Automated Nuclear Cartography Reveals Conserved Sperm Chromosome Territory Localization across 2 Million Years of Mouse Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Fluorescence In-Situ Hybridisation (FISH)
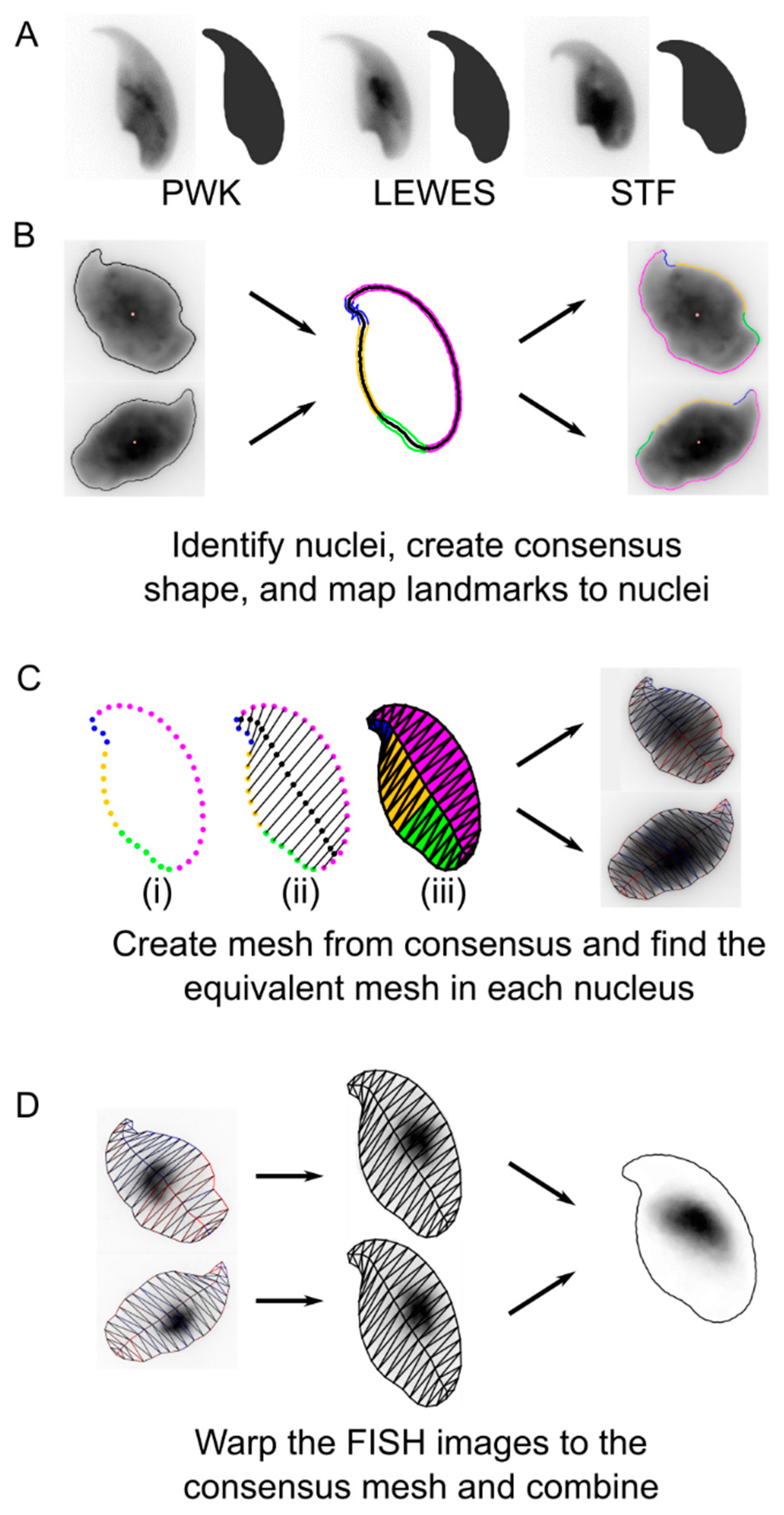
2.3. Image Analysis
3. Results
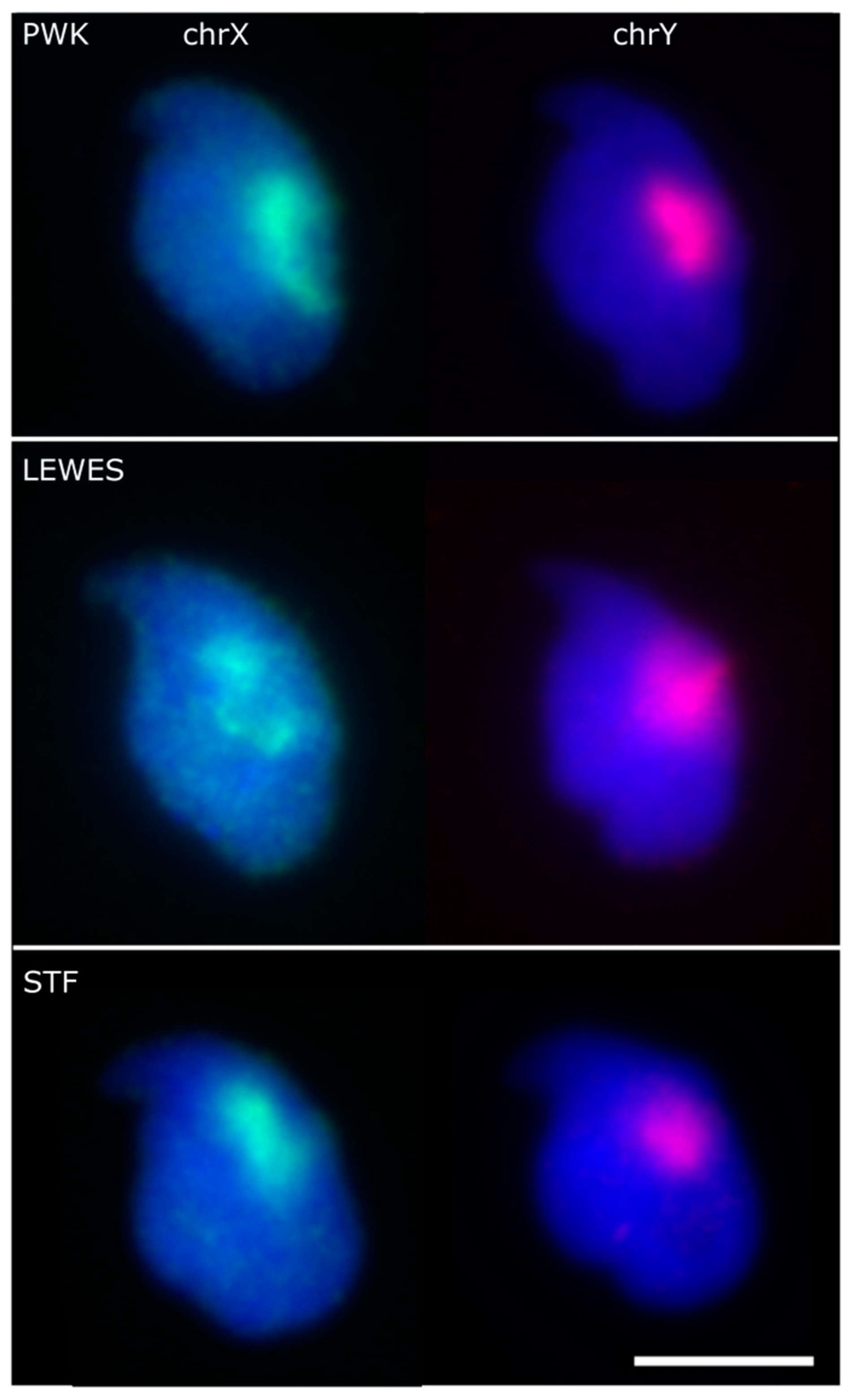
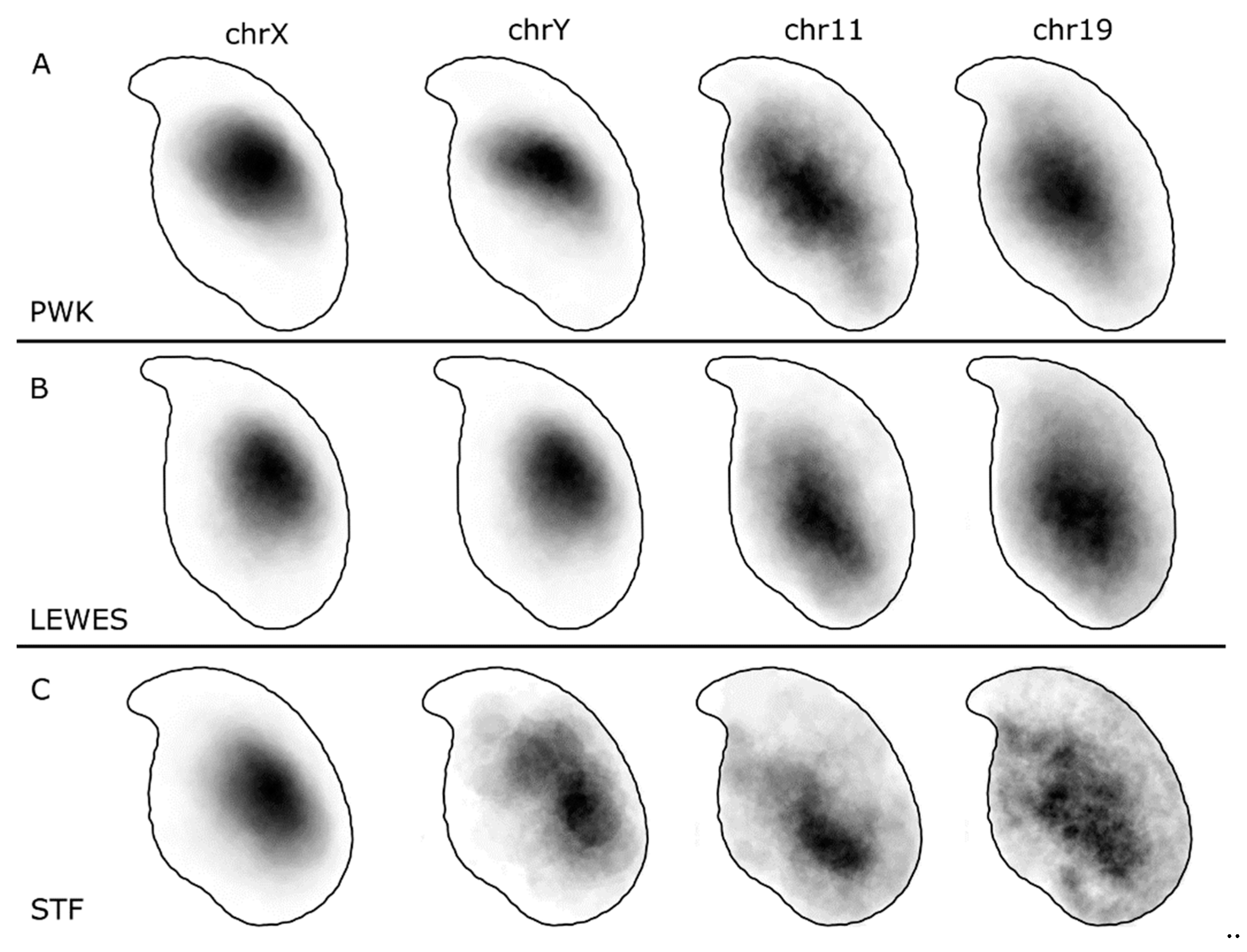
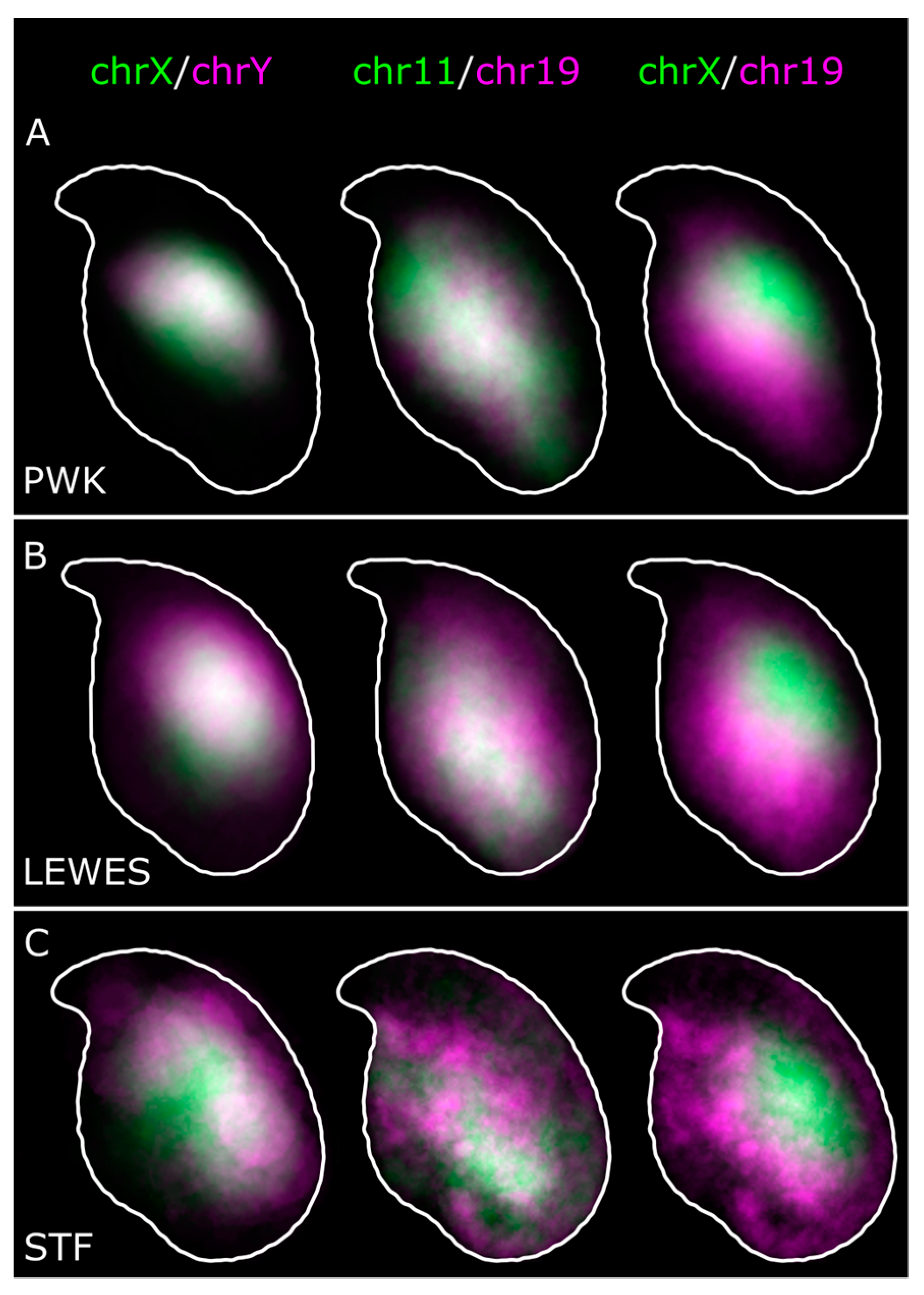
3.1. The Sex Chromosomes Have Conserved Position in Mouse Sperm Nuclei
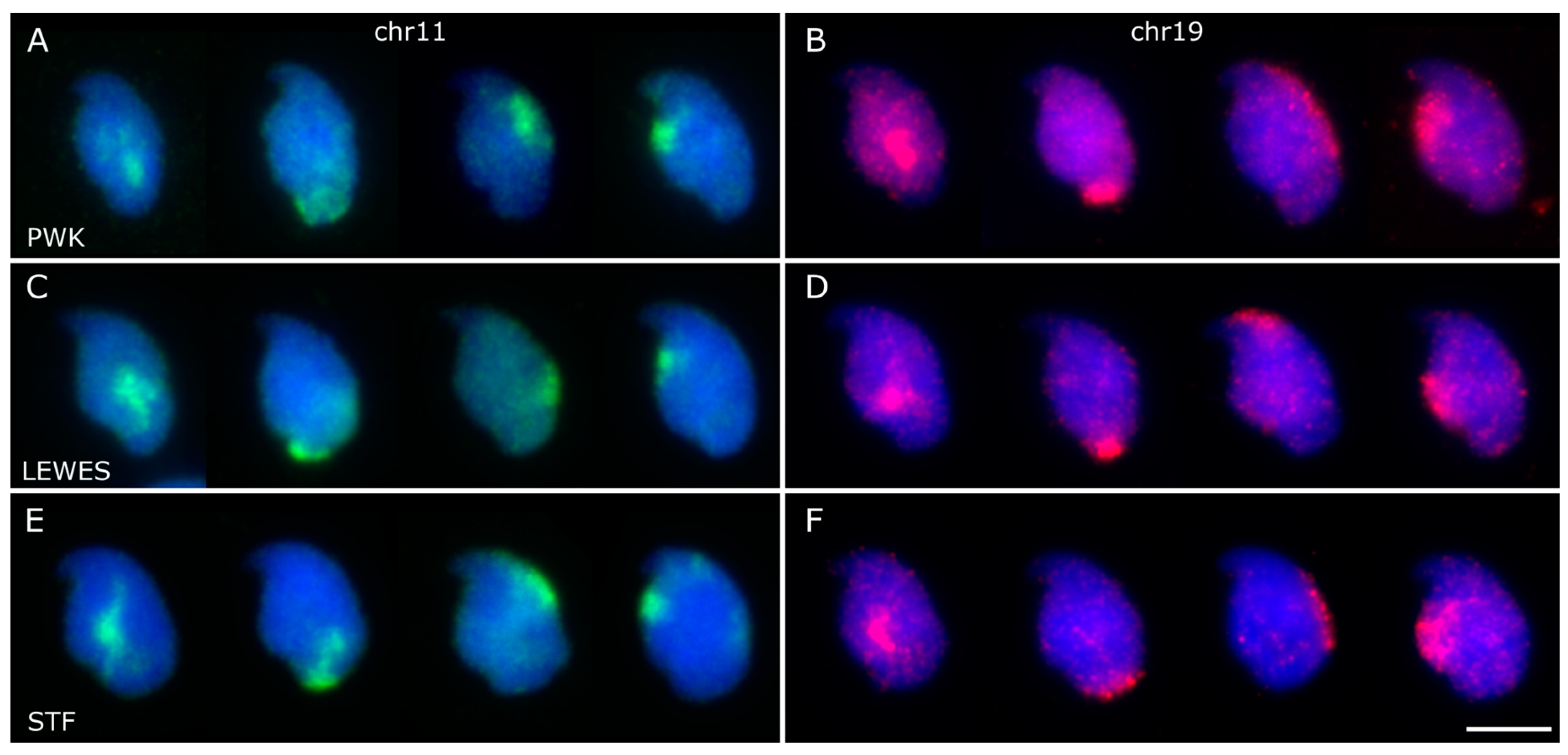
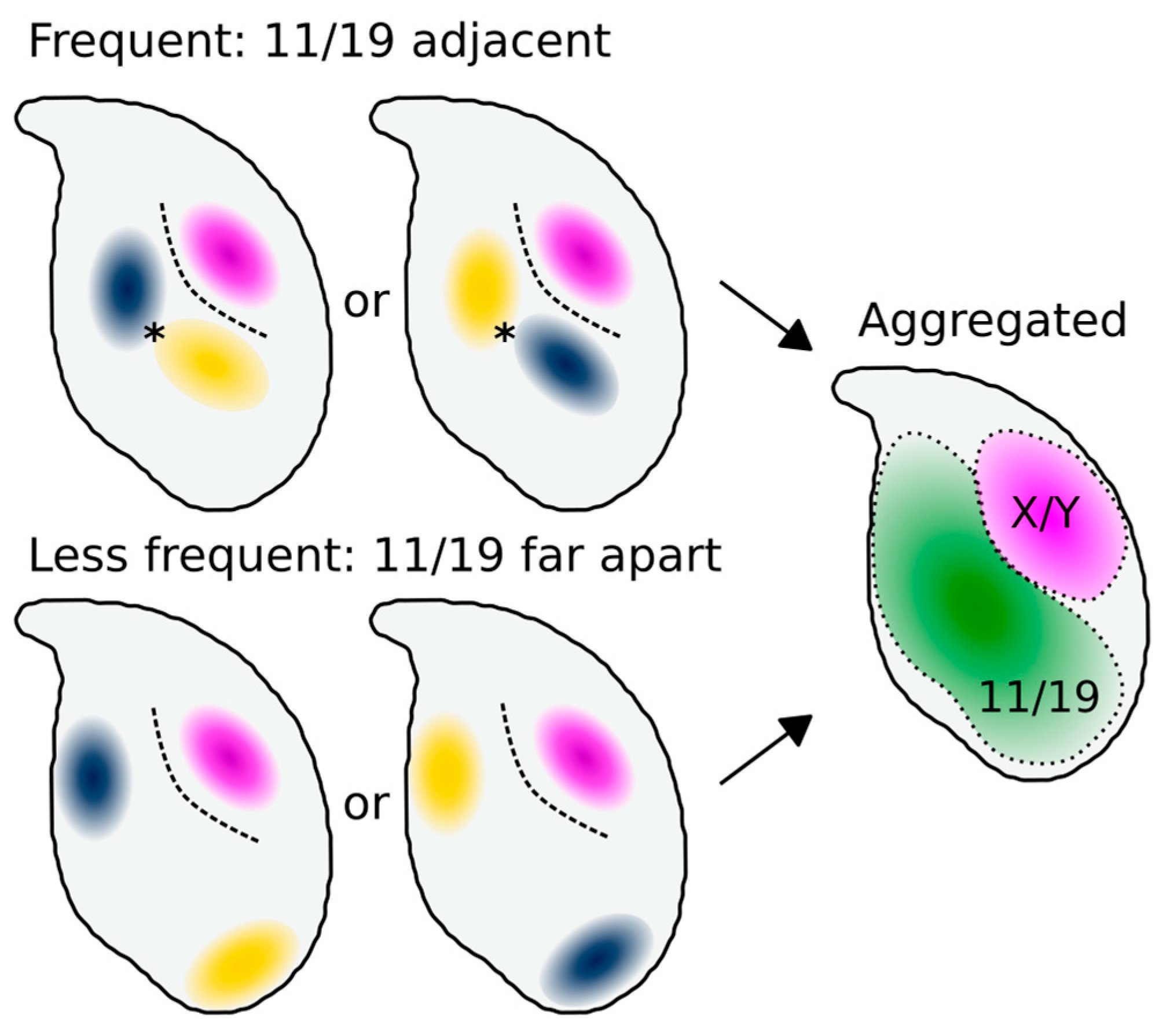
3.2. Chromosomes 11 and 19 Occupy Similar Nuclear Addresses
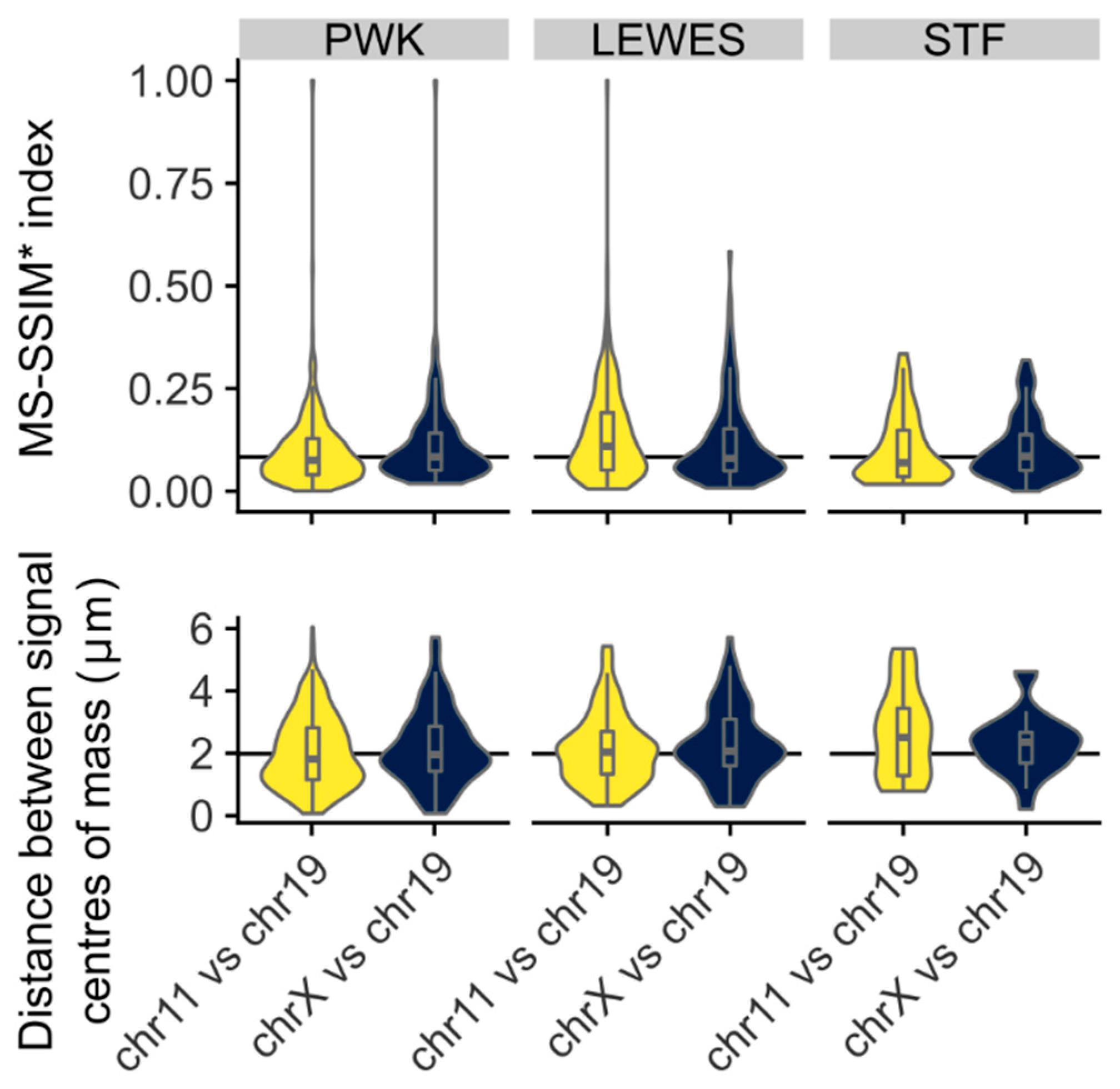
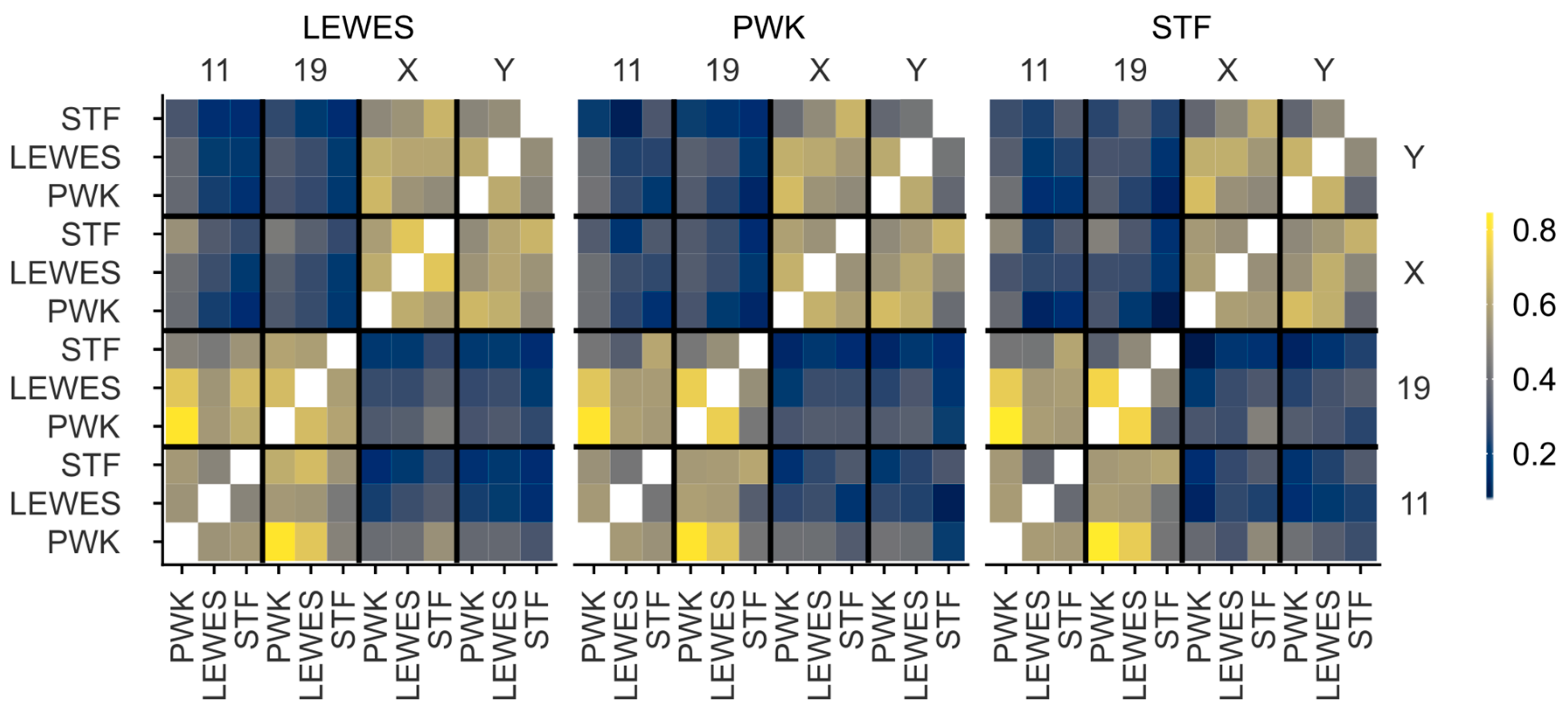
3.3. Quantification of Signal Positions Reveals Conserved Chromosome Organisation across Species
4. Discussion
4.1. Chromosomes X and Y Have a Conserved Dorsal/Sub-Acrosomal Position
4.2. Chromosomes 11 and 19 Have a Conserved Ventral/Basal Distribution
4.3. This Method Allows Rapid Screening of Large Numbers of Nuclei
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Skinner, B.M.; Volker, M.; Ellis, M.; Griffin, D.K. An appraisal of nuclear organisation in interphase embryonic fibroblasts of chicken, turkey and duck. Cytogenet. Genome Res. 2009, 126, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Foster, H.A.; Griffin, D.K.; Bridger, J.M. Interphase