Neural Ganglia Transcriptome and Peptidome Associated with Sexual Maturation in Female Pacific Abalone (Haliotis discus hannai)
Abstract
:1. Introduction
2. Materials and Methods
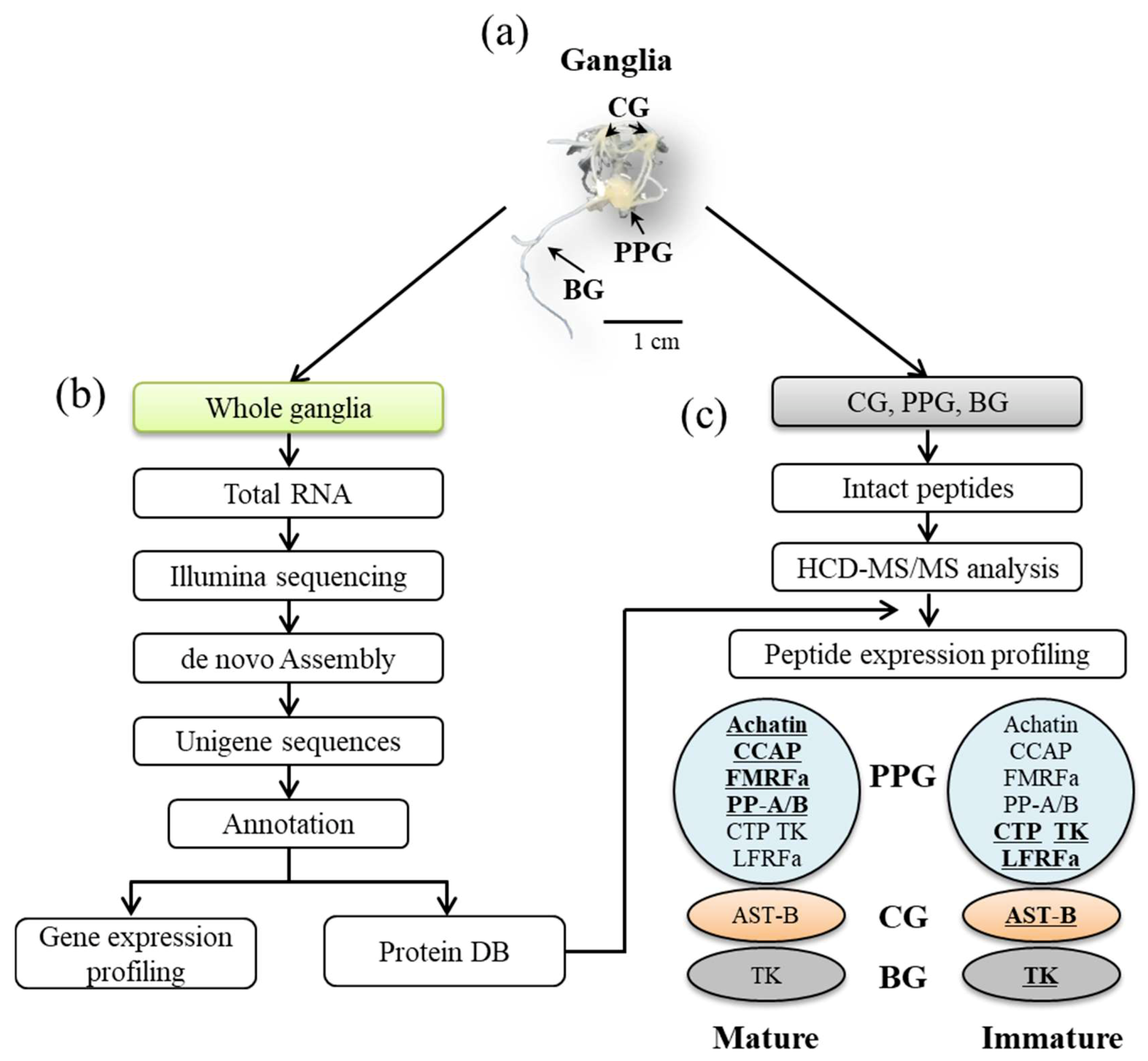
2.1. Animals and Tissue Collection
2.2. mRNA Library and Sequencing
2.3. De novo Assembly and Annotation
2.4. Differentially Expressed Genes (DEGs) Analysis
2.5. Sample Preparation and Peptide Extraction
2.6. LC-MS/MS
2.7. Peptide Identification and Quantification by MaxQunat
2.8. Neuropeptide Prediction
2.9. Quantitative Real-Time RT-PCR
3. Results
3.1. Transcriptome Assembly and Annotation
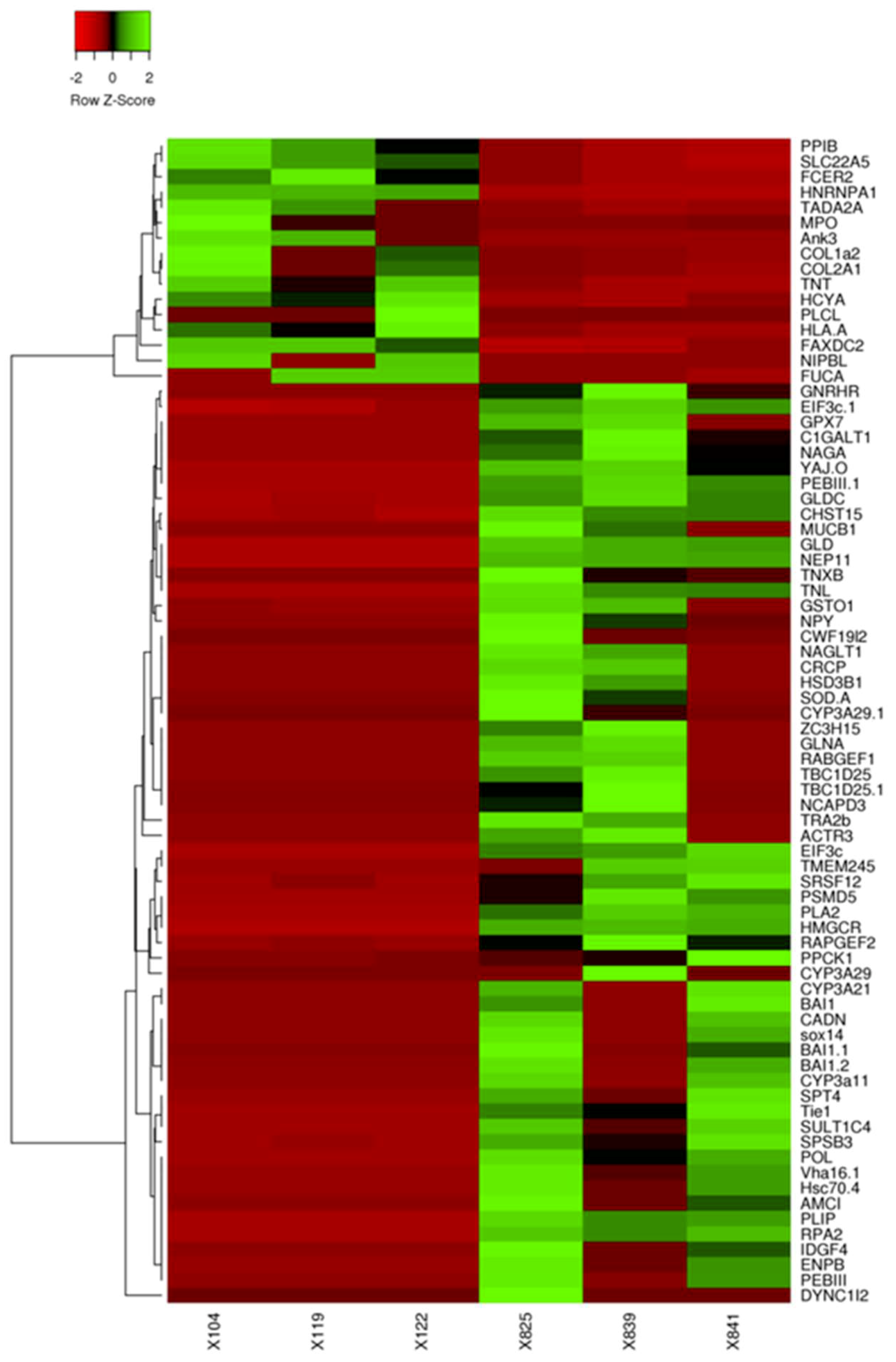
3.2. DEGs in the Ganglia of Immature and Mature Abalone
3.3. Transcriptome Analysis Identified Putative Genes Involved in Sexual Maturation
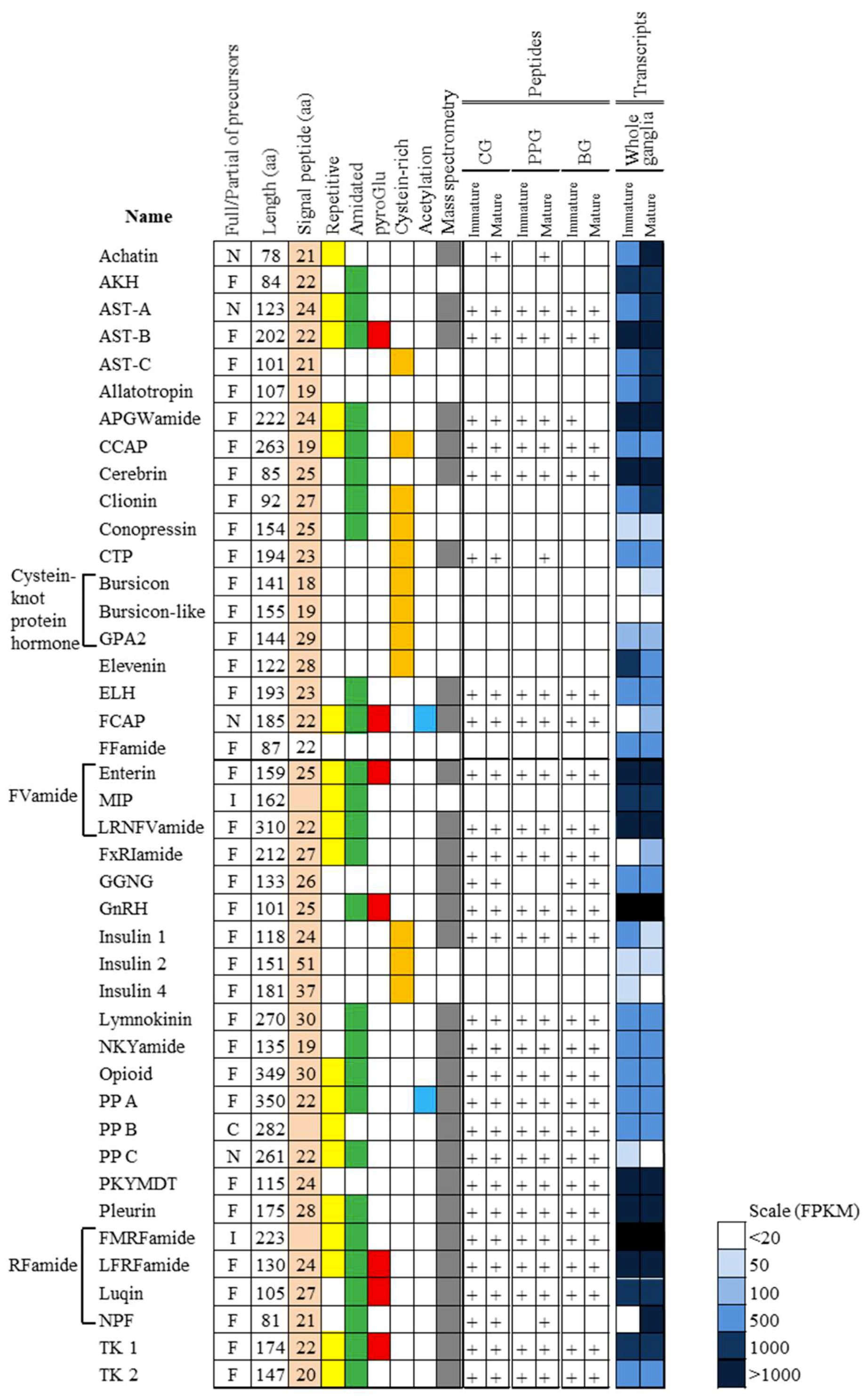
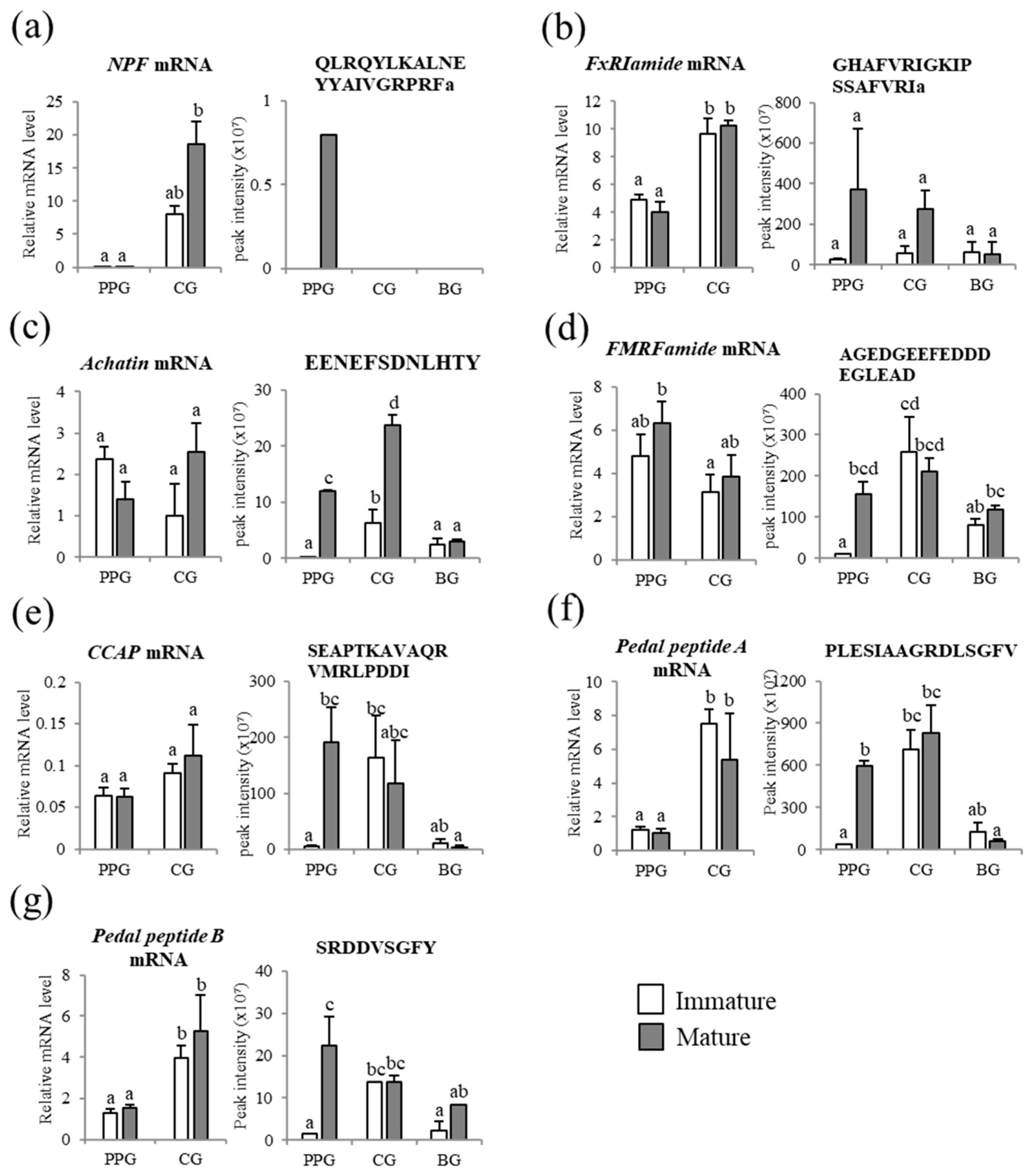
3.4. Peptidomic Analysis-Based Identification and Quantification of Neuropeptides
3.5. Comparison of Neuropeptides at the Transcript and Peptide Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Blázquez, M.; Bosma, P.T.; Fraser, E.J.; Van Look, K.J.W.; Trudeau, V.L. Fish as models for the neuroendocrine regulation of reproduction and growth. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 1998, 119, 345–364. [Google Scholar] [CrossRef]
- Hartenstein, V. The neuroendocrine system of invertebrates: a developmental and evolutionary perspective. J. Endocrinol. 2006, 190, 555–570. [Google Scholar] [CrossRef]
- Seeburg, P.H.; Mason, A.J.; Stewart, T.A.; Nikolics, K. The mammalian GnRH gene and its pivotal role in reproduction. In In Recent Progress in Hormone Research; Clark, J.H., Ed.; Academic Press Inc.: Cambridge, MA, USA, 1987; pp. 69–98. [Google Scholar]
- Nagaraju, G.P.C. Reproductive regulators in decapod crustaceans: An overview. J. Exp. Biol. 2011, 214, 3–16. [Google Scholar] [CrossRef]
- Van Wielendaele, P.; Badisco, L.; Broeck, J.V. Neuropeptidergic regulation of reproduction in insects. Gen. Comp. Endocrinol. 2013, 188, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Roubos, E.W.; van Heumen, W.R. Peptide processing and release by the neuroendocrine caudodorsal cells of Lymnaea stagnalis during an egg-laying cycle. Brain Res. 1994, 644, 83–89. [Google Scholar] [CrossRef]
- Ram, J.L.; Gallardo, C.S.; Ram, M.L.; Croll, R.P. Reproduction-associated immunoreactive peptides in the nervous systems of prosobranch gastropods. Biol. Bull. 1998, 195, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Henry, J.P.; Favrel, P.; Boucaud-Camou, E. Isolation and identification of a novel Ala-Pro-Gly-Trp-amide-related peptide inhibiting the motility of the mature oviduct in the cuttlefish, Sepia officinalis. Peptides 1997, 18, 1469–1474. [Google Scholar] [CrossRef]
- Li, K.W.; Smit, A.B.; Geraerts, W.P.M. Structural and functional characterization of neuropeptides involved in the control of male mating behavior of Lymnaea stagnalis. Peptides 1992, 13, 633–638. [Google Scholar] [CrossRef]
- Di Cristo, C.; Di Cosmo, A. Neuropeptidergic control of Octopus oviducal gland. Peptides 2007, 28, 163–168. [Google Scholar] [CrossRef]
