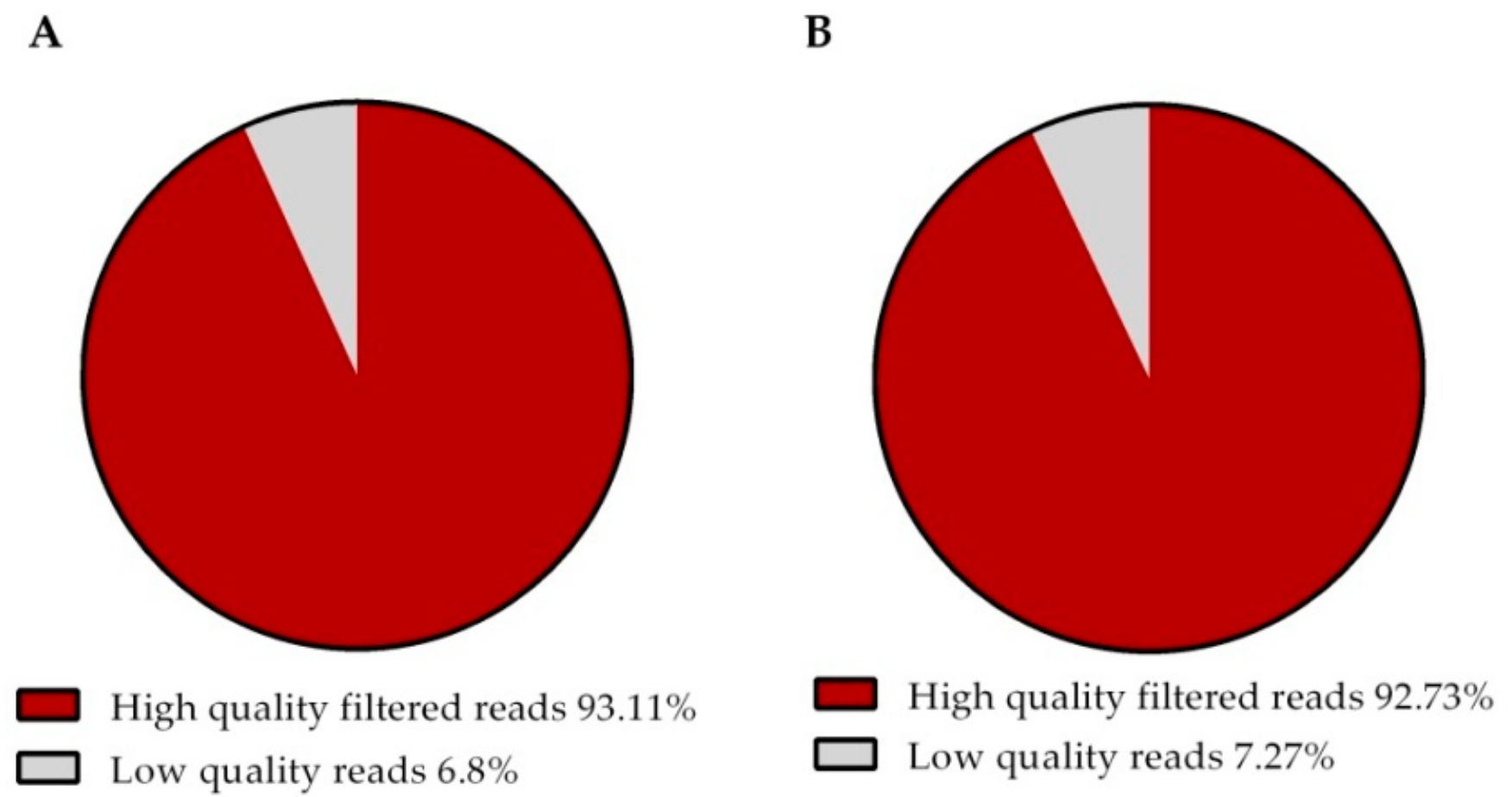
| Number total of HQ reads | 60,530,690 | 57,444,313 |
| Percentage of HQ reads (%) | 93.60 | 93.60 |
| Total number of HQ bases in HQ reads | 4,364,547,403 | 4,148,052,967 |
| Number of primer/Adaptor contaminate HQ reads | 1,452 | 1,332 |
| Total number of HQ filtered reads | 60,528,631 | 57,442,547 |
| Percentage of HQ filtered reads (%) | 93.11 | 92.24 |
| Gene Name | Log2 *FC | p-Value | FPKM | FDR | Up/Down-Regulated | Description | |
|---|---|---|---|---|---|---|---|
| M | U | ||||||
| FGF18 | inf | 0.00005 | 0.891434 | 0 | 0.000345978522902 | Up | Fibroblast Growth Factor 18 |
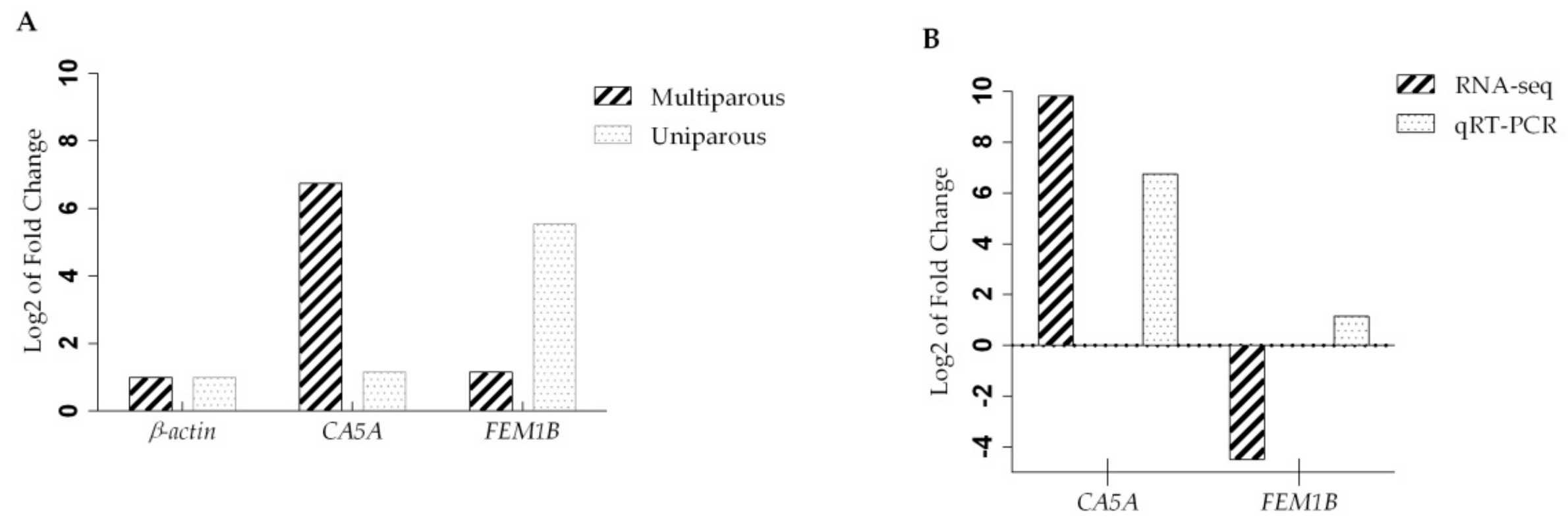
| CA5A | 9.82667 | 0.00005 | 126.35 | 0.13914 | 0.000345978522902 | Up | Carbonic anhydrase 5A |
| SVIP | 5.61939 | 0.00005 | 25.8765 | 0.526384 | 0.000345978522902 | Up | small VCP interacting protein |
| CXCL14 | 2.67744 | 0.00005 | 23.8323 | 3.72543 | 0.000345978522902 | Up | C-X-C motif chemokine ligand 14 |
| FST | 1.32408 | 0.00005 | 259.25 | 103.546 | 0.000345978522902 | Up | Follistatin |
| RENT | –inf | 0.00005 | 6.84467 | 0 | 0.000345978522902 | Down | Resistin |
| FEM1B | –4.48487 | 0.00005 | 557.61 | 24.9029 | 0.000345978522902 | Down | Fem-1-Like Death Receptor-Binding Protein α |
