Transcriptomic Analysis of Coding Genes and Non-Coding RNAs Reveals Complex Regulatory Networks Underlying the Black Back and White Belly Coat Phenotype in Chinese Wuzhishan Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Sample Collection and RNA Isolation
2.2. Library Preparation for Long Non-Coding RNA Sequencing and Data Analysis
2.3. Library Preparation for Micro RNA Sequencing and Data Analysis
2.4. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis
2.5. Construction of LncRNA–miRNA–Gene Regulatory Networks
2.6. Quantitative Polymerase Chain Reaction
2.7. Statistical Analysis
3. Results
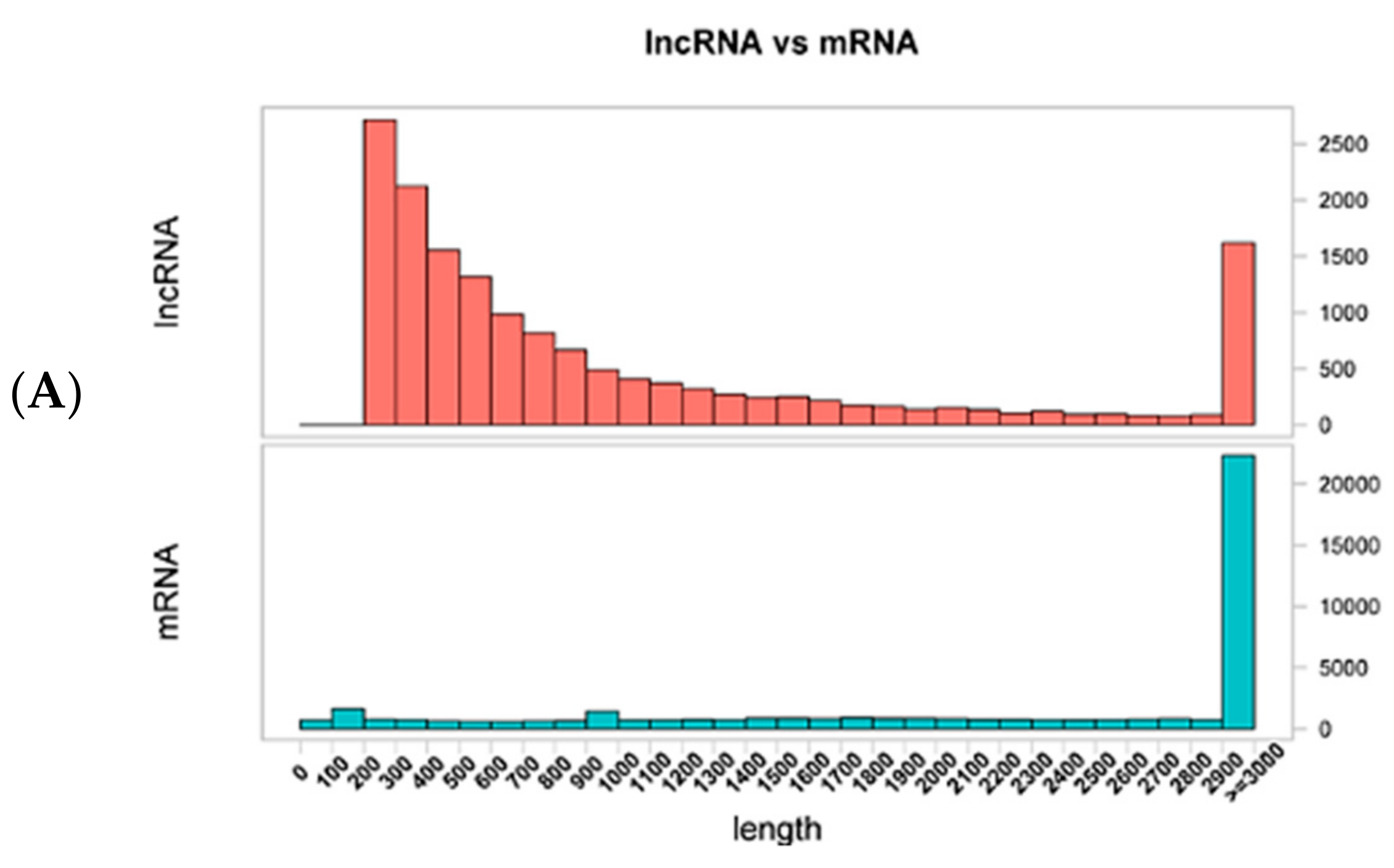
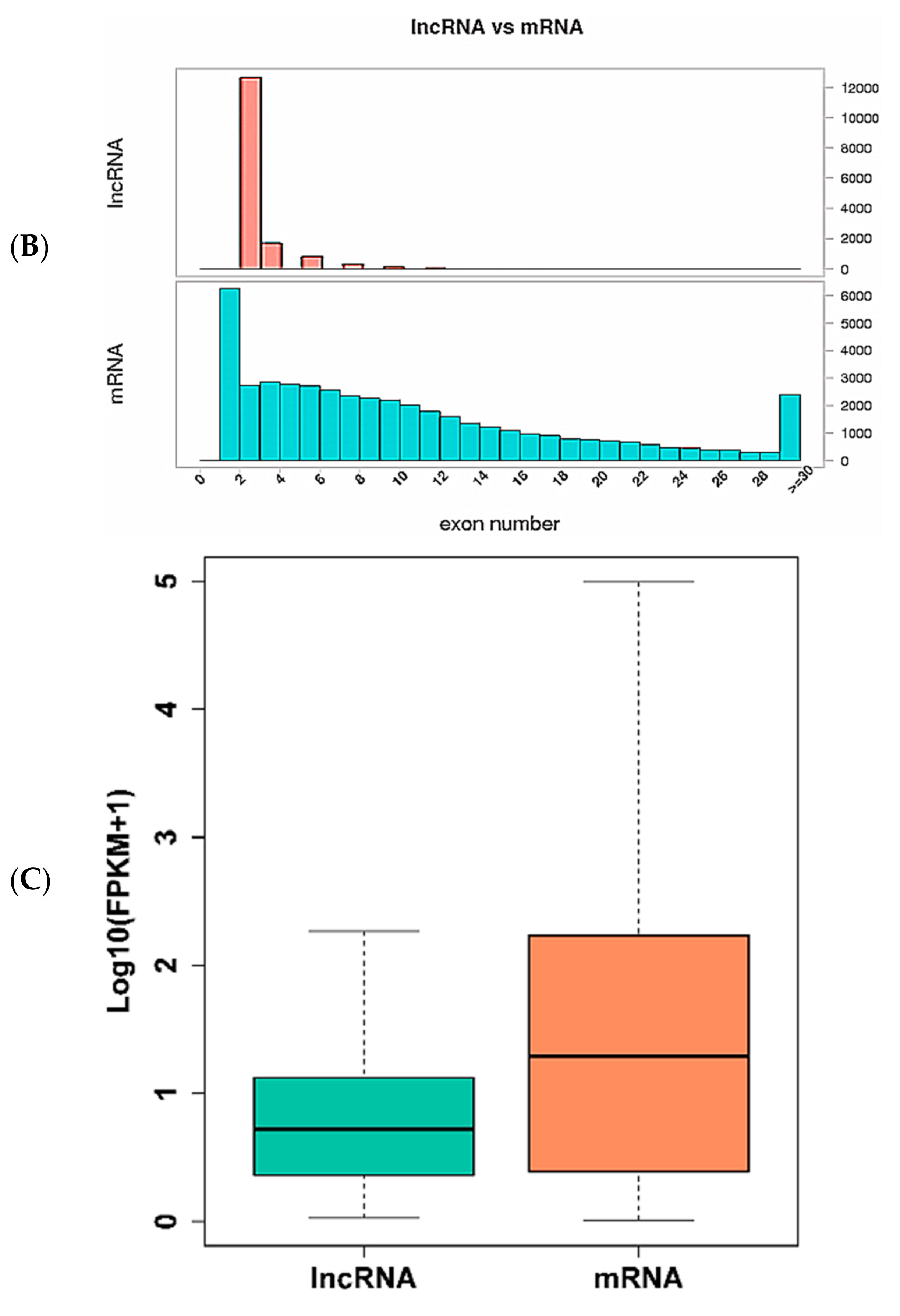
3.1. Identification of Long Non-Coding RNAs, Messenger RNAs and Micro RNAs in Wuzhishan Pig Skin by RNA-Sequencing
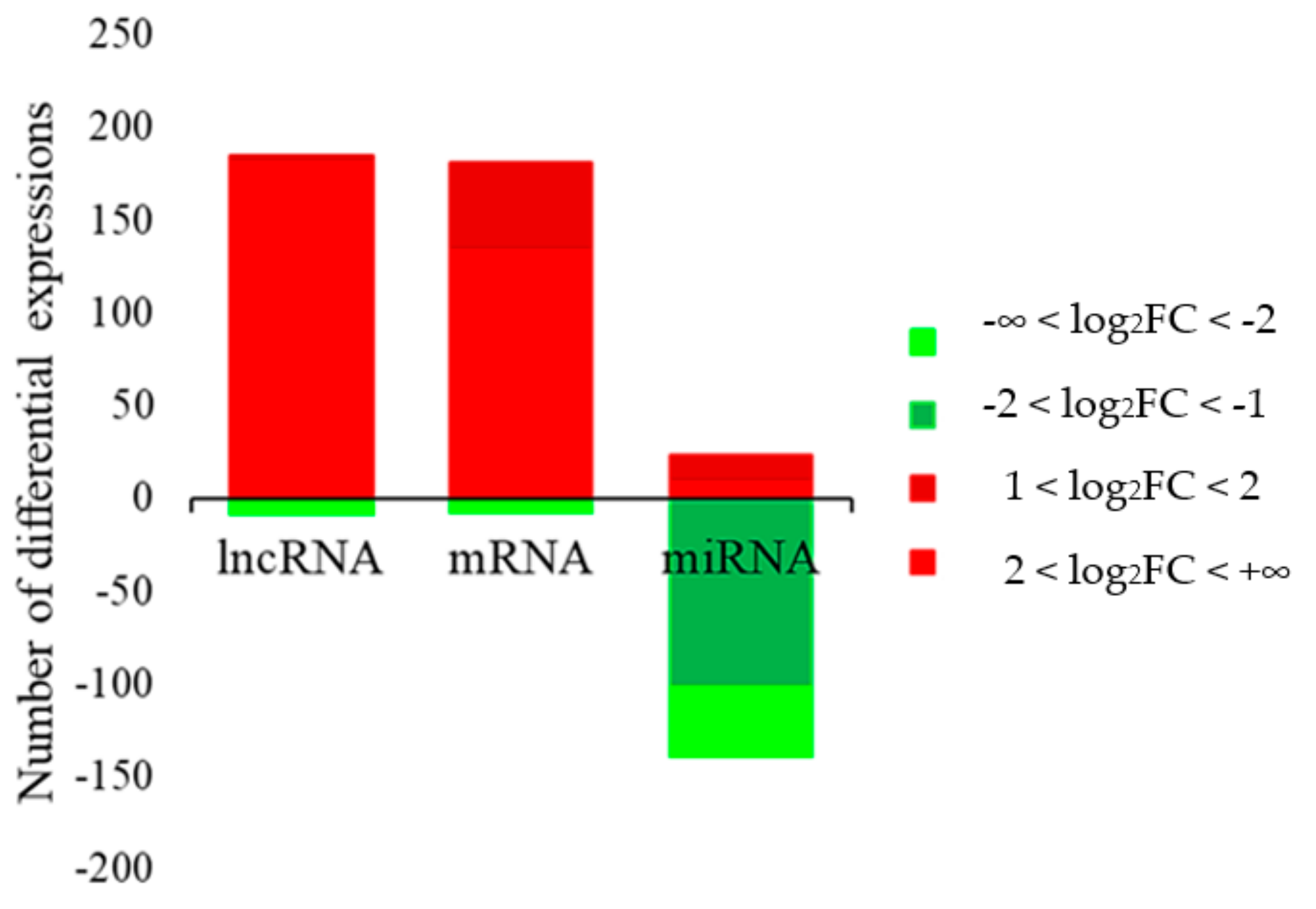
3.2. Differential Expression of Genes and Non-Coding RNAs (LncRNA and miRNA) in Skin Tissues
3.3. Functional Analysis of Differentially Expressed Transcripts
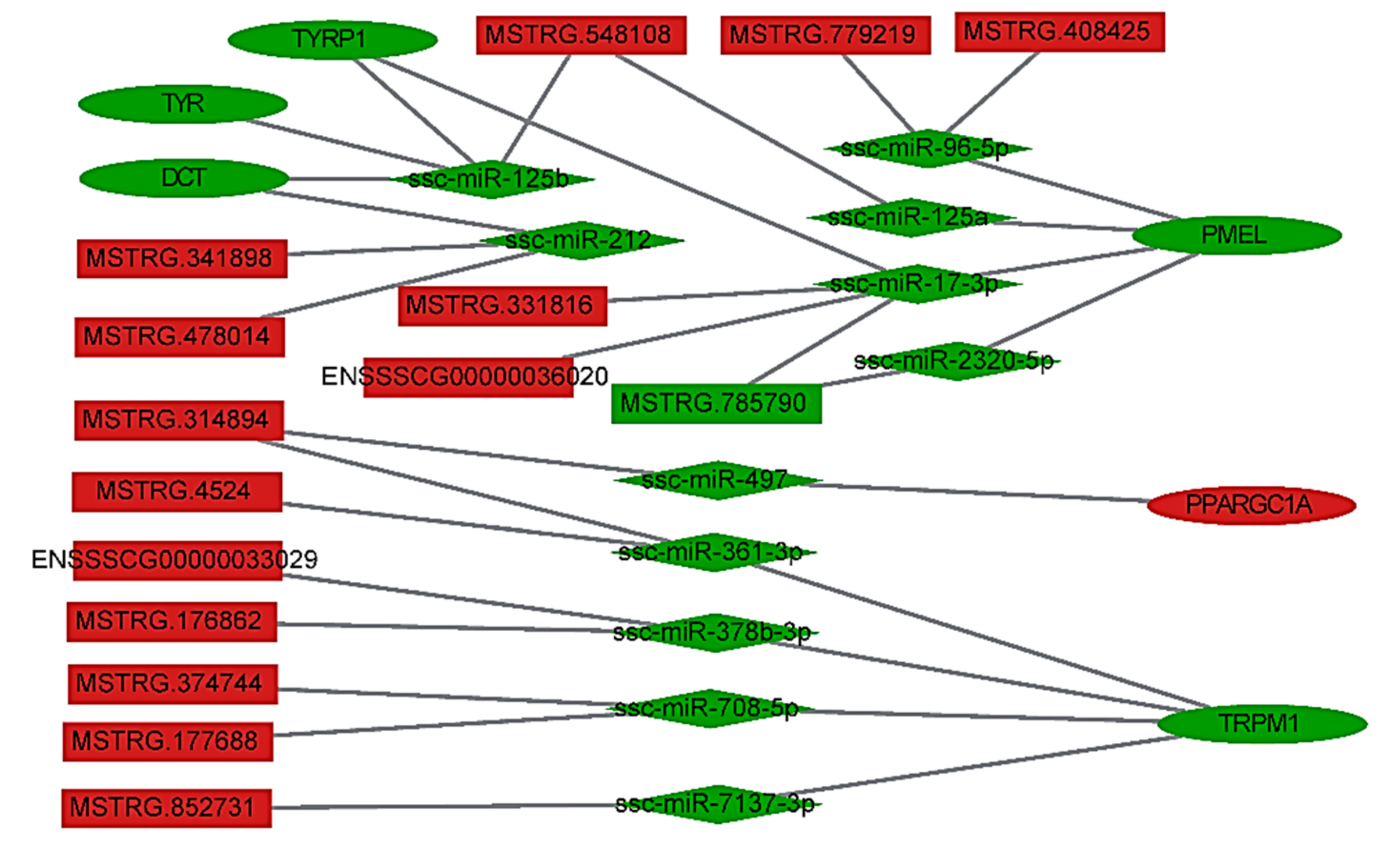
3.4. LncRNA–miRNA–Gene Interaction Network Construction
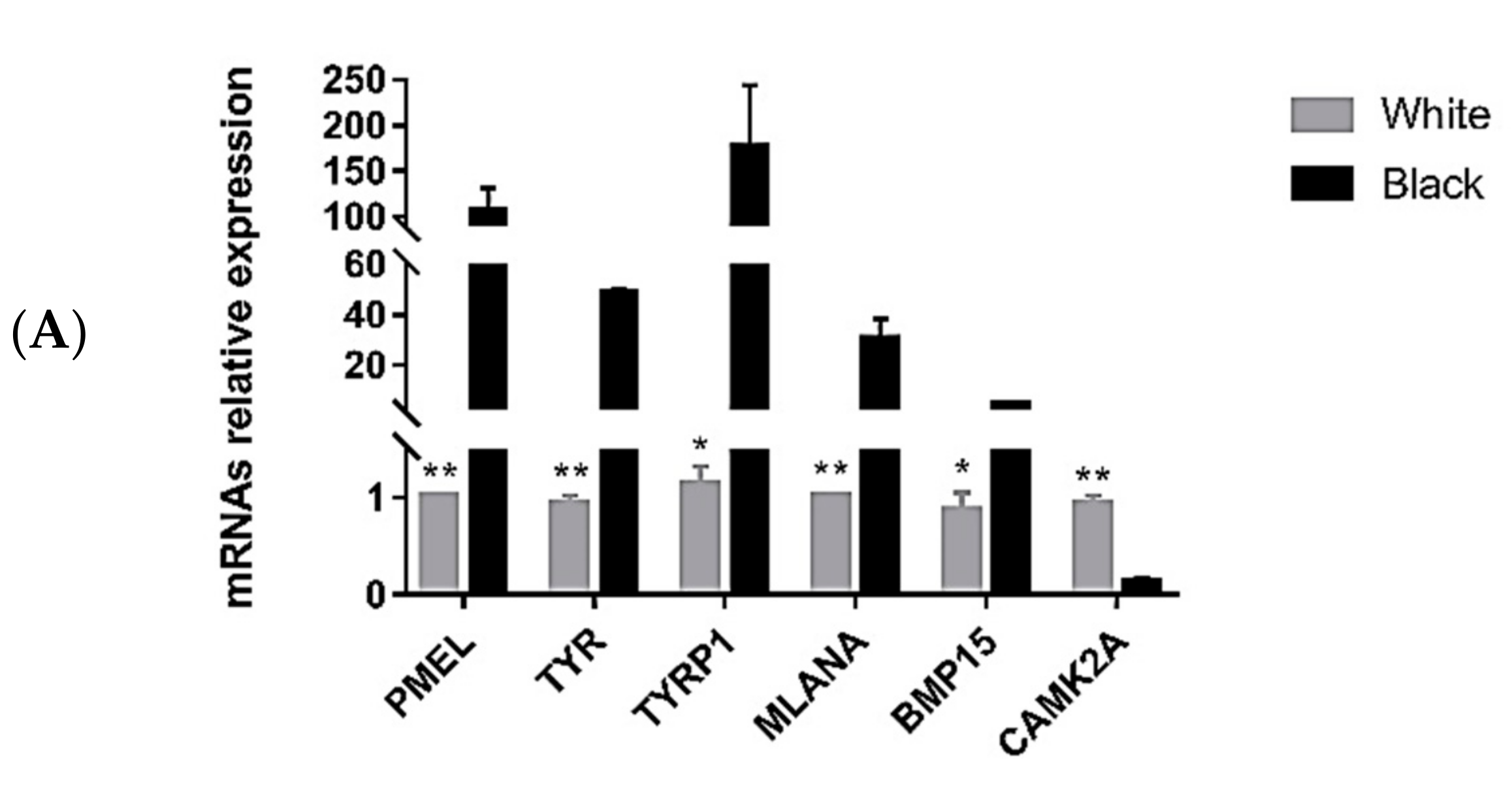
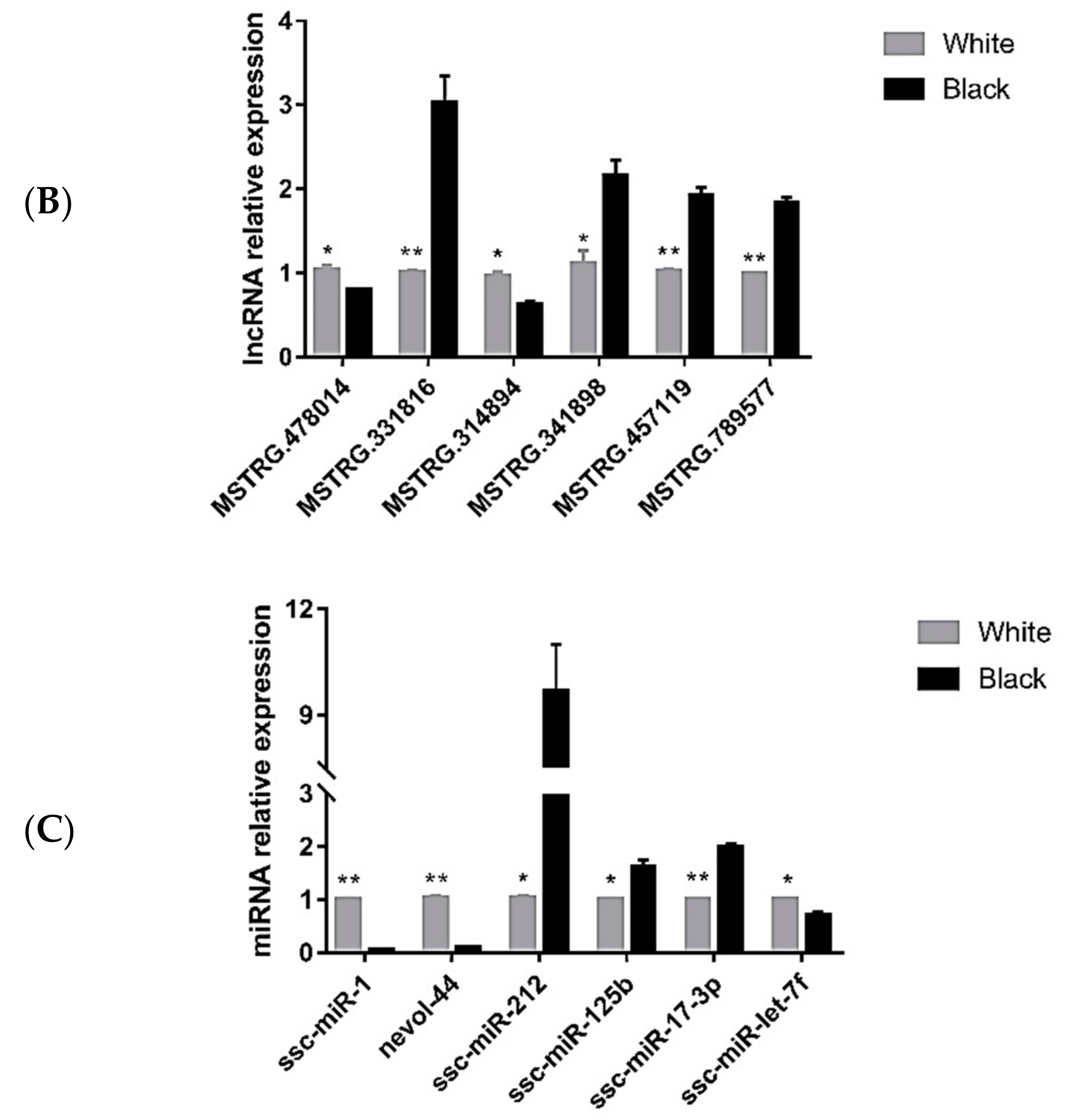
3.5. Quantitative Polymerase Chain Reaction Validation of Differentially Expressed Transcripts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Spillman, W.J. Inheritance of color coat in swine. Science 1906, 24, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Spillman, W.J. Inheritance of the belt in hampshire swine. Science 1907, 25, 541–543. [Google Scholar] [CrossRef] [PubMed]
- Solano, F. Melanins: Skin pigments and much more—types, structural models, biological functions and formation routes. New J. Sci. 2014, 2014. [Google Scholar] [CrossRef]
- Andersson, L.; Plastow, G.; Rothschild, M.; Ruvinsky, A. Molecular genetics of coat colour variation. Genet. Pig 2011, 38. [Google Scholar] [CrossRef]
- Fontanesi, L.; Russo, V. Molecular genetics of coat colour in pigs. Acta Agric. Slov. 2013, 4, 16. [Google Scholar]
- Kaelin, C.B.; Barsh, G.S.; Ostrander, E.A.; Ruvinsky, A. Molecular genetics of coat colour, texture and length in the dog. In Genetics of the Dog; CABI: Wallingford, UK, 2012; Volume 2, pp. 57–82. [Google Scholar] [CrossRef]