- Koene, J.M. Neuro-endocrine control of reproduction in hermaphroditic freshwater snails: mechanisms and evolution. Front. Behav. Neurosci. 2010, 4, 167. [Google Scholar]
- In, V.V.; Ntalamagka, N.; O’Connor, W.; Wang, T.; Powell, D.; Cummins, S.F.; Elizur, A. Reproductive neuropeptides that stimulate spawning in the Sydney rock oyster (Saccostrea glomerata). Peptides 2016, 82, 109–119. [Google Scholar] [CrossRef]
- Zatylny-Gaudin, C.L.; Cornet, V.; Leduc, A.; Zanuttini, B.; Corre, E.; Le Corguillé, G.; Bernay, B.; Garderes, J.; Kraut, A.; Couté, Y.; Henry, J. Neuropeptidome of the cephalopod Sepia officinalis: Identification, tissue mapping, and expression pattern of neuropeptides and neurohormones during egg laying. J. Proteome Res. 2015, 15, 48–67. [Google Scholar] [CrossRef]
- York, P.S.; Cummins, S.F.; Degnan, S.M.; Woodcroft, B.J.; Degnan, B.M. Marked changes in neuropeptide expression accompany broadcast spawnings in the gastropod Haliotis asinina. Front. Zool. 2012, 9, 9. [Google Scholar] [CrossRef]
- Stewart, M.J.; Favrel, P.; Rotgans, B.A.; Wang, T.; Zhao, M.; Sohail, M.; O’Connor, W.A.; Elizur, A.; Henry, J.; Cummins, S.F. Neuropeptides encoded by the genomes of the Akoya pearl oyster Pinctata fucata and Pacific oyster Crassostrea gigas: a bioinformatic and peptidomic survey. BMC Genomics 2014, 15, 840. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, Y.; Li, Y.; Li, W.; Li, R.; Xie, X.; Wang, S.; Hu, X.; Zhang, L.; Bao, Z. Identification and characterization of neuropeptides by transcriptome and proteome analyses in a aivalve mollusc Patinopecten yessoensis. Front. Genet. 2018, 9, 197. [Google Scholar] [CrossRef] [PubMed]
- Cook, P.A. The worldwide abalone industry. Modern Economy 2014, 5, 1181. [Google Scholar] [CrossRef]
- Nam, B.-H.; Kwak, W.; Kim, Y.O.; Kim, D.G.; Kong, H.J.; Kim, W.J.; Kang, J.H.; Park, J.Y.; An, C.M.; Moon, J.Y.; et al. Genome sequence of pacific abalone (Haliotis discus hannai): the first draft genome in family Haliotidae. GigaScience 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Di, G.; Luo, X.; Huang, M.; Chen, J.; Kong, X.; Miao, X.; Ke, C. Proteomic profiling of eggs from a hybrid abalone and its parental lines: Haliotis discus hannai Ino and Haliotis gigantea. Anim. Genet. 2015, 46, 646–654. [Google Scholar] [CrossRef]
- Nam, B.-H.; Jung, M.; Subramaniyam, S.; Yoo, S.I.; Markkandan, K.; Moon, J.Y.; Kim, Y.O.; Kim, D.G.; An, C.M.; Shin, Y.; et al. Transcriptome analysis revealed changes of multiple genes involved in Haliotis discus hannai innate immunity during Vibrio parahemolyticus infection. PLoS ONE 2016, 11, e0153474. [Google Scholar] [CrossRef]
- Choi, M.J.; Kim, G.D.; Kim, J.M.; Lim, H.K. Differentially-expressed genes associated with faster growth of the Pacific abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2015, 16, 27520–27534. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.A.; Rhee, J.S.; Kim, T.H.; Lee, J.S.; Choi, A.Y.; Choi, B.S.; Choi, I.Y.; Sohn, Y.C. Alternative splicing profile and sex-preferential gene expression in the female and male Pacific Abalone Haliotis discus hannai. Genes (Basel) 2017, 8, 99. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Porras, O.; Botwright, N.A.; McWilliam, S.M.; Cook, M.T.; Harris, J.O.; Wijffels, G.; Colgrave, .M.L. Exploiting genomic data to identify proteins involved in abalone reproduction. J. Proteomics 2014, 108, 337–353. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, M.A.; Kim, K.S.; Kim, J.W.; Lim, H.K.; Lee, J.S.; Sohn, Y.C. Characterization and spatiotemporal expression of gonadotropin-releasing hormone in the Pacific abalone, Haliotis discus hannai. Comp. Biochem. Physiol. A Mol. integr. Physiol. 2017, 209, 1–9. [Google Scholar] [CrossRef]