chromosome positioning in in vitro porcine cells and ex vivo porcine tissues. BMC Cell Biol. 2012, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, R.E.; Kiazim, L.; Skinner, B.; Fonseka, G.; Joseph, S.; Jennings, R.; Larkin, D.M.; Griffin, D.K. Patterns of microchromosome organization remain highly conserved throughout avian evolution. Chromosoma 2018. [Google Scholar] [CrossRef] [PubMed]
- Foster, H.A.; Abeydeera, L.R.; Griffin, D.K.; Bridger, J.M. Non-random chromosome positioning in mammalian sperm nuclei, with migration of the sex chromosomes during late spermatogenesis. J. Cell Sci. 2005, 118, 1811–1820. [Google Scholar] [CrossRef] [PubMed]
- Millan, N.M.; Lau, P.; Hann, M.; Ioannou, D.; Hoffman, D.; Barrionuevo, M.; Maxson, W.; Ory, S.; Tempest, H.G. Hierarchical radial and polar organisation of chromosomes in human sperm. Chromosome Res. 2012, 20, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, D.; Millan, N.M.; Jordan, E.; Tempest, H.G. A new model of sperm nuclear architecture following assessment of the organization of centromeres and telomeres in three-dimensions. Sci. Rep. 2017, 7, 41585. [Google Scholar] [CrossRef] [PubMed]
- Zalenskaya, I.A.; Zalensky, A.O. Non-random positioning of chromosomes in human sperm nuclei. Chromosome Res. 2004, 12, 163–173. [Google Scholar] [CrossRef]
