Determination of Genetic Effects of LIPK and LIPJ Genes on Milk Fatty Acids in Dairy Cattle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Population and Phenotypic Data
2.2. SNPs Identification and Genotyping
2.3. Estimation of linkage disequilibrium (LD)
2.4. Association Analyses
2.5. Protein Structure and Function Prediction
2.6. Predication of Transcription Factors Binding Sites (TFBSs)
2.7. Gene Expressions Assay of LIPK and LIPJ
3. Results
3.1. SNPs Identification
3.2. Associations between SNPs/Haplotype Blocks and Milk FAs
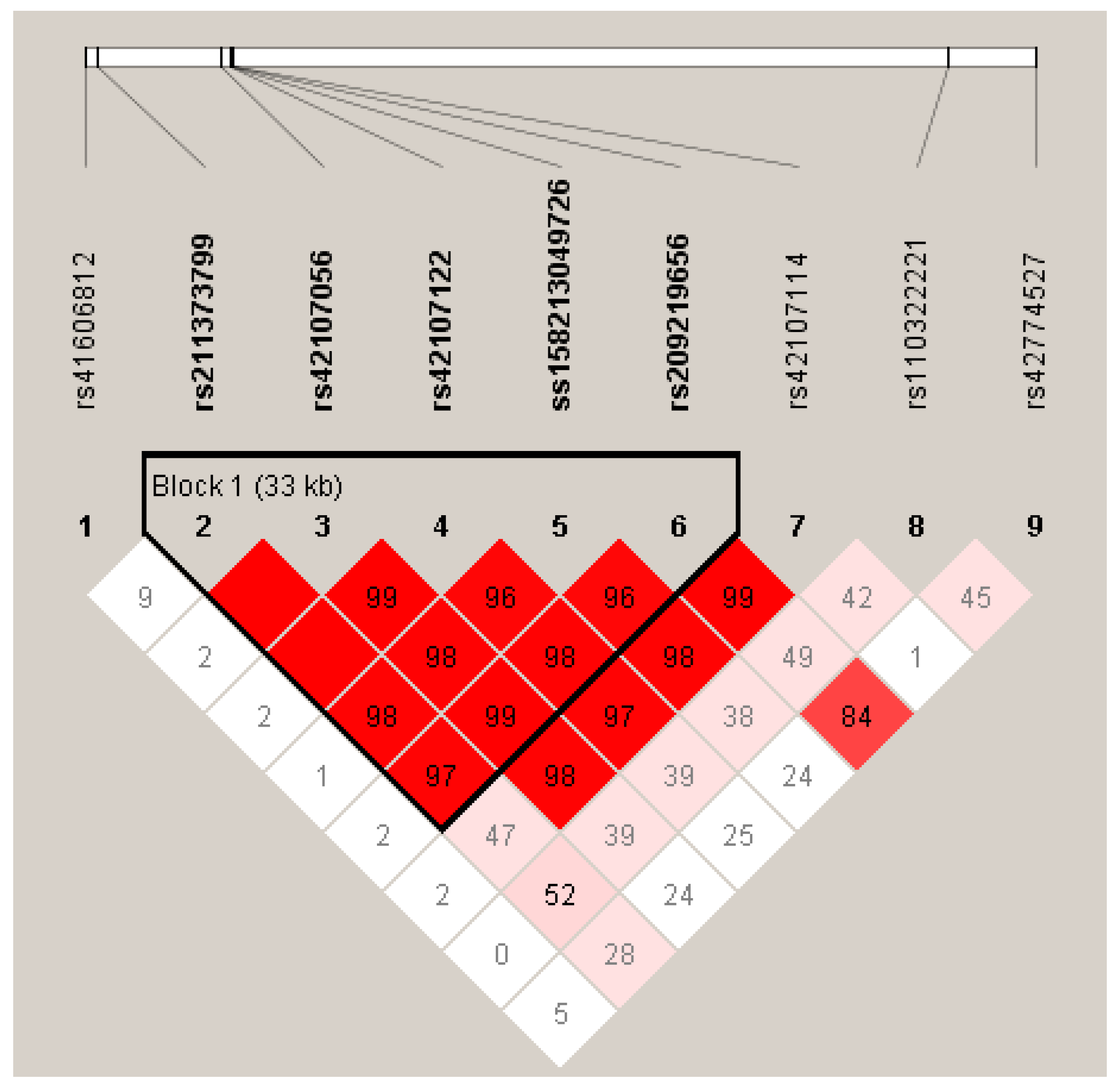
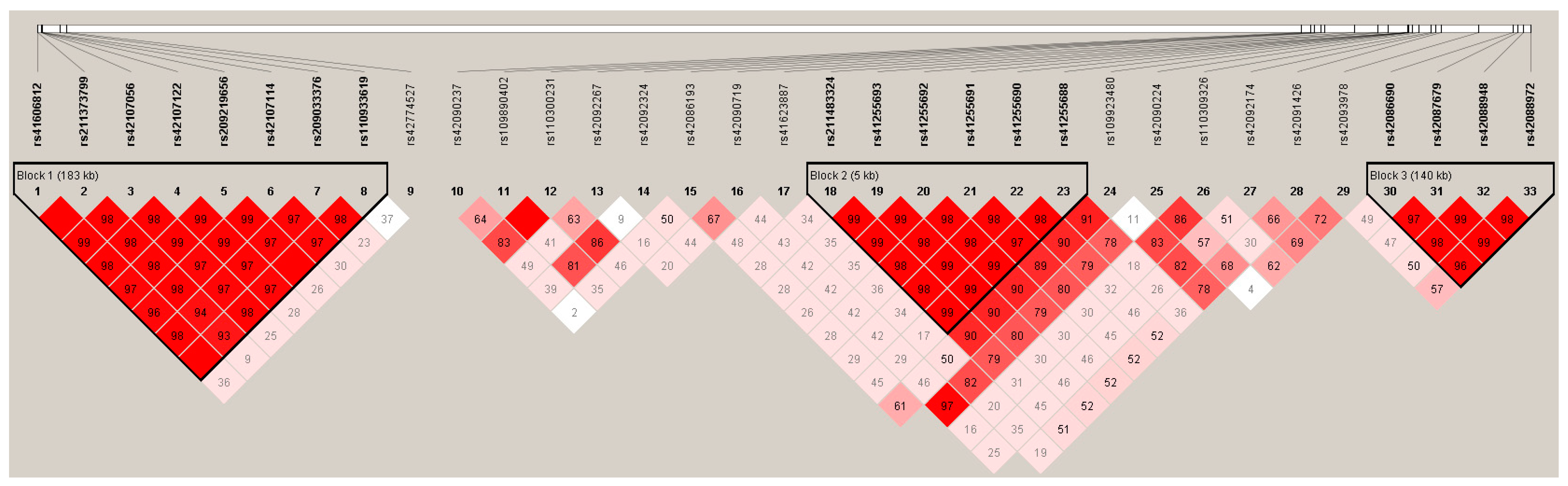
3.3. Linkage disequilibrium among the SNPs of LIPK, LIPJ and SCD Genes
3.4. rs42774527 Caused the Changes of the LIPK Protein Structure and Function
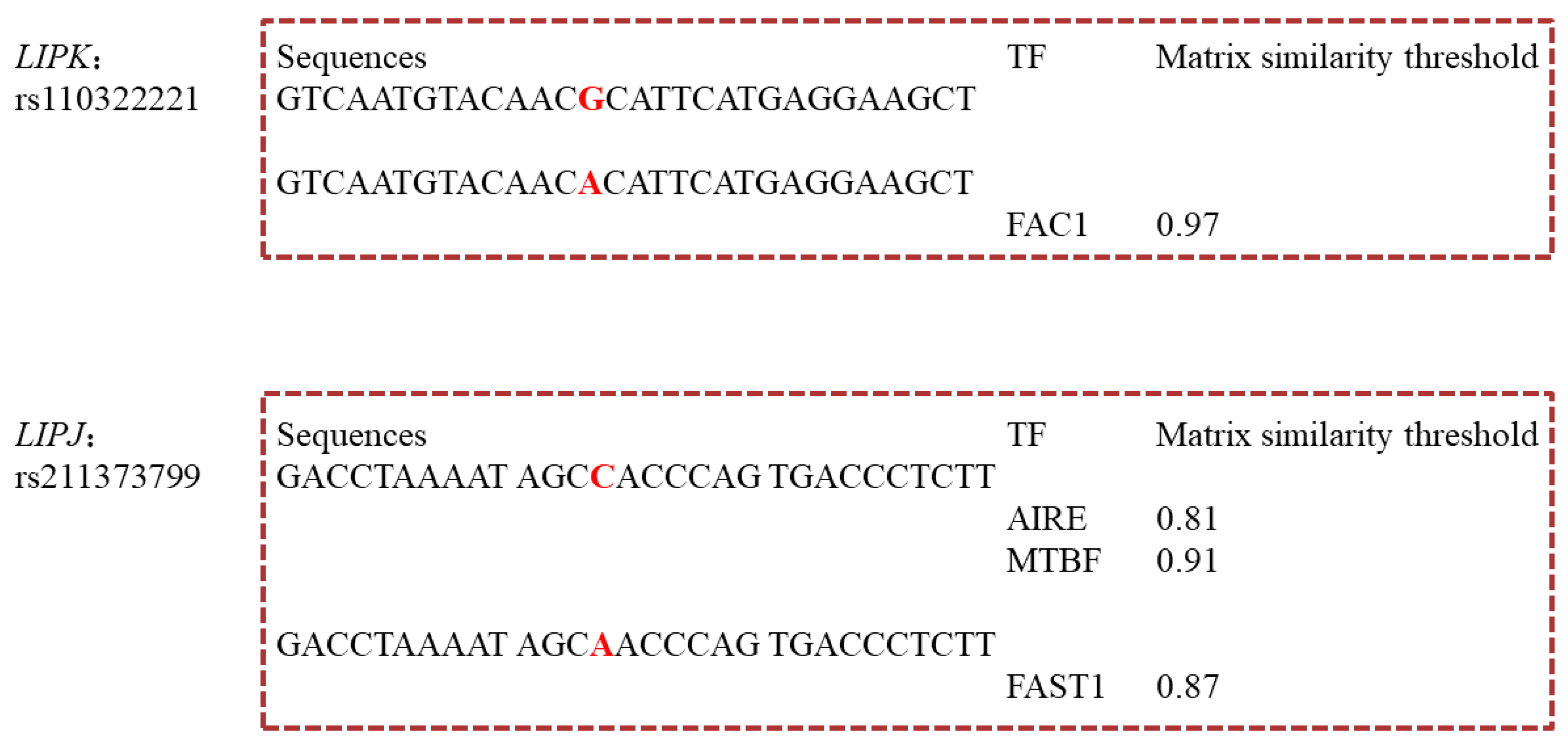
3.5. Transcription Factors Binding Sites (TFBSs) Changed by rs110322221 and rs211373799
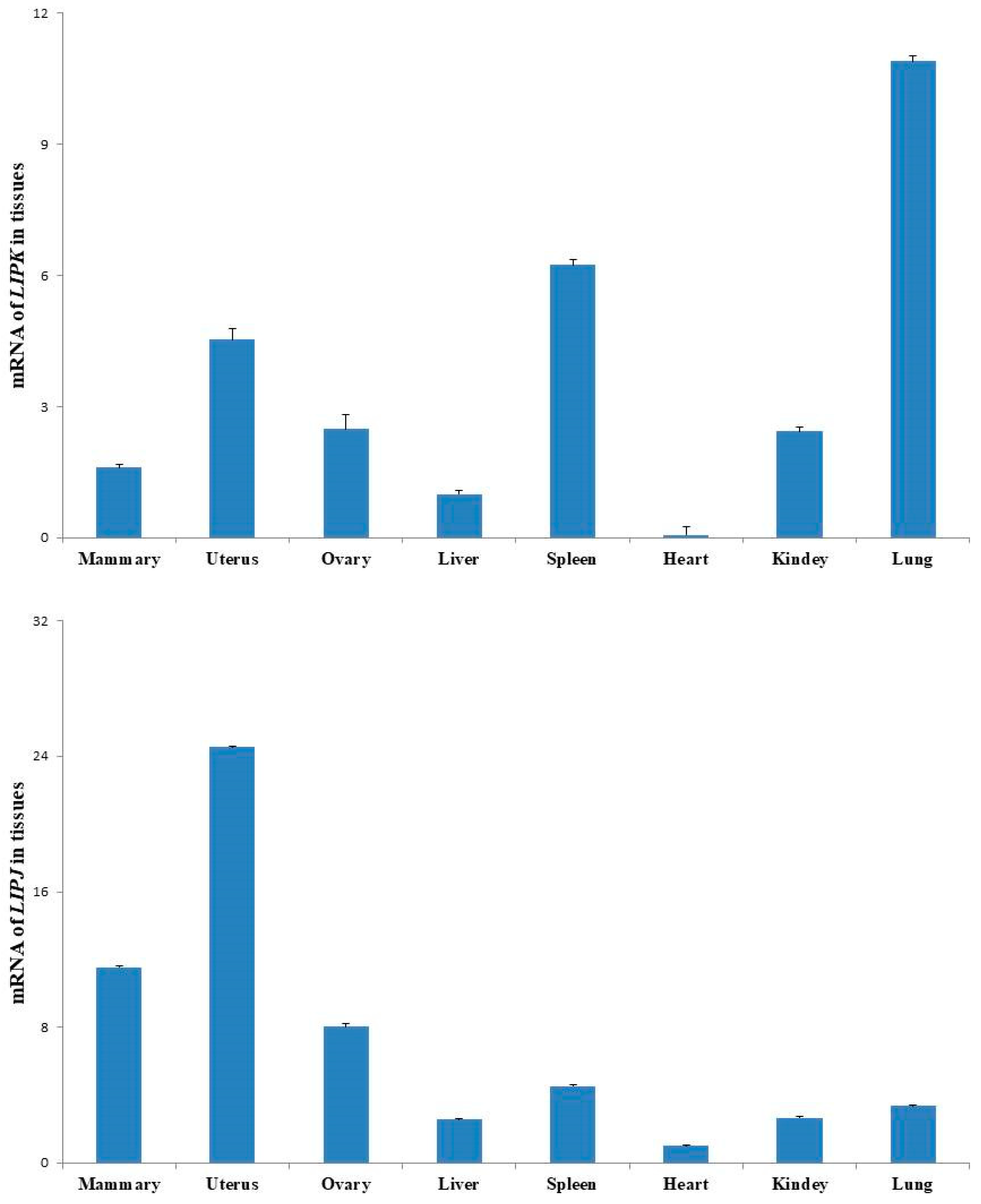
3.6. Expressions of LIPK and LIPJ Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of interest
Ethics Approval
References
- Turek, K.; Domagala, J.; Wszolek, M. Fatty acid profile and oxidation tests of fat extracted from yogurt using rose hip seed oil. Acta Sci. Polonorum Technol. Aliment. 2018, 17, 51–58. [Google Scholar] [CrossRef]
- Leamy, A.K.; Egnatchik, R.A.; Young, J.D. Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease. Prog. Lipid Res. 2013, 52, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.G.; Cao, H.X. Role of diet and nutritional management in non-alcoholic fatty liver disease. J Gastroenterol. Hepatol. 2013, 28 (Suppl. 4), 81–87. [Google Scholar] [CrossRef]
- Rolo, A.P.; Teodoro, J.S.; Palmeira, C.M. Role of oxidative stress in the pathogenesis of nonalcoholic steatohepatitis. Free Radic. Biol. Med. 2012, 52, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Kliem, K.E.; Humphries, D.J.; Grandison, A.S.; Morgan, R.; Livingstone, K.M.; Givens, D.I.; Reynolds, C.K. Effect of a whey protein and rapeseed oil gel feed supplement on milk fatty acid composition of Holstein cows. J. Dairy Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bauman, D.E.; Griinari, J.M. Regulation and nutritional manipulation of milk fat. Low-fat milk syndrome. Adv. Exp. Med. Biol. 2000, 480, 209–216. [Google Scholar] [CrossRef]