- Wang, T.; Nuurai, P.; McDougall, C.; York, P.S.; Bose, U.; Degnan, B.M.; Cummins, S.F. Identification of a female spawn-associated Kazal-type inhibitor from the tropical abalone Haliotis asinina. J. Pept. Sci. 2016, 22, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Chansela, P.; Saitongdee, P.; Stewart, P.; Soonklang, N.; Stewart, M.; Suphamungmee, W.; Poomtong, T.; Sobhon, P. Existence of APGWamide in the testis and its induction of spermiation in Haliotis asinina Linnaeus. Aquaculture 2008, 279, 142–149. [Google Scholar] [CrossRef]
- Kim, K.S.; Kim, T.H.; Kim, M.A.; Lee, J.S.; Sohn, Y.C. Expression profile and reproductive regulation of APGWamide in Pacific abalone (Haliotis discus hannai). Comp. Biochem. Physiol. A Mol. integr. Physiol. 2017, 222, 26–35. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk (accessed on 16 June 2017).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Gentleman, R. R Programming for Bioinformatics; Chapman & Hall/CRC: Boca Raton, FL, USA, 2008; pp. 119–182. [Google Scholar]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Kuehn, H.; Gould, J.; Tamayo, P.; Mesirov, J.P. GSEA-P: a desktop application for Gene Set Enrichment Analysis. Bioinformatics 2007, 23, 3251–3253. [Google Scholar] [CrossRef] [PubMed]
- Secher, A.; Kelstrup, C.D.; Conde-Frieboes, K.W.; Pyke, C.; Raun, K.; Wulff, B.S.; Olsen, J.V. Analytic framework for peptidomics applied to large-scale neuropeptide identification. Nat. Commun. 2016, 7, 11436. [Google Scholar] [CrossRef] [PubMed]
- Veenstra, J.A. Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. Gen. Comp. Endocrinol. 2010, 167, 86–103. [Google Scholar] [CrossRef]
- Adamson, K.J.; Wang, T.; Zhao, M.; Bell, F.; Kuballa, A.V.; Storey, K.B.; Cummins, S.F. Molecular insights into land snail neuropeptides through transcriptome and comparative gene analysis. BMC Genomics 2015, 16, 308. [Google Scholar] [CrossRef]
- Mirabeau, O.; Joly, J.S. Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci. USA 2013, 110, E2028–E2037. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Southey, B.R.; Amare, A.; Zimmerman, T.A.; Rodriguez-Zas, S.L.; Sweedler, J.V. NeuroPred: A tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res. 2006, 34, W267–W272. [Google Scholar] [CrossRef] [PubMed]
- Bose, U.; Suwansa-Ard, S.; Maikaeo, L.; Motti, C.A.; Hall, M.R.; Cummins, S.F. Neuropeptides encoded within a neural transcriptome of the giant triton snail Charonia tritonis, a Crown-of-Thorns Starfish predator. Peptides 2017, 98, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Kehoe, J.; Buldakova, S.; Acher, F.; Dent, J.; Bregestovski, P.; Bradley, J. Aplysia cys-loop glutamate-gated chloride channels reveal convergent evolution of ligand specificity. J. Mol. Evol. 2009, 69, 125–141. [Google Scholar] [CrossRef]