- Brenig, B.; Beck, J.; Floren, C.; Bornemann Kolatzki, K.; Wiedemann, I.; Hennecke, S.; Swalve, H.; Schütz, E. Molecular genetics of coat colour variations in White Galloway and White Park cattle. Anim. Genet. 2013, 44, 450–453. [Google Scholar] [CrossRef] [PubMed]
- Ruvinsky, A. Molecular genetics of coat colour variation. Genet. Cattle 2014, 67. [Google Scholar] [CrossRef]
- Koseniuk, A.; Ropka-Molik, K.; Rubiś, D.; Smołucha, G. Genetic background of coat colour in sheep. Arch. Anim. Breed. 2018, 61, 173–178. [Google Scholar] [CrossRef]
- Fan, R.; Xie, J.; Bai, J.; Wang, H.; Tian, X.; Bai, R.; Jia, X.; Yang, L.; Song, Y.; Herrid, M.; et al. Skin transcriptome profiles associated with coat color in sheep. BMC Genom. 2013, 14, 389. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, S.; Xu, J.; Feng, J.; Mahboob, S.; Al-Ghanim, K.A.; Sun, X.; Xu, P. Comparative transcriptome analysis reveals the genetic basis of skin color variation in common carp. PLoS ONE 2014, 9, e108200. [Google Scholar] [CrossRef] [PubMed]
- Brant, J.O.; Lopez, M.C.; Baker, H.V.; Barbazuk, W.B.; Maden, M. A comparative analysis of gene expression profiles during skin regeneration in Mus and Acomys. PLoS ONE 2015, 10, e142931. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xu, C.; Liu, Z.; Yue, Z.; Liu, L.; Yang, T.; Cong, B.; Yang, F. Comparative transcriptome analysis of mink (Neovison vison) skin reveals the key genes involved in the melanogenesis of black and white coat colour. Sci. Rep. 2017, 7, 12461. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; He, X.; Zhao, Y.; Bai, D.; Shiraigo, W.; Zhao, Q.; Manglai, D. Regulatory pathway analysis of coat color genes in Mongolian horses. Hereditas 2018, 155, 13. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Fu, Y.; Cao, J.; Yu, M.; Tang, X.; Zhao, S. Identification of differentially expressed miRNAs between white and black hair follicles by RNA-sequencing in the goat (Capra hircus). Int. J. Mol. Sci. 2014, 15, 9531–9545. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.X.; Wu, R.B.; Qiao, X.; Zhang, Y.J.; Wang, R.J.; Su, R.; Wu, J.H.; Dong, Y.; Li, J.Q. Hair follicle transcriptome profiles during the transition from anagen to catagen in Cashmere goat (Capra hircus). Genet. Mol. Res. 2015, 14, 17904–17915. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ge, W.; Luo, Z.; Guo, Y.; Jiao, B.; Qu, L.; Zhang, Z.; Wang, X. Integrated analysis of coding genes and non-coding RNAs during hair follicle cycle of cashmere goat (Capra hircus). BMC Genom. 2017, 18, 767. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, C.; Yu, W.; Zhao, S.; Gong, Y. Identification of genes related to white and black plumage formation by RNA-seq from white and black feather bulbs in ducks. PLoS ONE 2012, 7, e36592. [Google Scholar] [CrossRef] [PubMed]
- Apopo, S.; Liu, H.; Jing, L.; Du, X.; Xie, S.; Gong, Y.; Xu,, R.; Li,, S. Identification and profiling of microRNAs associated with white and black plumage pigmentation in the white and black feather bulbs of ducks by RNA sequencing. Anim. Genet. 2015, 46, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, G.; Liao, J.; Tang, M.; Sun, W. Transcriptome Profile analysis of mechanisms of black and white plumage determination in black-bone chicken. Cell Physiol. Biochem. 2018, 46, 2373–2384. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, F.; Cao, J.; Liu, X. Skin transcriptome profiles associated with skin color in chickens. PLoS ONE 2015, 10, e127301. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Jiang, J.; Fan, R.; Wang, H.; Meng, X.; He, X.; He, J.; Li, H.; Geng, J.; Yu, X.; et al. Identification and characterization of microRNAs in white and brown alpaca skin. BMC Genom. 2012, 13, 555. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xiao, H.; Li, H.; Zhao, Y.; Lai, S.; Yu, X.; Cai, T.; Du, C.; Zhang, W.; Li, J. Identification of conserved and novel microRNAs in cashmere goat skin by deep sequencing. PLoS ONE 2012, 7, e50001. [Google Scholar] [CrossRef]
- Novikova, I.V.; Hennelly, S.P.; Tung, C.S.; Sanbonmatsu, K.Y. Rise of the RNA machines: exploring the structure of long non-coding RNAs. J. Mol. Biol. 2013, 425, 3731–3746. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.C.; Wang, K.C. Long noncoding RNA: Significance and potential in skin biology. Cold Spring Harb. Perspect. Med. 2014, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Weikard, R.; Hadlich, F.; Kuehn, C. Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genom. 2013, 14, 789. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Wang, G.; Chen, L.; Jiang, J.; Liu, L.; Li, N.; Zhao, J.; Sun, X.; Zhou, P. Genome-wide analysis of long non-coding RNAs at early stage of skin pigmentation in goats (Capra hircus). BMC Genom. 2016, 17, 67. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Method. 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily conserved elements in vertebrate, insect, worm and yeast genomes. Genome Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Bennett, D.C.; Lamoreux, M.L. The color loci of mice–A genetic century. Pigment. Cell Res. 2003, 16, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Mao, H.; Zhang, Z.; Xiao, S.; Ding, N.; Huang, L. A 6-bp deletion in the TYRP1 gene causes the brown colouration phenotype in Chinese indigenous pigs. Heredity (Edinb.) 2011, 106, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Bin, B.H.; Kim, J.; Dong, S.E.; Park, P.J.; Choi, H.; Kim, B.J.; Yu, S.J.; Kang, H.; Kang, H.H.; et al. Novel inhibitory function of miR-125b in melanogenesis. Pigment Cell Melanoma. Res. 2014, 27, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, T.R.; Cho, E.G. SH3BP4, a novel pigmentation gene, is inversely regulated by miR-125b and MITF. Exp. Mol. Med. 2017, 49, e367. [Google Scholar] [CrossRef] [PubMed]
- Jian, Q.; An, Q.; Zhu, D.; Hui, K.; Liu, Y.; Chi, S.; Li, C. MicroRNA 340 is involved in UVB-induced dendrite formation through the regulation of RhoA expression in melanocytes. Mol. Cell. Biol. 2014, 34, 3407–3420. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.; Tarapore, R.S.; Poenitzsch Strong, A.M.; TeSlaa, J.J.; Grinblat, Y.; Setaluri, V.; Spiegelman, V.S. MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor (MITF) mRNA is inhibited by coding region determinant-binding protein (CRD-BP). J. Biol. Chem. 2015, 290, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Satzger, I.; Mattern, A.; Kuettler, U.; Weinspach, D.; Niebuhr, M.; Kapp, A.; Gutzmer, R. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp. Dermatol. 2012, 21, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Dynoodt, P.; Mestdagh, P.; Van Peer, G.; Vandesompele, J.; Goossens, K.; Peelman, L.J.; Geusens, B.; Speeckaert, R.M.; Lambert, J.L.; van Gele, M.J. Identification of miR-145 as a key regulator of the pigmentary process. J. Investig. Dermatol. 