| MMP9 | –3.14351 | 0.00005 | 46.1391 | 5.22128 | 0.000345978522902 | Down | Matrix Metalloproteinase 9 |
| HOXA9 | –2.26451 | 5E-05 | 9.80922 | 2.0415 | 0.000345978522902 | Down | Homeobox protein HOX-A9 |
| CYP2S1 | –2.93691 | 0.00005 | 0.31288 | 2.39599 | 0.000664855759107 | Down | Cytochrome P450, Family 2, Subfamily S, Polypeptide 1 |
| Pathway ID | Number of Genes | KEEG Pathway | Up-Regulate | Down-Regulate |
|---|---|---|---|---|
| oas04020 | 6 | Calcium-signaling pathway | ATP2A3; ERBB3; MYLK4; PLCB2; PTGFR; PRKCB | |
| oas04350 | 2 | TGF-β-signaling pathway | FST; INHBA | |
| oas04910 | 4 | Insulin-signaling pathway | CBL; SOCS3 | SHC4; PRKAR1A |
| oas04151 | 9 | PI3K-Akt-signaling pathway | FGF18; NR4A1 | COL27A1; CSF1R; ITGB7; IL6R; IL7R; SPP1; SYK |
| oas04915 | 4 | Estrogen-signaling pathway | SHC4; HSPA1A; MMP9; PLCB2 | |
| oas04024 | 3 | cAMP-signaling pathway | HTR1B; FOS; HHIP | |
| oas04917 | 3 | Prolactin-signaling pathway | FOS; SOCS3 | SHC4 |
| oas04010 | 7 | MAPK-signaling pathway | FOS; DUSP4; FGF18; NR4A1 | HSPA1A; IL1A; PRKCB |
| oas04015 | 10 | Rap1-signaling pathway | FGF18; P2RY1 | FYB; CSF1R; ITGAL; ITGAM; ITGB2; PLCB2; PRKCB; SKAP1 |