- Krag, K.; Poulsen, N.A.; Larsen, M.K.; Larsen, L.B.; Janss, L.L.; Buitenhuis, B. Genetic parameters for milk fatty acids in Danish Holstein cattle based on SNP markers using a Bayesian approach. BMC Genet. 2013, 14, 79. [Google Scholar] [CrossRef] [PubMed]
- Petrini, J.; Iung, L.H.; Rodriguez, M.A.; Salvian, M.; Pertille, F.; Rovadoscki, G.A.; Cassoli, L.D.; Coutinho, L.L.; Machado, P.F.; Wiggans, G.R.; et al. Genetic parameters for milk fatty acids, milk yield and quality traits of a Holstein cattle population reared under tropical conditions. J. Anim. Breed. Genet. 2016, 133, 384–395. [Google Scholar] [CrossRef] [PubMed]
- Narayana, S.G.; Schenkel, F.S.; Fleming, A.; Koeck, A.; Malchiodi, F.; Jamrozik, J.; Johnston, J.; Sargolzaei, M.; Miglior, F. Genetic analysis of groups of mid-infrared predicted fatty acids in milk. J. Dairy Sci. 2017, 100, 4731–4744. [Google Scholar] [CrossRef] [PubMed]
- Stoop, W.M.; Schennink, A.; Visker, M.H.; Mullaart, E.; van Arendonk, J.A.; Bovenhuis, H. Genome-wide scan for bovine milk-fat composition. I. Quantitative trait loci for short- and medium-chain fatty acids. J. Dairy Sci. 2009, 92, 4664–4675. [Google Scholar] [CrossRef] [PubMed]
- Schennink, A.; Stoop, W.M.; Visker, M.H.; van der Poel, J.J.; Bovenhuis, H.; van Arendonk, J.A. Short communication: Genome-wide scan for bovine milk-fat composition. II. Quantitative trait loci for long-chain fatty acids. J. Dairy Sci. 2009, 92, 4676–4682. [Google Scholar] [CrossRef] [PubMed]
- Palombo, V.; Milanesi, M.; Sgorlon, S.; Capomaccio, S.; Mele, M.; Nicolazzi, E.; Ajmone-Marsan, P.; Pilla, F.; Stefanon, B.; D’Andrea, M. Genome-wide association study of milk fatty acid composition in Italian Simmental and Italian Holstein cows using single nucleotide polymorphism arrays. J. Dairy Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.X.; Zhang, S.L.; Wang, S.; Wu, X.P.; Zhang, Q.; Liu, L.; Li, Y.H.; Qiao, L. Genome Wide Association Study Identifies 20 Novel Promising Genes Associated with Milk Fatty Acid Traits in Chinese Holstein. PLoS ONE 2014, 9, e96186. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Buitenhuis, A.J.; Lund, M.S.; Li, C.; Sun, D.; Zhang, Q.; Poulsen, N.A.; Su, G. Joint genome-wide association study for milk fatty acid traits in Chinese and Danish Holstein populations. J. Dairy Sci. 2015, 98, 8152–8163. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, G.; Iannaccone, M.; Auzino, B.; Macciotta, N.P.P.; Kovitvadhi, A.; Nicolae, I.; Pauciullo, A. Remarkable genetic diversity detected at river buffalo prolactin receptor (PRLR) gene and association studies with milk fatty acid composition. Anim. Genet. 2018, 49, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.X.; Zhang, S.L.; Yang, S.H.; Alim, M.A.; Zhang, Q.; Li, Y.H.; Liu, L. Genetic effects of FASN, PPARGC1A, ABCG2 and IGF1 revealing the association with milk fatty acids in a Chinese Holstein cattle population based on a post genome-wide association study. BMC Genet. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sun, D.; Zhang, S.; Liu, L.; Alim, M.A.; Zhang, Q. A post-GWAS confirming the SCD gene associated with milk medium- and long-chain unsaturated fatty acids in Chinese Holstein population. Anim. Genet. 2016, 47, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.K.; Jang, W.H.; Ko, J.H.; Kang, J.S.; Noh, M.J.; Yoo, O.J. Lipase and its modulator from Pseudomonas sp strain KFCC 10818: Proline-to-glutamine substitution at position 112 induces formation of enzymatically active lipase in the absence of the modulator. J. Bacteriol. 2001, 183, 5937–5941. [Google Scholar] [CrossRef]
- Holmes, R.S.; Cox, L.A.; VandeBerg, J.L. Comparative studies of mammalian acid lipases: Evidence for a new gene family in mouse and rat (Lipo). Comp. Biochem. Physiol. Part D Genom. Proteom. 2010, 5, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, S.; Agarwal, P.; Krishnan, M.Y. Use of an adipocyte model to study the transcriptional adaptation of Mycobacterium tuberculosis to store and degrade host fat. Int. J. Mycobacteriol. 2016, 5, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Kelsey, J.A.; Corl, B.A.; Collier, R.J.; Bauman, D.E. The effect of breed, parity, and stage of lactation on conjugated linoleic acid (CLA) in milk fat from dairy cows. J. Dairy Sci. 2003, 86, 2588–2597. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Capitan, A.; Pausch, H.; Stothard, P.; Van Binsbergen, R.; Brondum, R.F.; Liao, X.P.; Djari, A.; Rodriguez, S.C.; Grohs, C.; et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat. Genet. 2014, 46, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics. Pearon, 1996; 4 dition. ISBN -10: 0582243025. [Google Scholar]