- Kocer, A.; Henry-Berger, J.; Noblanc, A.; Champroux, A.; Pogorelcnik, R.; Guiton, R.; Janny, L.; Pons-Rejraji, H.; Saez, F.; Johnson, G.D.; et al. Oxidative DNA damage in mouse sperm chromosomes: Size matters. Free Radic. Biol. Med. 2015, 89, 993–1002. [Google Scholar] [CrossRef]
- Champroux, A.; Damon-Soubeyrand, C.; Goubely, C.; Bravard, S.; Henry-Berger, J.; Guiton, R.; Saez, F.; Drevet, J.; Kocer, A. Nuclear Integrity but not topology of mouse sperm chromosome is affected by oxidative DNA damage. Genes 2018, 9, 501. [Google Scholar] [CrossRef]
- Namekawa, S.H.; Park, P.J.; Zhang, L.-F.; Shima, J.E.; McCarrey, J.R.; Griswold, M.D.; Lee, J.T. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 2006, 16, 660–667. [Google Scholar] [CrossRef]
- Finch, K.A.; Fonseka, K.G.L.; Abogrein, A.; Ioannou, D.; Handyside, A.H.; Thornhill, A.R.; Hickson, N.; Griffin, D.K. Nuclear organization in human sperm: Preliminary evidence for altered sex chromosome centromere position in infertile males. Hum. Reprod. 2008, 23, 1263–1270. [Google Scholar] [CrossRef]
- Olszewska, M.; Wiland, E.; Kurpisz, M. Positioning of chromosome 15, 18, X and Y centromeres in sperm cells of fertile individuals and infertile patients with increased level of aneuploidy. Chromosome Res. 2008, 16, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, D.; Griffin, D.K. Male fertility, chromosome abnormalities, and nuclear organization. Cytogenet. Genome Res. 2010, 133, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Greaves, I.K.; Rens, W.; Ferguson-Smith, M.A.; Griffin, D.; Marshall Graves, J.A. Conservation of chromosome arrangement and position of the X in mammalian sperm suggests functional significance. Chromosome Res. 2003, 11, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Stevens, T.J.; Lando, D.; Basu, S.; Atkinson, L.P.; Cao, Y.; Lee, S.F.; Leeb, M.; Wohlfahrt, K.J.; Boucher, W.; O’Shaughnessy-Kirwan, A.; et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature 2017, 544, 59. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Lubling, Y.; Stevens, T.J.; Schoenfelder, S.; Yaffe, E.; Dean, W.; Laue, E.D.; Tanay, A.; Fraser, P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013, 502, 59. [Google Scholar] [CrossRef]
- Battulin, N.; Fishman, V.S.; Mazur, A.M.; Pomaznoy, M.; Khabarova, A.A.; Afonnikov, D.A.; Prokhortchouk, E.B.; Serov, O.L. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 2015, 16, 77. [Google Scholar] [CrossRef]
- Skinner, B.M.; Rathje, C.C.; Bacon, J.; Johnson, E.E.P.; Larson, E.L.; Kopania, E.E.K.; Good, J.M.; Yousafzai, G.; Affara, N.A.; Ellis, P.J.I. A high-throughput method for unbiased quantitation and categorisation of nuclear morphology. bioRxiv 2018, 312470. [Google Scholar] [CrossRef]