- Ness, G.C. Physiological feedback regulation of cholesterol biosynthesis: Role of translational control of hepatic HMG-CoA reductase and possible involvement of oxylanosterols. Biochim. Biophys. Acta 2015, 1851, 667–673. [Google Scholar] [CrossRef]
- Rasmussen, M.K.; Ekstrand, B.; Zamaratskaia, G. Regulation of 3beta-hydroxysteroid dehydrogenase/Delta(5)-Delta(4) isomerase: A review. Int. J. Mol. Sci. 2013, 14, 17926–17942. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhu, D.; Li, Y.; Qiu, X.; Cui, X.; Tang, J. Hemolymph levels of methyl farnesoate during ovarian development of the swimming crab Portunus trituberculatus, and its relation to transcript levels of HMG-CoA reductase and farnesoic acid O-methyltransferase. Biol. Bull. 2015, 228, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Di Cristo, C.; Di Donato, P.; Palumbo, A.; d’Ischia, M.; Paolucci, M.; Di Cosmo, A. Steroidogenesis in the brain of Sepia officinalis and Octopus vulgaris. Front. Biosci. (Elite Ed) 2010, 2, 673–683. [Google Scholar]
- Kirkpatrick, R.B.; Matico, R.E.; McNulty, D.E.; Strickler, J.E.; Rosenberg, M. An abundantly secreted glycoprotein from Drosophila melanogaster is related to mammalian secretory proteins produced in rheumatoid tissues and by activated macrophages. Gene 1995, 153, 147–154. [Google Scholar] [CrossRef]
- Hipfner, D.R.; Cohen, S.M. New growth factors for imaginal discs. Bioessays 1999, 21, 718–720. [Google Scholar] [CrossRef]
- Zimmerman, S.G.; Merrihew, G.E.; MacCoss, M.J.; Berg, C.A. Proteomics analysis identifies orthologs of human chitinase-like proteins as inducers of tube morphogenesis defects in Drosophila melanogaster. Genetics 2017, 206, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Flanagan, J.U.; Jowsey, I.R. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 51–88. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.G.; Ahn, C.S.; Kim, S.H.; Bae, Y.A.; Kwon, N.Y.; Kang, I.; Yang, H.J.; Sohn, W.M.; Kong, Y. Clonorchis sinensis omega-class glutathione transferases play major roles in the protection of the reproductive system during maturation and the response to oxidative stress. Parasit Vectors 2016, 9, 337. [Google Scholar] [CrossRef]
- de Jong-Brink, M.; Reid, C.N.; Tensen, C.P.; ter Maat, A. Parasites flicking the NPY gene on the host’s switchboard: why NPY? FASEB J. 1999, 1, 1972–1984. [Google Scholar] [CrossRef]
- Ahn, S.J.; Martin, R.; Rao, S.; Choi, M.Y. Neuropeptides predicted from the transcriptome analysis of the gray garden slug Deroceras reticulatum. Peptides 2017, 93, 51–65. [Google Scholar] [CrossRef]
- El Filali, Z.; Van Minnen, J.; Liu, W.K.; Smit, A.B.; Li, K.W. Peptidomics analysis of neuropeptides involved in copulatory behavior of the mollusk Lymnaea stagnalis. J. Proteome Res. 2006, 5, 1611–1617. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Fang, Y.; Feng, M.; Hu, H.; Qi, Y.; Huo, X.; Meng, L.; Wu, B.; Li, J. Quantitative neuropeptidome analysis reveals neuropeptides are correlated with social behavior regulation of the honeybee workers. J. Proteome Res. 2015, 14, 4382–4393. [Google Scholar] [CrossRef] [PubMed]