- Lengi, A.J.; Corl, B.A. Identification and characterization of a novel bovine stearoyl-CoA desaturase isoform with homology to human SCD5. Lipids 2007, 42, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Ulbricht, T.L.; Southgate, D.A. Coronary heart disease: Seven dietary factors. Lancet 1991, 338, 985–992. [Google Scholar] [CrossRef]
- Simopoulos, A.P. Omega-3 fatty acids in health and disease and in growth and development. Am. J. Clin. Nutr. 1991, 54, 438–463. [Google Scholar] [CrossRef] [PubMed]
- Skonieczna-Zydecka, K.; Grochans, E.; Maciejewska, D.; Szkup, M.; Schneider-Matyka, D.; Jurczak, A.; Loniewski, I.; Kaczmarczyk, M.; Marlicz, W.; Czerwinska-Rogowska, M.; et al. Faecal Short Chain Fatty Acids Profile is Changed in Polish Depressive Women. Nutrients 2018, 10. [Google Scholar] [CrossRef]
- Martinez, L.; Torres, S.; Baulies, A.; Alarcon-Vila, C.; Elena, M.; Fabrias, G.; Casas, J.; Caballeria, J.; Fernandez-Checa, J.C.; Garcia-Ruiz, C. Myristic acid potentiates palmitic acid-induced lipotoxicity and steatohepatitis associated with lipodystrophy by sustaning de novo ceramide synthesis. Oncotarget 2015, 6, 41479–41496. [Google Scholar] [CrossRef]
- Kwon, J.O.; Jin, W.J.; Kim, B.; Kim, H.H.; Lee, Z.H. Myristoleic acid inhibits osteoclast formation and bone resorption by suppressing the RANKL activation of Src and Pyk2. Eur. J. Pharmacol. 2015, 768, 189–198. [Google Scholar] [CrossRef]
- Li, Y.; Wang, C.; Huang, Y.; Fu, R.; Zheng, H.; Zhu, Y.; Shi, X.; Padakanti, P.K.; Tu, Z.; Su, X.; et al. Elegans Fatty Acid Two-Hydroxylase Regulates Intestinal Homeostasis by Affecting Heptadecenoic Acid Production. Cell Physiol. Biochem. 2018, 49, 947–960. [Google Scholar] [CrossRef]
- Medeiros-de-Moraes, I.M.; Goncalves-de-Albuquerque, C.F.; Kurz, A.R.M.; Oliveira, F.M.J.; de Abreu, V.H.P.; Torres, R.C.; Carvalho, V.F.; Estato, V.; Bozza, P.T.; Sperandio, M.; et al. Omega-9 Oleic Acid, the Main Compound of Olive Oil, Mitigates Inflammation during Experimental Sepsis. Oxid. Med. Cell Longev. 2018, 2018, 6053492. [Google Scholar] [CrossRef] [PubMed]