2013, 133, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, H.; Wu, Y.; Zhou, J.; Yang, G.; Wang, W. lncRNA MEG3 promotes hepatic insulin resistance by serving as a competing endogenous RNA of miR-214 to regulate ATF4 expression. Int. J. Mol. Med. 2019, 43, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, L.; Lu, C.; Shen, Q.; Su, Y.; Zhi, Z.; Wu, F.; Zhang, H.; Wen, Z.; Chen, G.; et al. Long non-coding RNA FAL1 functions as a ceRNA to antagonize the effect of miR-637 on the down-regulation of AKT1 in Hirschsprung’s disease. Cell. Prolif. 2018, 51, e12489. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Yang, R.; Zheng, X.; Zhang, L.; Jiang, R.; Chen, J. LncRNA RP11-79H23.3 Functions as a Competing Endogenous RNA to Regulate PTEN Expression through Sponging hsa-miR-107 in the Development of Bladder Cancer. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Maronas, O.; Sochtig, J.; Ruiz, Y.; Phillips, C.; Carracedo, A.; Lareu, M.V. The genetics of skin, hair and eye color variation and its relevance to forensic pigmentation predictive tests. Forensic Sci. Rev. 2015, 27, 13–40. [Google Scholar] [PubMed]
- Montagna, W.; Carlisle, K. The architecture of black and white facial skin. J. Am. Acad. Dermatol. 1991, 24, 929–937. [Google Scholar] [CrossRef]
- Ozeki, H.; Ito, S.; Wakamatsu, K.; Hirobe, T. Chemical characterization of hair melanins in various coat-color mutants of mice. J. Investig. Dermatol. 1995, 105, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Sponenberg, D.P.; Ito, S.; Eng, L.A.; Schwink, K. Pigment types of various color genotypes of horses. Pigment Cell Res. 1988, 1, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Aliev, G.; Rachkovsky, M.; Ito, S.; Wakamatsu, K.; Ivanov, A. Pigment types in selected color genotypes of Asiatic sheep. Pigment Cell Res. 1990, 3, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Cecchi, T.; Cozzali, C.; Passamonti, P.; Ceccarelli, P.; Pucciarelli, F.; Gargiulo, A.M.; Frank, E.N.; Renieri, C. Melanins and melanosomes from llama (Lama glama L.). Pigment Cell Res. 2004, 17, 307–311. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Wakamatsu, K. Human hair melanins: what we have learned and have not learned from mouse coat color pigmentation. Pigment Cell Melanoma Res. 2011, 24, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Andl, T.; Botchkareva, N.V. MicroRNAs (miRNAs) in the control of HF development and cycling: The next frontiers in hair research. Exp. Dermatol. 2015, 24, 821–826. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, T.; O’Connor, M.N.; Seabra, M.C.; Cutler, D.F.; Futter, C.E. Regulation of melanosome number, shape and movement in the zebrafish retinal pigment epithelium by OA1 and PMEL. J. Cell Sci. 2015, 128, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Kwon, B.S.; Halaban, R.; Ponnazhagan, S.; Kim, K.; Chintamaneni, C.; Bennett, D.; Pickard, R.T. Mouse silver mutation is caused by a single base insertion in the putative cytoplasmic domain of Pmel 17. Nucleic Acids Res. 1995, 23, 154–158. [Google Scholar] [CrossRef] [PubMed]
- Kerje, S.; Sharma, P.; Gunnarsson, U.; Kim, H.; Bagchi, S.; Fredriksson, R.; Schutz, K.; Jensen, P.; von Heijne, G.; Okimoto, R.; et al. The dominant white, dun and smoky color variants in chicken are associated with insertion/deletion polymorphisms in the PMEL17 gene. Genetics 2004, 168, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.A.; Wahl, J.M.; Rees, C.A.; Murphy, K.E. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc. Natl. Acad. Sci. USA 2006, 103, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
- De Maziere, A.M.; Muehlethaler, K.; van Donselaar, E.; Salvi, S.; Davoust, J.; Cerottini, J.C.; Levy, F.; Slot, J.W.; Rimoldi, D. The melanocytic protein Melan-A/MART-1 has a subcellular localization distinct from typical melanosomal proteins. Traffic 2002, 3, 678–693. [Google Scholar] [CrossRef] [PubMed]
- Hoashi, T.; Watabe, H.; Muller, J.; Yamaguchi, Y.; Vieira, W.D.; Hearing, V.J. MART-1 is required for the function of the melanosomal matrix protein PMEL17/GP100 and the maturation of melanosomes. J. Biol. Chem. 2005, 280, 14006–14016. [Google Scholar] [CrossRef] [PubMed]
- Giordano, F.; Bonetti, C.; Surace, E.M.; Marigo, V.; Raposo, G. The ocular albinism type 1 (OA1) G-protein-coupled receptor functions with MART-1 at early stages of melanogenesis to control melanosome identity and composition. Hum. Mol. Genet. 2009, 18, 4530–4545. [Google Scholar] [CrossRef] [PubMed]