- Kamatani, Y.; Minakata, H.; Kenny, P.T.; Iwashita, T.; Watanabe, K.; Funase, K.; Sun, X.P.; Yongsiri, A.; Kim, K.H.; Novales-Li, P.; et al. Achatin-I, an endogenous neuroexcitatory tetrapeptide from Achatina fulica Ferussac containing a D-amino acid residue. Biochem. Biophys. Res. Commun. 1989, 160, 1015–1020. [Google Scholar] [CrossRef]
- Vehovszky, A.; Agricola, H.J.; Elliott, C.J.; Ohtani, M.; Kárpáti, L.; Hernádi, L. Crustacean cardioactive peptide (CCAP)-related molluscan peptides (M-CCAPs) are potential extrinsic modulators of the buccal feeding network in the pond snail Lymnaea stagnalis. Neurosci. Lett. 2005, 373, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Fu, H.; Xiong, Y.; Jiang, S.; Zhang, W.; Sun, S.; Jin, S.; Gong, Y.; Wang, Y.; Shan, D.; et al. Molecular insights into reproduction regulation of female Oriental River prawns Macrobrachium nipponense through comparative transcriptomic analysis. Sci. Rep. 2017, 7, 12161. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.V.; Cummins, S.F.; Elizur, A.; Ventura, T. Transcriptomic characterization and curation of candidate neuropeptides regulating reproduction in the eyestalk ganglia of the Australian crayfish, Cherax quadricarinatus. Sci. Rep. 2016, 6, 38658. [Google Scholar] [CrossRef]
- Bao, C.; Yang, Y.; Huang, H.; Ye, H. Neuropeptides in the cerebral ganglia of the mud crab, Scylla paramamosain: transcriptomic analysis and expression profiles during vitellogenesis. Sci. Rep. 2015, 5, 17055. [Google Scholar] [CrossRef]
- Endress, M.; Zatylny-Gaudin, C.; Corre, E.; Le Corguillé, G.; Benoist, L.; Leprince, J.; Lefranc, B.; Bernay, B.; Leduc, A.; Rangama, J.; et al. Crustacean cardioactive peptides: Expression, localization, structure, and a possible involvement in regulation of egg-laying in the cuttlefish Sepia officinalis. Gen. Comp. Endocrinol. 2018, 260, 67–79. [Google Scholar] [CrossRef]
- Walker, R.J.; Papaioannou, S.; Holden-Dye, L. A review of FMRFamide- and RFamide-like peptides in metazoa. Invert. Neurosci. 2009, 9, 111–153. [Google Scholar] [CrossRef]
- Price, D.A.; Greenberg, M.J. Structure of a molluscan cardioexcitatory neuropeptide. Science 1977, 197, 670–671. [Google Scholar] [CrossRef]
- Di Cristo, C.; Paolucci, M.; Iglesias, J.; Sanchez, J.; Di Cosmo, A. Presence of two neuropeptides in the fusiform ganglion and reproductive ducts of Octopus vulgaris: FMRFamide and gonadotropin-releasing hormone (GnRH). J. Exp. Zool. 2002, 292, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Brussaard, A.B.; Kits, K.S.; Ter Maat, A.; Van Minnen, J.; Moed, P.J. Dual inhibitory action of FMRFamide on neurosecretory cells controlling egg laying behavior in the pond snail. Brain Res. 1988, 447, 35–51. [Google Scholar] [CrossRef]
- Roszer, T.; Kiss-Toth, E.D. FMRF-amide is a glucose-lowering hormone in the snail Helix aspersa. Cell Tissue Res. 2014, 358, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Kindy, M.S.; Srivatsan, M.; Peretz, B. Age-related differential expression of neuropeptide mRNAs in Aplysia. Neuroreport 1991, 2, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Di Cosmo, A.; Di Cristo, C. Neuropeptidergic control of the optic gland of Octopus vulgaris: FMRF-amide and GnRH immunoreactivity. J. Comp. Neurol. 1998, 398, 1–12. [Google Scholar] [CrossRef]