- Toulza, E.; Mattiuzzo, N.R.; Galliano, M.F.; Jonca, N.; Dossat, C.; Jacob, D.; de Daruvar, A.; Wincker, P.; Serre, G.; Guerrin, M. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. 2007, 8, R107. [Google Scholar] [CrossRef] [PubMed]
- Jordan-Sciutto, K.L.; Dragich, J.M.; Rhodes, J.L.; Bowser, R. Fetal Alz-50 clone 1, a novel zinc finger protein, binds a specific DNA sequence and acts as a transcriptional regulator. J. Biol. Chem. 1999, 274, 35262–35268. [Google Scholar] [CrossRef] [PubMed]
- Kalra, R.; Bhagyaraj, E.; Tiwari, D.; Nanduri, R.; Chacko, A.P.; Jain, M.; Mahajan, S.; Khatri, N.; Gupta, P. AIRE promotes androgen-independent prostate cancer by directly regulating IL-6 and modulating tumor microenvironment. Oncogenesis 2018, 7, 43. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Whitman, M. FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development 1999, 126, 5621–5634. [Google Scholar] [PubMed]
- Han, B.; Liang, W.; Liu, L.; Li, Y.; Sun, D. Determination of genetic effects of ATF3 and CDKN1A genes on milk yield and compositions in Chinese Holstein population. BMC Genet. 2017, 18, 47. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Liang, W.; Liu, L.; Li, Y.; Sun, D. Genetic association of the ACACB gene with milk yield and composition traits in dairy cattle. Anim. Genet. 2018, 49, 169–177. [Google Scholar] [CrossRef]
- Yang, S.; Gao, Y.; Zhang, S.; Zhang, Q.; Sun, D. Identification of Genetic Associations and Functional Polymorphisms of SAA1 Gene Affecting Milk Production Traits in Dairy Cattle. PLoS ONE 2016, 11, e0162195. [Google Scholar] [CrossRef]
- Alim, M.A.; Dong, T.; Xie, Y.; Wu, X.P.; Zhang, Y.; Zhang, S.; Sun, D.X. Effect of polymorphisms in the CSN3 (kappa-casein) gene on milk production traits in Chinese Holstein Cattle. Mol. Biol. Rep. 2014, 41, 7585–7593. [Google Scholar] [CrossRef]
- Alim, M.A.; Wang, P.; Wu, X.P.; Li, C.; Cui, X.G.; Zhang, S.L.; Zhang, Q.; Zhang, Y.; Sun, D.X. Effect of FASN gene on milk yield and milk composition in the Chinese Holstein dairy population. Anim. Genet. 2014, 45, 111–113. [Google Scholar] [CrossRef]
- Yang, S. Genetic Effects Analysis and Fucntinal Verification of TRIB3 and SAA Assocaited with Milk Production Traits in Chinese Holstein. Ph.D. Thesis, China Agricultural University, Beijing, China, 2015. [Google Scholar]
