Genome-Wide Assessment of Avocado Germplasm Determined from Specific Length Amplified Fragment Sequencing and Transcriptomes: Population Structure, Genetic Diversity, Identification, and Application of Race-Specific Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and RNA Extraction
2.2. SLAF-seq and SNP Analysis
2.3. Transcriptome Sequencing andSNP Analysis
2.4. Identification and Application of Race-Specific Markers
2.5. Data Analysis
3. Results
3.1. Sequence Assembly and SNP Detection
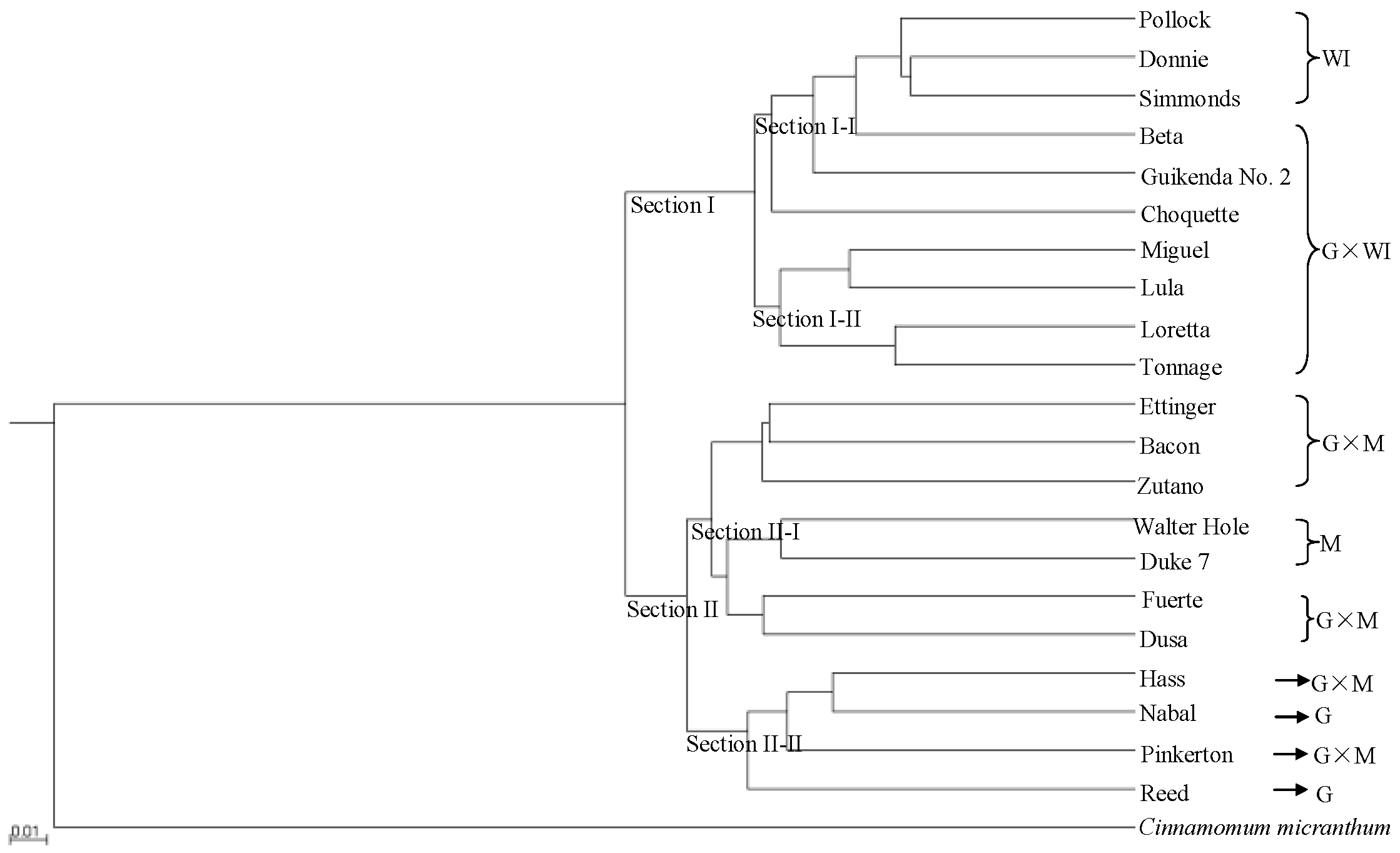
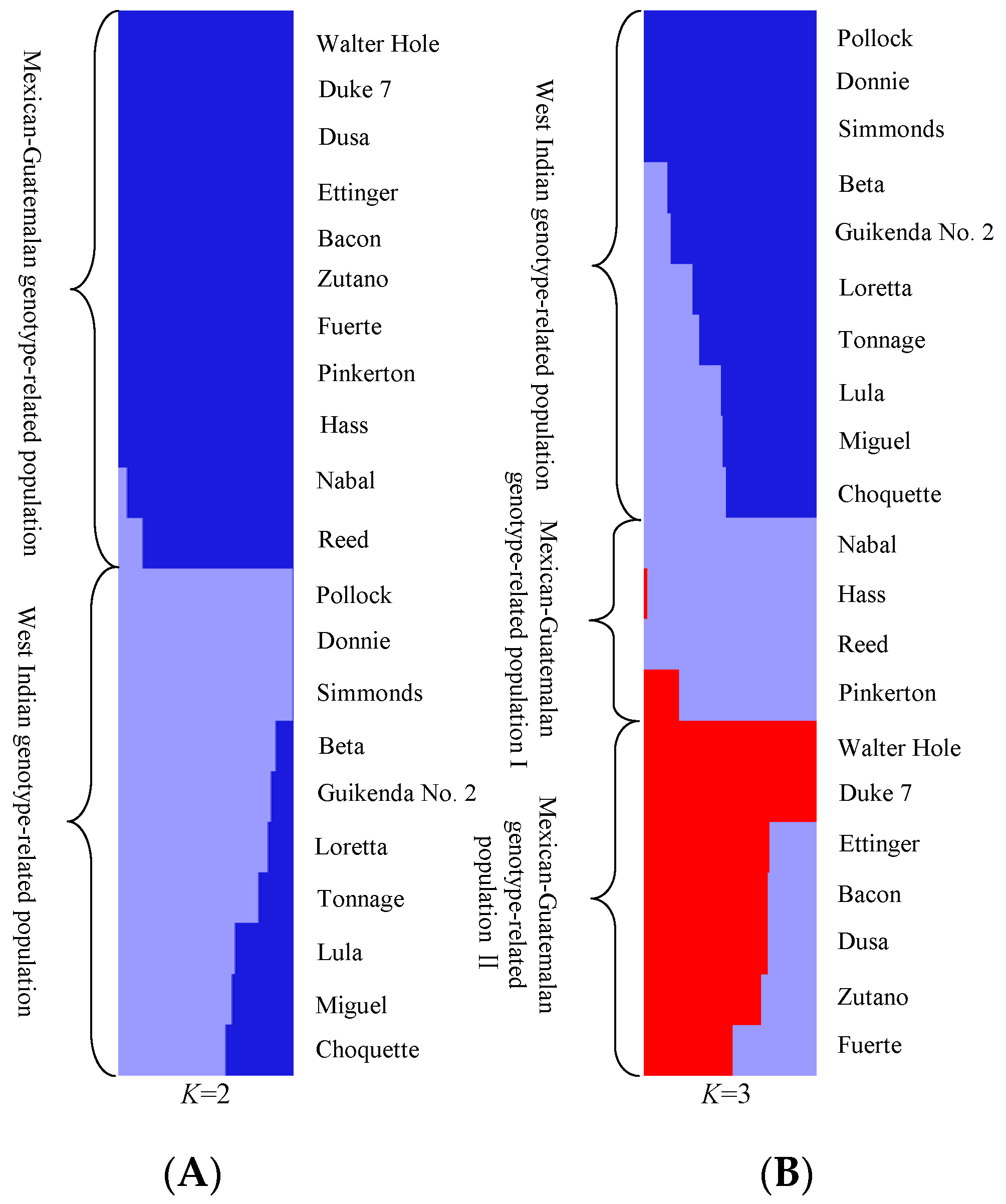
3.2. Phylogenomic and Population Analysis by SLAF-seq Based on SNP Data
3.3. Phylogenetic Analysis Using Transcriptome SNP Data
3.4. Genetic Diversity and Molecular Variance Analyses
3.5. Acquisition, Verification, and Application of Race-Specific SNPs Using SLAF-seq
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schaffer, B.; Wolstenholme, B.N.; Whiley, A.W. The Avocado: Botany, Production and Uses, 2nd ed.; CPI Group (UK) Ltd.: Croydon, UK, 2012. [Google Scholar]
- Galindo-Tovar, M.E.; Ogata-Aguilar, N.; Arzate-Fernandez, A.M. Some aspects of avocado (Perseaamericana Mill.) diversity and domestication in Mesoamerica. Genet. Resour. Crop Evol. 2008, 55, 441–450. [Google Scholar] [CrossRef]
- Berg, B.; Ellstrand, N. Taxonomy of the avocado. Calif. Avocado Soc. 1986, 70, 135–146. [Google Scholar]
- Williams, L.O. The avocados, a synopsis of the genus Persea, subg. Persea. Econ. Bot. 1977, 31, 315–320. [Google Scholar] [CrossRef]
- Bergh, B.O.; Scora, R.W.; Storey, W.B. A comparison of leaf terpenes in Persea subgenus Persea. Bot. Gaz. 1973, 134, 130–134. [Google Scholar] [CrossRef]
- Kopp, L.E. A taxonomic revision of the genus Persea in the western Hemisphere (Persea-Lauraceae). Mem. N. Y. Bot. Gard. 1966, 14, 1–120. [Google Scholar]
- Fiedler, J.; Bufler, G.; Bangerth, F. Genetic relationships of avocado (Perseaamericana Mill.) using RAPD markers. Euphytica 1998, 101, 249–255. [Google Scholar] [CrossRef]
- Mhameed, S.; Sharon, D.; Kaufman, D.; Lahav, E.; Hillel, J.; Degani, C.; Lavi, Y. Genetic relationships within avocado (Persea americana Mill.) cultivars and between Persea species. Theor. Appl. Genet. 1997, 94, 279–286. [Google Scholar] [CrossRef]
- Goldring, A.; Zamir, D.; Degani, C. Duplicated phosphoglucose isomerase genes in avocado. Theor. Appl. Genet. 1985, 71, 491–494. [Google Scholar] [CrossRef]