- Lloyd, P.E.; Connolly, C.M. Sequence of pedal peptide: a novel neuropeptide from the central nervous system of Aplysia. J. Neurosci. 1989, 9, 312–317. [Google Scholar] [CrossRef]
- Moroz, L.L.; Edwards, J.R.; Puthanveettil, S.V.; Kohn, A.B.; Ha, T.; Heyland, A.; Knudsen, B.; Sahni, A.; Yu, F.; Liu, L.; et al. Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell 2006, 127, 1453–1467. [Google Scholar] [CrossRef]
- Stay, B.; Tobe, S.S. The role of allatostatins in juvenile hormone synthesis in insects and crustaceans. Annu. Rev. Entomol. 2007, 52, 277–299. [Google Scholar] [CrossRef]
- Verlinden, H.; Gijbels, M.; Lismont, E.; Lenaerts, C.; Vanden Broeck, J.; Marchal, E. The pleiotropic allatoregulatory neuropeptides and their receptors: A mini-review. J. Insect Physiol. 2015, 80, 2–14. [Google Scholar] [CrossRef]
- Schoofs, L.; Holman, G.M.; Hayes, T.K.; Nachman, R.J.; De Loof, A. Isolation, identification and synthesis of locustamyoinhibiting peptide (LOM-MIP), a novel biologically active neuropeptide from Locusta migratoria. Regul. Pept. 1991, 36, 111–119. [Google Scholar] [CrossRef]
- Furuya, K.; Milchak, R.J.; Schegg, K.M.; Zhang, J.; Tobe, S.S.; Coast, G.M.; Schooley, D.A. Cockroach diuretic hormones: characterization of a calcitonin-like peptide in insects. Proc. Natl. Acad. Sci. USA 2000, 97, 6469–6474. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Kim, C.H.; Go, H.J.; Egertová, M.; Zampronio, C.G.; Jones, A.M.; Park, N.G.; Elphick, M.R. Biochemical, anatomical, and pharmacological characterization of calcitonin-type neuropeptides in starfish: Discovery of an ancient role as muscle relaxants. Front. Neurosci. 2018, 12, 382. [Google Scholar] [CrossRef] [PubMed]
- Hoek, R.M.; Li, K.W.; van Minnen, J.; Lodder, J.C.; de Jong-Brink, M.; Smit, A.B.; van Kesteren, R.E. LFRFamides: a novel family of parasitation-induced -RFamide neuropeptides that inhibit the activity of neuroendocrine cells in Lymnaea stagnalis. J Neurochem. 2005, 92, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Vanden Broeck, J.; Torfs, H.; Poels, J.; Van Poyer, W.; Swinnen, E.; Ferket, K.; De Loof, A. Tachykinin-like peptides and their receptors. A review. Ann. N. Y. Acad. Sci. 1999, 897, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Dubos, M.P.; Zels, S.; Schwartz, J.; Pasquier, J.; Schoofs, L.; Favrel, P. Characterization of a tachykinin signalling system in the bivalve mollusc Crassostrea gigas. Gen. Comp. Endocrinol. 2018, 266, 110–118. [Google Scholar] [CrossRef] [PubMed]

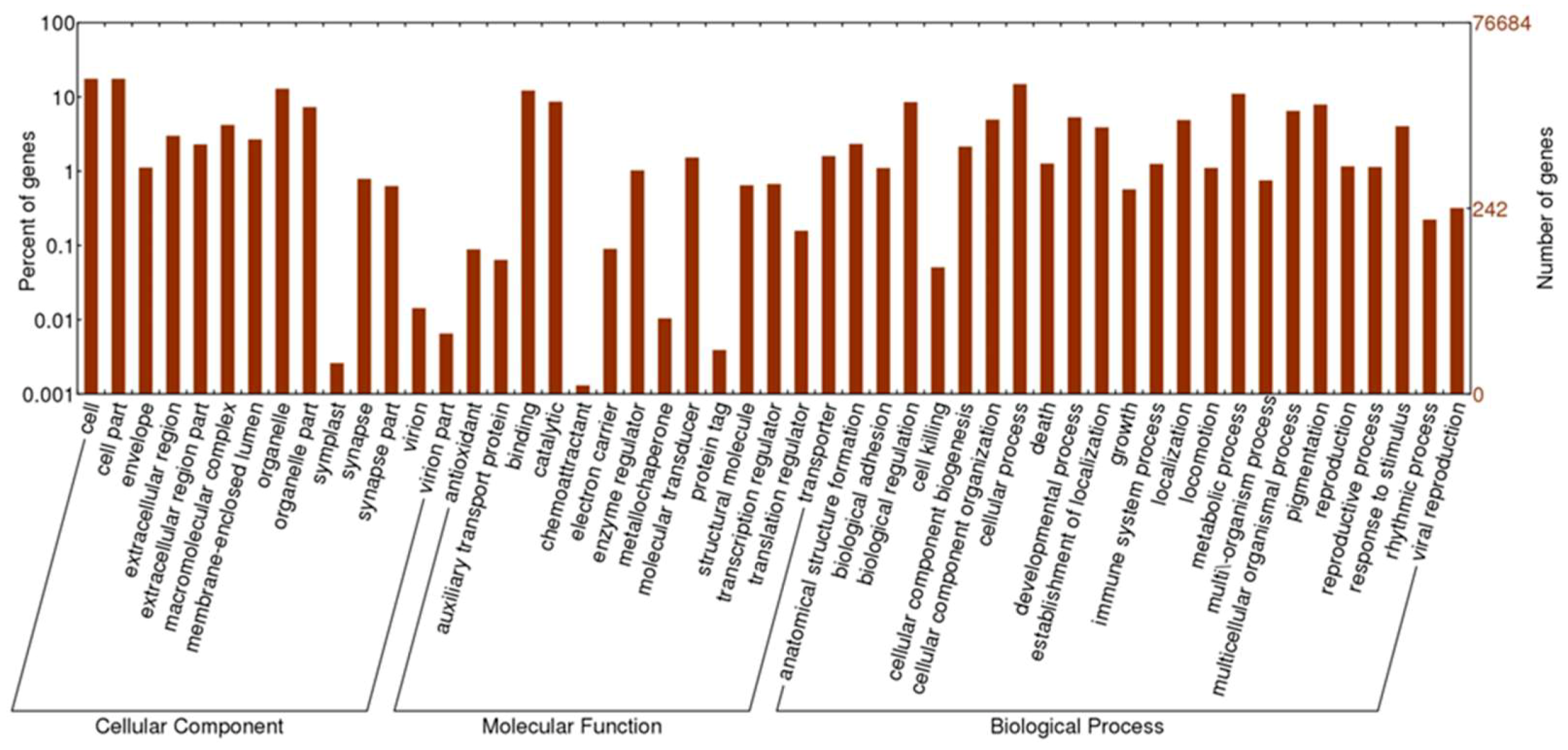
| Assembly | Total no. of raw reads | 329,064,126 |
| Total no. of clean reads | 305,530,780 (92.8%) | |
| Total no. of unigenes | 76,684 | |
| Average length of unigene (bp) | 741 | |
| >500 bp unigene | 32,477 | |
| >1000 bp unigene | 15,480 | |
| GC content (%) | 45.25 | |
| Proteins with complete ORF | 4379 | |
| Annotation | Blastx and Swiss-Prot Blastx only Swiss-Prot only | 21,891 5001 674 |
| GO | 15,585 | |
| KEGG | 11,728 |
| Putative Function ID no. | Gene Name | Description | Immature vs. Mature | |
|---|---|---|---|---|
| Log2FC | p-value | |||
| Neuropeptide signaling pathway | ||||
| TBIU004747 | NPF | Pro-neuropeptide F | –8.82 | 7.73 × 10−16 |