| Gene | SNP Name | Location | Position | GenBank no. | Significant Milk FAs | p-Interval | Allele | TFBS | Amino | Changes of Protein Secondary Structure | SIFT | PROVEAN | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (UMD 3.1) | Acid | Alpha Helix | Extended Strand | Beta Turn | Random Coil | ||||||||||
| LIPK | g.10428101G>A | 5′ UTR | Chr26: 10428101 | rs110322221 | C6:0, C8:0, C10:0, C14:0, C20:0, SFA, SFA/UFA, total index | <1.00 × 10−4~4.88 × 10−2 | G | ||||||||
| A | FAC1 | ||||||||||||||
| g.10449831C>A | Exon-11 | Chr26: 10449831 | rs42774527 | C6:0, C8:0, C10:0, C14:0, C17:0, C17:1, C18:1cis-9, C20:0, C17index, total index | <1.00 × 10−4~3.76 × 10−2 | C | Thr | 34.09% | 19.95% | 6.06% | 39.90% | 0.04 | -3.315 | ||
| A | Lys | 33.08% | 19.44% | 6.31% | 41.16% | ||||||||||
| LIPJ | g.10214117A>C | 5′ flanking region | Chr26: 10214117 | rs41606812 | C17:0, C17:1, C20:0 | <1.00 × 10−4~2.55 × 10−2 | A | ||||||||
| C | |||||||||||||||
| g.10217380C>A | 5′ UTR | Chr26: 10217380 | rs211373799 | C6:0, C8:0, C10:0, C14:0, C17:1, C20:0 | <1.00 × 10−4~3.41 × 10−2 | C | AIRE | ||||||||
| MTBF | |||||||||||||||
| A | FAST1 | ||||||||||||||
| g.10247997T>C | 3′ UTR | Chr26: 10247997 | rs42107056 | C6:0, C8:0, C14:0, C17:1, C14index, C16index, SFA, total index | 1.30 × 10−3~2.15 × 10−2 | T | |||||||||
| C | |||||||||||||||
| g.10250098C>T | 3′ flanking region | Chr26: 10250098 | rs42107122 | C6:0, C14:0, C14index, C16index, SFA, UFA, total index | 3.00 × 10−4~4.36 × 10−2 | C | |||||||||
| T | |||||||||||||||
| g.10250120A>G | 3′ flanking region | Chr26: 10250120 | ss158213049726 | C6:0, C10:0, C14:0, C17:1, C18:1cis-9, C14index, C16index, SFA, UFA, total index | <1.00 × 10−4~3.37 × 10−2 | A | |||||||||
| G | |||||||||||||||
| g.10251075G>T | 3′ flanking region | Chr26: 10251075 | rs209219656 | C6:0, C14:0, C17:1, C20:0, C14index, C16index | 1.00 × 10−3~3.68 × 10−2 | G | |||||||||
| T | |||||||||||||||
| g.10251111T>C | 3′ flanking region | Chr26: 10251111 | rs42107114 | C6:0, C8:0, C10:0, C17:1, C20:0, C14index, total index | <1.00 × 10−4~4.22 × 10−2 | T | |||||||||
| C | |||||||||||||||
| Haplotype Combination (No.) | C6:0 (%) | C8:0 (%) | C10:0 (%) | C11:0 (%) | C12:0 (%) | C13:0 (%) | C14:0 (%) | C14:1 (%) | C15:0 (%) | C16:0 (%) | C16:1 (%) | C17:0 (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1H1 (236–250) | 0.4915 ± 0.0138 A | 0.9582 ± 0.0120 Aa | 2.8777 ± 0.0356 Aa | 0.0607 ± 0.0028 | 3.0778 ± 0.0464 | 0.1002 ± 0.0035 | 10.3703 ± 0.0785 | 0.6266 ± 0.0209 | 0.9980 ± 0.0143 | 34.8257 ± 0.2024 | 1.2791 ± 0.0283 | 0.5652 ± 0.0036 |