- Furnier, G.R.; Cummings, M.P.; Clegg, M.T. Evolution of the avocados as revealed by DNA restriction site variation. J. Hered. 1990, 81, 183–188. [Google Scholar] [CrossRef]
- Ashworth, V.E.T.M.; Clegg, M.T. Microsatellite markers in avocado (Perseaamericana Mill.). genealogical relationships among cultivated avocado genotypes. J. Hered. 2003, 94, 407–415. [Google Scholar] [CrossRef]
- Schnell, R.J.; Brown, J.S.; Olano, C.T.; Power, E.J.; Krol, C.A.; Kuhn, D.N.; Motamayor, J.C. Evaluation of avocado germplasm using microsatellite markers. J. Am. Soc. Hortic. Sci. 2003, 128, 881–889. [Google Scholar] [CrossRef]
- Gross-German, E.; Viruel, M.A. Molecular characterization of avocado germplasm with a new set of SSR and EST-SSR markers: Genetic diversity, population structure, and identification of race-specific markers in a group of cultivated genotypes. Tree Genet. Genomes 2013, 9, 539–555. [Google Scholar] [CrossRef]
- Gupta, S.K.; Baek, J.; Carrasquilla-Garcia, N.; Penmetsa, R.V. Genome-wide polymorphism detection in peanut using next-generation restriction-site-associated DNA (RAD) sequencing. Mol. Breed. 2015, 35, 145. [Google Scholar] [CrossRef]
- Sun, X.W.; Liu, D.Y.; Zhang, X.F.; Li, W.B.; Liu, H.; Hong, W.G.; Jiang, C.B.; Guan, N.; Ma, C.X.; Zeng, H.P.; et al. SLAF-seq: An efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 2013, 8, e58700. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Wang, L.H.; Xin, H.G.; Li, D.H.; Ma, C.X.; Ding, X.; Hong, W.G.; Zhang, X.R. Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC Plant Biol. 2013, 13, 141. [Google Scholar] [CrossRef]
- Hu, X.H.; Zhang, S.Z.; Miao, H.R.; Cui, F.G.; Shen, Y.; Yang, W.Q.; Xu, T.T.; Chen, N.; Chi, X.Y.; Zhang, Z.M.; et al. High-density genetic map construction and identification of QTLs controlling oleic and linoleic acid in peanut using SLAF-seq and SSRs. Sci. Rep. 2018, 8, 5479. [Google Scholar] [CrossRef]
- Su, W.J.; Wang, L.J.; Lei, J.; Chai, S.S.; Liu, Y.; Yang, Y.Y.; Yang, X.S.; Jiao, C.H. Genome-wide assessment of population structure and genetic diversity and development of a core germplasm set for sweet potato based on specific length amplified fragment (SLAF) sequencing. PLoS ONE 2017, 12, e0172066. [Google Scholar] [CrossRef]
- Yin, J.L.; Fang, Z.W.; Sun, C.; Zhang, P.; Zhang, X.; Lu, C.; Wang, S.P.; Ma, D.F.; Zhu, Y.X. Rapid identification of a stripe rust resistant gene in a space-induced wheat mutant using specific length amplified fragment (SLAF) sequencing. Sci. Rep. 2018, 8, 3086. [Google Scholar] [CrossRef]