- Prieto, G.; Chevalier, M.; Guibelalde, E. MS_SSIM Index and MS_SSIM* Index as a Java Plugin for ImageJ; Department of Radiology, Faculty of Medicine, Universidad Complutense: Madrid, Spain, 2009. [Google Scholar]
- Rouse, D.M.; Hemami, S.S. Analyzing the role of visual structure in the recognition of natural image content with multi-scale SSIM. In Proceedings of the Human Vision and Electronic Imaging XIII, San Jose, CA, USA, 27–31 January 2008; International Society for Optics and Photonics: Bellingham, WA, USA, 2008; Volume 6806, p. 680615. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- R Core Team R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Nuñez, J.R.; Anderton, C.R.; Renslow, R.S. Optimizing colormaps with consideration for color vision deficiency to enable accurate interpretation of scientific data. PLoS ONE 2018, 13, e0199239. [Google Scholar] [CrossRef]
- Renieblas, G.P.; Nogués, A.T.; González, A.M.; Gómez-Leon, N.; del Castillo, E.G. Structural similarity index family for image quality assessment in radiological images. J. Med. Imaging 2017, 4, 035501. [Google Scholar] [CrossRef] [PubMed]
- Fabre, P.-H.; Hautier, L.; Dimitrov, D.; Douzery, E.J. A glimpse on the pattern of rodent diversification: A phylogenetic approach. BMC Evol. Biol. 2012, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Chevret, P.; Veyrunes, F.; Britton-Davidian, J. Molecular phylogeny of the genus Mus (Rodentia: Murinae) based on mitochondrial and nuclear data. Biol. J. Linn. Soc. 2005, 84, 417–427. [Google Scholar] [CrossRef]
- Silver, L.M. Mouse Genetics: Concepts and Applications; Oxford University Press: Oxford, NY, USA, 1995; ISBN 978-0-19-507554-0. [Google Scholar]
- Palomo, L.J.; Justo, E.R.; Vargas, J.M. Mus spretus (Rodentia: Muridae). Mamm. Species 2009, 840, 1–10. [Google Scholar] [CrossRef]
- Soh, Y.Q.S.; Alföldi, J.; Pyntikova, T.; Brown, L.G.; Graves, T.; Minx, P.J.; Fulton, R.S.; Kremitzki, C.; Koutseva, N.; Mueller, J.L.; et al. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 2014, 159, 800–813. [Google Scholar] [CrossRef] [PubMed]
- Ellis, P.J.I.; Bacon, J.; Affara, N.A. Association of Sly with sex-linked gene amplification during mouse evolution: A side effect of genomic conflict in spermatids? Hum. Mol. Genet. 2011, 20, 3010–3021. [Google Scholar]
- Rugarli, E.I.; Adler, D.A.; Borsani, G.; Tsuchiya, K.; Franco, B.; Hauge, X.; Disteche, C.; Chapman, V.; Ballabio, A. Different chromosomal localization of the Clcn4 gene in Mus spretus and C57BL/6J mice. Nat. Genet. 1995, 10, 466–471. [Google Scholar] [CrossRef]