| H1H2 (196–215) | 0.5079 ± 0.0143 A | 0.9214 ± 0.0126 Ab | 2.8373 ± 0.0368 A | 0.0607 ± 0.0029 | 3.0641 ± 0.0478 | 0.1020 ± 0.0036 | 10.2305 ± 0.0814 | 0.6608 ± 0.0219 | 0.9895 ± 0.0150 | 34.5606 ± 0.2108 | 1.3468 ± 0.0295 | 0.5725 ± 0.0038 |
| H1H3 (134–147) | 0.4066 ± 0.0163 B | 0.8758 ± 0.0139B | 2.6856 ± 0.0408 B | 0.0536 ± 0.0032 | 2.9704 ± 0.0526 | 0.0971 ± 0.0042 | 10.3702 ± 0.0897 | 0.6668 ± 0.0249 | 1.0024 ± 0.0170 | 34.9864 ± 0.2406 | 1.3309 ± 0.0333 | 0.5729 ± 0.0043 |
| H1H4 (84–91) | 0.4642 ± 0.0186 AB | 0.9134 ± 0.0159 ABb | 2.7806 ± 0.0463 | 0.0593 ± 0.0038 | 2.9892 ± 0.0611 | 0.0981 ± 0.0050 | 10.3107 ± 0.1029 | 0.6756 ± 0.0291 | 0.9988 ± 0.0200 | 34.7553 ± 0.2768 | 1.3364 ± 0.0384 | 0.5688 ± 0.0050 |
| H2H2 (94–99) | 0.5145 ± 0.0180 ACa | 0.9540 ± 0.0154 A | 2.7562 ± 0.0451 ABb | 0.0590 ± 0.0037 | 3.0134 ± 0.0584 | 0.0978 ± 0.0049 | 10.2163 ± 0.0996 | 0.6841 ± 0.0285 | 0.9919 ± 0.0192 | 34.7388 ± 0.2684 | 1.3455 ± 0.0376 | 0.5670 ± 0.0049 |
| H2H3 (52–56) | 0.5886 ± 0.0215 Cb | 0.9553 ± 0.0182A | 2.7167 ± 0.0526 ABb | 0.0551 ± 0.0044 | 2.9365 ± 0.0691 | 0.1012 ± 0.0059 | 10.1683 ± 0.1181 | 0.6606 ± 0.0340 | 0.9837 ± 0.0236 | 34.8336 ± 0.3231 | 1.3490 ± 0.0443 | 0.5638 ± 0.0059 |
| p | <1.00 × 10−4 ** | <1.00 × 10−4 ** | <1.00 × 10−4 ** | 2.50 × 10−1 | 8.57 × 10−2 | 9.06 × 10−1 | 1.74 × 10−1 | 2.91 × 10−1 | 9.65 × 10−1 | 6.11 × 10−1 | 1.75 × 10−1 | 2.64 × 10−1 |
| Haplotype combination (No.) | C17:1 (%) | C18:0 (%) | C18:1cis-9 (%) | C18index (%) | C20:0 (%) | C14index (%) | C16index (%) | C17index (%) | SFA (%) | UFA (%) | SFA/UFA (%) | Total index (%) |
| H1H1 (212–250) | 0.1877 ± 0.0028A | 14.0724 ± 0.1029 | 19.0132 ± 0.1380 | 57.1817 ± 0.3145 | 0.1671 ± 0.0020a | 5.7655 ± 0.1594 Aa | 3.5534 ± 0.0726 | 24.5170 ± 0.2489a | 68.2270 ± 0.1876 | 30.1878 ± 0.1709 | 2.3037 ± 0.0250 | 27.2671 ± 0.1630a |
| H1H2 (174–215) | 0.1955 ± 0.0029a | 14.0472 ± 0.1081 | 19.1074 ± 0.1447 | 57.2138 ± 0.3281 | 0.1737 ± 0.0021 Ab | 6.1702 ± 0.1694 | 3.7593 ± 0.0752 | 25.3334 ± 0.2578 b | 68.0475 ± 0.1961 | 30.3502 ± 0.1793 | 2.2650 ± 0.0260 | 27.5520 ± 0.1695 |