- Yang, J.H.; Zhang, C.T.; Zhao, N.; Zhang, L.L.; Hu, Z.Y.; Chen, S.; Zhang, M.F. Chinese root-type mustard provides phylogenomic insights into the evolution of the multi-use diversified allopolyploid Brassica juncea. Mol. Plant 2018, 11, 512–514. [Google Scholar] [CrossRef]
- Gramazio, P.; Prohens, J.; Borràs, D.; Plazas, M.; Herraiz, F.J.; Vilanova, S. Comparison of transcriptome-derived simple sequence repeat (SSR) and single nucleotide polymorphism (SNP) markers for genetic fingerprinting, diversity evaluation, and establishment of relationships in eggplants. Euphytica 2017, 213, 264. [Google Scholar] [CrossRef]
- Rasal, K.D.; Chakrapani, V.; Pandey, A.K.; Rasal, A.R.; Sundaray, J.K.; Ninawe, A.; Jayasankar, P. Status and future perspectives of single nucleotide polymorphisms (SNPs) markers in farmed fishes: Way ahead using next generation sequencing. Gene Rep. 2017, 6, 81–86. [Google Scholar] [CrossRef]
- Ge, Y.; Ramchiary, N.; Wang, T.; Liang, C.; Wang, N.; Wang, Z.; Choi, S.R.; Lim, Y.P.; Piao, Z.Y. Development and linkage mapping of unigene-derived microsatellite markers in Brassica rapa L. Breed. Sci. 2011, 61, 160–167. [Google Scholar] [CrossRef]
- Chen, Z.H.; Ge, Y.; Sun, P.G.; Sun, C.J.; Wu, Q. The effects of propagating material and culture conditions on the propagation coefficient of avocado seedlings. China Fruit 2017, 5, 61–64. [Google Scholar] [CrossRef]
- Alcaraz, M.L.; Hormaza, J.I. Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Hereditas 2007, 144, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H. Avocado Cultivation and Processing; Jindun Press: Beijing, China, 2009. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.M.; Barker, G.L.A.; Berry, S.T.; Coghill, J.A.; Gwilliam, R.; Kirby, S.; Robinson, P.; Brenchley, R.C.; D’Amore, R.; McKenzie, N.; et al. Transcript-specific, single nucleotide polymorphism discovery and linkage analysis in hexaploid bread wheat (Triticum aestivum L.). Plant Biotechnol. J. 2011, 9, 1086–1099. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver. 3.0: An intergrated software package for population genetics data analysis. Evol.Bioinform. 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Chen, H.F.; Morrell, P.L.; Ashworth, V.E.T.M.; De la Cruz, M.; Clegg, M. Tracing the geographic origins of major avocado cultivars. J. Hered. 2009, 100, 56–65. [Google Scholar] [CrossRef]