- Oancea, E.; Vriens, J.; Brauchi, S.; Jun, J.; Splawski, I.; Clapham, D.E. TRPM1 forms ion channels associated with melanin content in melanocytes. Sci. Signal. 2009, 2, a21. [Google Scholar] [CrossRef] [PubMed]
- Devi, S.; Kedlaya, R.; Maddodi, N.; Bhat, K.M.; Weber, C.S.; Valdivia, H.; Setaluri, V. Calcium homeostasis in human melanocytes: Role of transient receptor potential melastatin 1 (TRPM1) and its regulation by ultraviolet light. Am. J. Physiol. Cell Physiol. 2009, 297, C679–C687. [Google Scholar] [CrossRef] [PubMed]

| Genes | Gene ID | Gene Name | White vs. Black log2FC | p-Value |
|---|---|---|---|---|
| lncRNAs | MSTRG.789577 | –5.51 | 0.0019 | |
| MSTRG.478014 | 2.20 | 0.0346 | ||
| MSTRG.331816 | 2.92 | 0.0317 | ||
| MSTRG.314894 | 2.71 | 0.0236 | ||
| MSTRG.341898 | 3.35 | 1.09E–04 | ||
| MSTRG.457119 | 5.82 | 1.89E–06 | ||
| genes | ENSSSCG00000026409 | TYR | –2.03 | 0.0428 |
| ENSSSCG00000005206 | MLANA | –2.68 | 0.0487 | |
| ENSSSCG00000000371 | PMEL | –3.84 | 5.64E–09 | |
| ENSSSCG00000005193 | TYRP1 | –4.93 | 1.58E–04 | |
| ENSSSCG00000014443 | CAMK2A | 2.80 | 5.29E–04 | |
| ENSSSCG00000012310 | BMP15 | 8.16 | 5.46E–12 | |
| microRNAs | ssc-miR-125b | –1.76 | 0.0000 | |
| ssc-miR-17-3p | –3.17 | 0.0057 | ||
| ssc-miR-212 | –4.76 | 1.14E–04 | ||
| ssc-let-7f | 1.47 | 0.0000 | ||
| ssc-miR-1 | 2.48 | 0.0000 | ||
| Novel-44 | 15.37 | 0.0000 |
| Sample Name | Raw Reads | Clean Reads | Error Rate (%) | Q20 (%) | Q30 (%) | % of Mapped Reads | Uniquely Mapped Reads |
|---|---|---|---|---|---|---|---|
| WW1 | 97269174 | 85519188 | 0.02 | 96.78 | 92.01 | 88.89 | 74229023 |
| WW2 | 102600722 | 83351954 | 0.02 | 96.94 | 92.43 | 90.72 | 73653848 |
| WW3 | 84105516 | 82637176 | 0.02 | 97.14 | 92.52 | 95.13 | 76182061 |
| WB1 | 98557820 | 81182410 | 0.02 | 96.70 | 91.90 | 90.64 | 71722643 |
| WB2 | 113729814 | 100498792 | 0.02 | 96.70 | 91.89 | 90.78 | 89001334 |
| WB3 | 81046142 | 79,101,022 | 0.02 | 97.48 | 92.66 | 95.09 | 72977205 |
| Category | Terms | DEGs No. | p-Value | Genes |
|---|---|---|---|---|
| GO | GO:0042438-melanin biosynthetic process | 4 | 1.38E–05 | PMEL, TYR, TYRP1, DCT |