| oas04911 | 2 | Insulin secretion | PRKCB; PLCB2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Montiel, W.; Collí-Dula, R.C.; Ramón-Ugalde, J.P.; Martínez-Núñez, M.A.; Zamora-Bustillos, R. RNA-seq Transcriptome Analysis in Ovarian Tissue of Pelibuey Breed to Explore the Regulation of Prolificacy. Genes 2019, 10, 358. https://doi.org/10.3390/genes10050358
Hernández-Montiel W, Collí-Dula RC, Ramón-Ugalde JP, Martínez-Núñez MA, Zamora-Bustillos R. RNA-seq Transcriptome Analysis in Ovarian Tissue of Pelibuey Breed to Explore the Regulation of Prolificacy. Genes. 2019; 10(5):358. https://doi.org/10.3390/genes10050358
Chicago/Turabian StyleHernández-Montiel, Wilber, Reyna Cristina Collí-Dula, Julio Porfirio Ramón-Ugalde, Mario Alberto Martínez-Núñez, and Roberto Zamora-Bustillos. 2019. "RNA-seq Transcriptome Analysis in Ovarian Tissue of Pelibuey Breed to Explore the Regulation of Prolificacy" Genes 10, no. 5: 358. https://doi.org/10.3390/genes10050358
APA StyleHernández-Montiel, W., Collí-Dula, R. C., Ramón-Ugalde, J. P., Martínez-Núñez, M. A., & Zamora-Bustillos, R. (2019). RNA-seq Transcriptome Analysis in Ovarian Tissue of Pelibuey Breed to Explore the Regulation of Prolificacy. Genes, 10(5), 358. https://doi.org/10.3390/genes10050358