- Davis, J.; Henderson, D.; Kobayashi, M.; Clegg, M.T. Genealogical relationships among cultivated avocado as revealed through RFLP analyses. J. Hered. 1998, 89, 319–323. [Google Scholar] [CrossRef]


| Accession | Origin | Source | Race | Type |
|---|---|---|---|---|
| Walter Hole | California, USA | GVTC, Guangxi, China | M [11] | C |
| Duke7 | California, USA | CATAS-SSCRI, Guangdong, China | M [1,11] | RS |
| Nabal | Antigua, Guatemala | GVTC, Guangxi, China | G [11] | C |
| Reed | California, USA | CATAS-SSCRI, Guangdong, China | G [1] | C |
| Pollock | Florida, US | CATAS-SSCRI, Guangdong, China | WI [1] | C |
| Donnie | Florida, USA | CATAS-SSCRI, Guangdong, China | WI [1] | C |
| Simmonds | Florida, USA | CATAS-SSCRI, Guangdong, China | WI [1] | C |
| Bacon | California, USA | GVTC, Guangxi, China | G × M [1,11] | C |
| Hass | California, USA | GVTC, Guangxi, China | G × M [1,11,12,13] | C |
| Pinkerton | California, USA | GVTC, Guangxi, China | G × M [11,13,14,15,16,17,18,19,20,21,22,23,24,25] | C |
| Zutano | California, USA | GVTC, Guangxi, China | G × M [1,11] | C |
| Ettinger | KefarMalal, Israel | GVTC, Guangxi, China | G × M [1,11,12,13] | C |
| Fuerte | Puebla, Mexico | CATAS-SSCRI, Guangdong, China | G × M [1,11,12,13] | C |
| Dusa | Westfalia Estate, South Africa | CATAS-SSCRI, Guangdong, China | G × M [11] | RS |
| Miguel | Florida, USA | CATAS-SSCRI, Guangdong, China | G × WI [1] | C |
| Loretta | Florida, USA | CATAS-SSCRI, Guangdong, China | G × WI [1] | C |
| Beta | Florida, USA | CATAS-SSCRI, Guangdong, China | G × WI [1] | C |
| Choquette | Florida, USA | CATAS-SSCRI, Guangdong, China | G × WI [1,12] | C |
| Lula | Florida, USA | CATAS-SSCRI, Guangdong, China | G × WI [1,12] | C |
| Tonnage | Florida, USA | CATAS-SSCRI, Guangdong, China | G × WI [1] | C |
| Guikenda No. 2 | Guangxi, China | GVTC, Guangxi, China | G × WI [26] | C |
| Guikenda No. 3 | Guangxi, China | GVTC, Guangxi, China | Unknown | LS |
| Guikenda No. 4 | Guangxi, China | GVTC, Guangxi, China | Unknown | LS |
| Guiyan No. 8 | Guangxi, China | GVTC, Guangxi, China | Unknown | LS |
| Guiyan No. 10 | Guangxi, China | GVTC, Guangxi, China | Unknown | LS |
| Qiongken No. 1 | Guangxi, China | GVTC, Guangxi, China | Unknown | LS |
| Qiongken No. 2 | Guangxi, China | GVTC, Guangxi, China | Unknown | LS |