| H1H3 (119–148) | 0.1944 ± 0.0033 | 14.0278 ± 0.1234 | 18.9345 ± 0.1647 | 56.9697 ± 0.3745 | 0.1696 ± 0.0024 | 6.1980 ± 0.1838 | 3.6901 ± 0.0849 | 24.8310 ± 0.2861 | 68.0476 ± 0.2240 | 30.3144 ± 0.2052 | 2.2995 ± 0.0298 | 27.2367 ± 0.1857 a |
| H1H4 (79–91) | 0.1960 ± 0.0039 | 14.1424 ± 0.1452 | 19.4779 ± 0.1965 | 57.8644 ± 0.4411 | 0.1619 ± 0.0028 B | 6.3962 ± 0.2167 b | 3.7332 ± 0.0982 | 25.1307 ± 0.3321 | 67.5861 ± 0.2651 | 30.7677 ± 0.2399 | 2.2465 ± 0.0350 | 27.9861 ± 0.2200 b |
| H2H2 (86–99) | 0.2014 ± 0.0037 B | 13.8249 ± 0.1418 | 19.3393 ± 0.1880 | 57.8175 ± 0.4261 | 0.1680 ± 0.0027 | 6.5399 ± 0.2096 | 3.7366 ± 0.0958 | 25.2834 ± 0.3248 | 67.5006 ± 0.2558 | 30.8790 ± 0.2348 | 2.2421 ± 0.0337 | 27.9063 ± 0.2131 b |
| H2H3 (41–56) | 0.1822 ± 0.0045 Ab | 13.9019 ± 0.1738 | 19.0366 ± 0.2300 | 57.2853 ± 0.5135 | 0.1605 ± 0.0035 B | 6.1413 ± 0.2510 B | 3.7223 ± 0.1123 | 24.1729 ± 0.3870 a | 67.9823 ± 0.3101 | 30.4244 ± 0.2834 | 2.2738 ± 0.0414 | 27.5623 ± 0.2559 |
| p | 2.00 × 10−4 ** | 4.92 × 10−1 | 1.05 × 10−1 | 3.13 × 10−1 | <1.00 × 10−4 ** | 1.90 × 10−3 ** | 7.34 × 10−2 | 2.40 × 10−3 ** | 5.06 × 10−2 | 3.98 × 10−2 * | 3.32 × 10−1 | 9.00 × 10−4 ** |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, L.; Han, B.; Liu, L.; Lv, X.; Ma, Z.; Li, C.; Xu, L.; Li, Y.; Zhao, F.; Yang, Y.; et al. Determination of Genetic Effects of LIPK and LIPJ Genes on Milk Fatty Acids in Dairy Cattle. Genes 2019, 10, 86. https://doi.org/10.3390/genes10020086
Shi L, Han B, Liu L, Lv X, Ma Z, Li C, Xu L, Li Y, Zhao F, Yang Y, et al. Determination of Genetic Effects of LIPK and LIPJ Genes on Milk Fatty Acids in Dairy Cattle. Genes. 2019; 10(2):86. https://doi.org/10.3390/genes10020086
Chicago/Turabian StyleShi, Lijun, Bo Han, Lin Liu, Xiaoqing Lv, Zhu Ma, Cong Li, Lingna Xu, Yanhua Li, Feng Zhao, Yuze Yang, and et al. 2019. "Determination of Genetic Effects of LIPK and LIPJ Genes on Milk Fatty Acids in Dairy Cattle" Genes 10, no. 2: 86. https://doi.org/10.3390/genes10020086
APA StyleShi, L., Han, B., Liu, L., Lv, X., Ma, Z., Li, C., Xu, L., Li, Y., Zhao, F., Yang, Y., & Sun, D. (2019). Determination of Genetic Effects of LIPK and LIPJ Genes on Milk Fatty Acids in Dairy Cattle. Genes, 10(2), 86. https://doi.org/10.3390/genes10020086