| TBIU007725 | FxRI | FMRFamide | –2.01 | 0.001 |
| TBIU006586 | Sort1 | Sortilin | –2.11 | 0.013 |
| TBIU017900 | Glra2 | Glycine receptor subunit alpha-2 | –3.29 | 0.0099 |
| TBIU032133 | Pkd1l2 | Polycystic kidney disease protein 1-like 2 | –9.48 | 0.054 |
| G-protein coupled receptor activity | ||||
| TBIU024402 | GNRHR | Gonadotropin-releasing hormone receptor | –7.28 | 3.23 × 10−5 |
| G-protein coupled receptor signaling pathway | ||||
| TBIU011409 | RAPGEF2 | Rap guanine nucleotide exchange factor 2 | –4.91 | 0.0001 |
| TBIU007260 | CELSR2 | Cadherin EGF LAG seven-pass G-type receptor 2 | –5.09 | 0.00034 |
| Oxidoreductase activity | ||||
| TBIU021351 | Gsto1 | Glutathione S-transferase omega-1 | –2.85 | 4.57 × 10−5 |
| Steroid biosynthesis | ||||
| TBIU020046 | HSD3B1 | 3 beta-hydroxysteroid dehydrogenase/Delta 5-->4-isomerase type 1 | –13.8 | 3.26 × 10−5 |
| Cholesterol/Isoprenoid biosynthesis | ||||
| TBIU019957 | hmgcr | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | –13.8 | 2.42 × 10−10 |
| Lipid catabolic process | ||||
| TBIU001813 | MtsPLA2 | Phospholipase A2 | –11.9 | 5.66 × 10−5 |
| Gluconeogenesis | ||||
| TBIU022277 | PPCK1 | Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | –3.74 | 6.87 × 10−5 |
| Carbohydrate metabolic process | ||||
| TBIU014745 | Idgf4 | Chitinase-like protein Idgf4 | –12.9 | 9.53 × 10−6 |
| Translational initiation | ||||
| TBIU023683 | eif3c | Eukaryotic translation initiation factor 3 subunit C | –2.59 | 1.57 × 10−5 |
| Other pathways | ||||
| TBIU021499 | Cyp3a11 | Cytochrome P450 3A11 | –13.2 | 0.0001 |
| TBIU019312 | Tie1 | Tyrosine-protein kinase receptor Tie-1 | –12.9 | 2.91 × 10−6 |
| TBIU016501 | Bai1 | Brain-specific angiogenesis inhibitor 1 | –13.2 | 0.00016 |
| Neuropeptide Family | Log FCa (Mature vs Immature) | p-value | Peptide Sequence and PTMb | Mass (Da) | Maturec | Immaturec | p-valuef |
|---|---|---|---|---|---|---|---|
| Branchial ganglion (BG) | |||||||
| MAP-1 | 4.15 | 7.09 × 10−3 | SVILTSILLQERRYDRMS | 2179.18 | 0.92 ± 0.18 | 0.42 ± 0.04 | n.s. |
| Tachykinin 1 | −3.06 | 4.75 × 10−2 | EALDDNTAASLYKLLQPAYQSTVAEd | 2710.33 | ND | 20.60 ± 4.89 | N/A |
| pQPHFGFHGVRamide | 1162.58 | 3.29 | 4.57 ± 0.75 | n.s. | |||
| TELGFGYVGSRamide | 1183.60 | 10.30 ± 2.91 | 17.64 ± 1.72 | n.s. | |||
| MAP-2 | −1.35 | 4.61 × 10−2 | GGGFGPASNPDSWSEVYRTGN | 2153.94 | 0.26 ± 0.00 | 0.21 ± 0.01 | n.s. |
| MAP-3 | −2.60 | 4.42 × 10−2 | AGIANQVTRILPIQVLSPDDLM(O)d | 2379.28 | ND | 0.21 ± 0.14 | N/A |
| Cerebral ganglion (CG) | |||||||
| Allatostatin B (=WWamide) | −2.74 | 6.01 × 10−4 | QWSNFHSWamided | 1089.48 | ND | 0.10 | N/A |
| AGWDNGFASWamide | 1108.47 | 5.78 ± 1.42 | 4.11 ± 0.47 | n.s. | |||
| NWNQFITWamide | 1106.53 | 4.14 ± 0.56 | 2.56 ± 1.29 | n.s. | |||
| WGNFGTSGKKWASSDFPAWamide | 2127.00 | 7.21 ± 2.93 | 8.47 ± 0.07 | n.s. | |||
| MAP-4 | −1.69 | 4.18 × 10−2 | YVHFNIGNDHQVS | 1528.71 | 0.52 ± 0.15 | 0.80 ± 0.15 | n.s |
| Pleuro-pedal ganglion (PPG) | |||||||
| Achatin | 2.41 | 3.83 × 10−2 | EENEFSDNLHTY | 1496.61 | 1.19 ± 0.02 | 0.02 | n.s. |
| GFGDKRGFGDe | 1054.48 | 0.08 | ND | N/A | |||
| FMRFamide | 1.57 | 3.09 × 10−2 | AGEDGEEFEDDDEGLEAD | 1940.69 | 15.59 ± 1.68 | 1.41 ± 0.27 | * |
| DGEDEKRFMRFamide | 1427.66 | 0.47 ± 0.15 | 0.42 ± 0.01 | n.s | |||