| GO:0048066-developmental pigmentation | 4 | 4.7E–04 | PMEL, TYR, TYRP1, DCT | |
| GO:0006582-melanin metabolic process | 4 | 1.66E–05 | PMEL, TYR, TYRP1, DCT | |
| GO:0043473-pigmentation | 4 | 0.00456 | PMEL, TYR, TYRP1, DCT | |
| GO:0042440-pigment metabolic process | 5 | 1.1E–04 | PMEL, TYR, TYRP1,PPARGC1A, DCT | |
| GO:0033162-melanosome membrane | 3 | 6.52E–05 | TYRP1, TYR, DCT | |
| GO:0006570-tyrosine metabolic process | 2 | 0.00243 | TYR, DCT | |
| GO:0042470-melanosome | 5 | 8.7E–04 | PMEL, MLANA, TYR, TYRP1, DCT | |
| GO:0048770-pigment granule | 6 | 8.7E–04 | PMEL, MLANA, TYR, TYRP1, DCT, TRPM1 | |
| GO:0046148-pigment biosynthetic process | 4 | 5.0E–04 | PMEL, TYR, TYRP1, DCT | |
| KEGG pathway | hsa00350-Tyrosine metabolism | 3 | 0.0022 | TYRP1, TYR, DCT |
| hsa04916-Melanogenesis | 4 | 0.0056 | TYRP1, TYR, DCT, CAMK2A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Q.; Liu, X.; Chao, Z.; Wang, K.; Wang, J.; Tang, Q.; Luo, Y.; Zheng, J.; Tan, S.; Fang, M. Transcriptomic Analysis of Coding Genes and Non-Coding RNAs Reveals Complex Regulatory Networks Underlying the Black Back and White Belly Coat Phenotype in Chinese Wuzhishan Pigs. Genes 2019, 10, 201. https://doi.org/10.3390/genes10030201
Xu Q, Liu X, Chao Z, Wang K, Wang J, Tang Q, Luo Y, Zheng J, Tan S, Fang M. Transcriptomic Analysis of Coding Genes and Non-Coding RNAs Reveals Complex Regulatory Networks Underlying the Black Back and White Belly Coat Phenotype in Chinese Wuzhishan Pigs. Genes. 2019; 10(3):201. https://doi.org/10.3390/genes10030201
Chicago/Turabian StyleXu, Qiao, Ximing Liu, Zhe Chao, Kejun Wang, Jue Wang, Qiguo Tang, Yabiao Luo, Jie Zheng, Shuyi Tan, and Meiying Fang. 2019. "Transcriptomic Analysis of Coding Genes and Non-Coding RNAs Reveals Complex Regulatory Networks Underlying the Black Back and White Belly Coat Phenotype in Chinese Wuzhishan Pigs" Genes 10, no. 3: 201. https://doi.org/10.3390/genes10030201
APA StyleXu, Q., Liu, X., Chao, Z., Wang, K., Wang, J., Tang, Q., Luo, Y., Zheng, J., Tan, S., & Fang, M. (2019). Transcriptomic Analysis of Coding Genes and Non-Coding RNAs Reveals Complex Regulatory Networks Underlying the Black Back and White Belly Coat Phenotype in Chinese Wuzhishan Pigs. Genes, 10(3), 201. https://doi.org/10.3390/genes10030201