| Daling No. 2 | Guangxi, China | GVTC, Guangxi, China | Unknown | LS |
| Daling No. 4 | Guangxi, China | GVTC, Guangxi, China | Unknown | LS |
| Population | Subpopulation | Ho | He | Nei | I | PIC | MAF |
|---|---|---|---|---|---|---|---|
| Population (K = 2) | Mexican–Guatemalan genotype-related population | 0.21 | 0.31 | 0.33 | 0.48 | 0.26 | 0.23 |
| West Indian genotype-related population | 0.21 | 0.30 | 0.32 | 0.47 | 0.25 | 0.22 | |
| Population (K = 3) | Mexican–Guatemalan genotype-related population I | 0.29 | 0.39 | 0.48 | 0.58 | 0.31 | 0.31 |
| Mexican–Guatemalan genotype-related population II | 0.25 | 0.33 | 0.37 | 0.50 | 0.27 | 0.24 | |
| West Indian genotype-related population | 0.21 | 0.30 | 0.32 | 0.47 | 0.25 | 0.22 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, Y.; Zhang, T.; Wu, B.; Tan, L.; Ma, F.; Zou, M.; Chen, H.; Pei, J.; Liu, Y.; Chen, Z.; et al. Genome-Wide Assessment of Avocado Germplasm Determined from Specific Length Amplified Fragment Sequencing and Transcriptomes: Population Structure, Genetic Diversity, Identification, and Application of Race-Specific Markers. Genes 2019, 10, 215. https://doi.org/10.3390/genes10030215
Ge Y, Zhang T, Wu B, Tan L, Ma F, Zou M, Chen H, Pei J, Liu Y, Chen Z, et al. Genome-Wide Assessment of Avocado Germplasm Determined from Specific Length Amplified Fragment Sequencing and Transcriptomes: Population Structure, Genetic Diversity, Identification, and Application of Race-Specific Markers. Genes. 2019; 10(3):215. https://doi.org/10.3390/genes10030215
Chicago/Turabian StyleGe, Yu, Teng Zhang, Bin Wu, Lin Tan, Funing Ma, Minghong Zou, Haihong Chen, Jinli Pei, Yuanzheng Liu, Zhihao Chen, and et al. 2019. "Genome-Wide Assessment of Avocado Germplasm Determined from Specific Length Amplified Fragment Sequencing and Transcriptomes: Population Structure, Genetic Diversity, Identification, and Application of Race-Specific Markers" Genes 10, no. 3: 215. https://doi.org/10.3390/genes10030215
APA StyleGe, Y., Zhang, T., Wu, B., Tan, L., Ma, F., Zou, M., Chen, H., Pei, J., Liu, Y., Chen, Z., Xu, Z., & Wang, T. (2019). Genome-Wide Assessment of Avocado Germplasm Determined from Specific Length Amplified Fragment Sequencing and Transcriptomes: Population Structure, Genetic Diversity, Identification, and Application of Race-Specific Markers. Genes, 10(3), 215. https://doi.org/10.3390/genes10030215