- Nguyen, D.K.; Yang, F.; Kaul, R.; Alkan, C.; Antonellis, A.; Friery, K.F.; Zhu, B.; de Jong, P.J.; Disteche, C.M. Clcn4-2 genomic structure differs between the X locus in Mus spretus and the autosomal locus in Mus musculus: AT motif enrichment on the X. Genome Res. 2011, 21, 402–409. [Google Scholar] [CrossRef]
- Tsend-Ayush, E.; Dodge, N.; Mohr, J.; Casey, A.; Himmelbauer, H.; Kremitzki, C.L.; Schatzkamer, K.; Graves, T.; Warren, W.C.; Grützner, F. Higher-order genome organization in platypus and chicken sperm and repositioning of sex chromosomes during mammalian evolution. Chromosoma 2009, 118, 53–69. [Google Scholar] [CrossRef]
- Veron, A.; Lemaitre, C.; Gautier, C.; Lacroix, V.; Sagot, M.-F. Close 3D proximity of evolutionary breakpoints argues for the notion of spatial synteny. BMC Genom. 2011, 12, 303. [Google Scholar] [CrossRef]
- Dai, Z.; Xiong, Y.; Dai, X. Neighboring genes show interchromosomal colocalization after their separation. Mol. Biol. Evol. 2014, 31, 1166–1172. [Google Scholar] [CrossRef]
- Kishikawa, H.; Tateno, H.; Yanagimachi, R. Chromosome analysis of BALB/c mouse spermatozoa with normal and abnormal head morphology. Biol. Reprod. 1999, 61, 809–812. [Google Scholar] [CrossRef] [PubMed]