| DGQDKRFMRFamidee | 1297.64 | 0.06 ± 0.01 | ND | N/A | |||
| FMRFGKSGEEE | 1315.59 | 0.90 ± 0.21 | 0.29 ± 0.03 | * | |||
| SGDDEKRFMRFamide | 1385.65 | 0.17 ± 0.03 | 0.12 ± 0.10 | n.s. | |||
| CCAP | 2.94 | 1.57 × 10−3 | SEAPTKAVAQRVMRLPDDI | 2096.11 | 19.14 ± 3.59 | 0.56 ± 0.16 | * |
| Pedal peptide A (PP-A) | 1.29 | 1.22 × 10−4 | SFDSINKGSGLSGFM | 1545.71 | 13.82 ± 1.09 | 1.63 ± 0.30 | * |
| PFDSISSGGGMAGFAe | 1399.61 | 4.75 ± 0.56 | ND | N/A | |||
| PFDSIASGRGIAGFA | 1464.74 | 24.22 ± 11.65 | 2.74 ± 0.04 | n.s. | |||
| PLESIAAGRDLSGFV | 1530.80 | 59.38 ± 3.57 | 3.69 ± 0.52 | * | |||
| SFDSINAGSGLSGFA | 1428.65 | 5.67 ± 1.63 | 0.95 ± 0.12 | n.s. | |||
| PFDSISGSSAFSDFA | 1533.66 | 3.53 ± 0.25 | 0.69 ± 0.25 | * | |||
| PFDSIASGGGMAGFA | 1383.61 | 4.31 ± 0.65 | 0.29 ± 0.07 | * | |||
| PFDSIASGRGIAGFA | 1464.74 | 24.22 ± 11.65 | 2.74 ± 0.04 | n.s. | |||
| Pedal peptide B (PP-B) | 1.67 | 4.50 × 10−4 | SPENDLSSFY | 1044.45 | 2.24 ± 0.68 | 0.17 | n.s. |
| SNEDLSGFY | 1030.42 | 6.24 ± 1.44 | 0.17 | n.s. | |||
| SRGDGLSNFYe | 1114.50 | 2.97 ± 0.74 | ND | N/A | |||
| MAP-5 | 9.73 | 1.29 × 10−5 | RSLSLDDTYWVDNVD | 1796.82 | 76.19 ± 13.23 | 4.84 ± 2.49 | * |
| MAP-6 | 2.61 | 1.73 × 10−2 | MGIGKGAQSFGDGQNNDQ | 1822.79 | 0.61 ± 0.21 | 0.28 ± 0.02 | n.s. |
| MAP-7 | 2.83 | 7.90 × 10−3 | STIFDRMGRFYamide | 1390.68 | 3.69 ± 1.31 | 1.57 ± 0.41 | n.s. |
| MAP-8 | 3.05 | 8.73 × 10−3 | TETRAAFWDNAAGRQPY | 1952.91 | 1.12 ± 0.55 | 0.04 ± 0.02 | n.s. |
| CTP | −1.189 | 1.93 × 10−2 | AVLANTQRIIKAE | 1425.83 | 1.95 ± 0.46 | 1.53 ± 0.25 | n.s. |
| LFRFamide | −1.72 | 2.90 × 10−4 | AKEEQDSEAAVVAPSEHH | 1989.90 | 21.92 ± 1.66 | 22.93 ± 4.51 | n.s. |
| NFHWGRETEE | 1303.56 | 29.83 ± 6.31 | 47.67 ± 3.33 | * | |||
| Tachykinin 1 | −1.63 | 2.16 × 10−4 | EALDDNTAASLYKLLQPAYQSTVAE | 2710.33 | 8.13 ± 2.44 | 14.17 ± 4.98 | n.s. |
| pQPHFGFHGVRamide | 1162.58 | 5.40 ± 0.78 | 6.68 ± 1.60 | n.s. | |||
| Sensorin-A | −1.57 | 1.05 × 10−3 | RSLDQSERRLVA | 1428.78 | 34.12 ± 6.33 | 31.79 ± 3.58 | n.s. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.A.; Markkandan, K.; Han, N.-Y.; Park, J.-M.; Lee, J.S.; Lee, H.; Sohn, Y.C. Neural Ganglia Transcriptome and Peptidome Associated with Sexual Maturation in Female Pacific Abalone (Haliotis discus hannai). Genes 2019, 10, 268. https://doi.org/10.3390/genes10040268
Kim MA, Markkandan K, Han N-Y, Park J-M, Lee JS, Lee H, Sohn YC. Neural Ganglia Transcriptome and Peptidome Associated with Sexual Maturation in Female Pacific Abalone (Haliotis discus hannai). Genes. 2019; 10(4):268. https://doi.org/10.3390/genes10040268
Chicago/Turabian StyleKim, Mi Ae, Kesavan Markkandan, Na-Young Han, Jong-Moon Park, Jung Sick Lee, Hookeun Lee, and Young Chang Sohn. 2019. "Neural Ganglia Transcriptome and Peptidome Associated with Sexual Maturation in Female Pacific Abalone (Haliotis discus hannai)" Genes 10, no. 4: 268. https://doi.org/10.3390/genes10040268
APA StyleKim, M. A., Markkandan, K., Han, N.-Y., Park, J.-M., Lee, J. S., Lee, H., & Sohn, Y. C. (2019). Neural Ganglia Transcriptome and Peptidome Associated with Sexual Maturation in Female Pacific Abalone (Haliotis discus hannai). Genes, 10(4), 268. https://doi.org/10.3390/genes10040268