- Cassuto, N.G.; Le Foll, N.; Chantot-Bastaraud, S.; Balet, R.; Bouret, D.; Rouen, A.; Bhouri, R.; Hyon, C.; Siffroi, J.P. Sperm fluorescence in situ hybridization study in nine men carrying a Robertsonian or a reciprocal translocation: Relationship between segregation modes and high-magnification sperm morphology examination. Fertil. Steril. 2011, 96, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Acloque, H.; Bonnet-Garnier, A.; Mompart, F.; Pinton, A.; Yerle-Bouissou, M. Sperm Nuclear architecture is locally modified in presence of a Robertsonian Translocation t(13;17). PLoS ONE 2013, 8, e78005. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Ficca, M.; Muller-Navia, J.; Scherthan, H. Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a well defined genome architecture in the sperm nucleus. J. Cell Sci. 1998, 111, 1363–1370. [Google Scholar] [PubMed]
- Choi, G.P.T.; Mahadevan, L. Planar morphometrics using Teichmüller maps. Proc. R. Soc. A Math. Phys. Eng. Sci. 2018, 474, 20170905. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skinner, B.M.; Bacon, J.; Rathje, C.C.; Larson, E.L.; Kopania, E.E.K.; Good, J.M.; Affara, N.A.; Ellis, P.J.I. Automated Nuclear Cartography Reveals Conserved Sperm Chromosome Territory Localization across 2 Million Years of Mouse Evolution. Genes 2019, 10, 109. https://doi.org/10.3390/genes10020109
Skinner BM, Bacon J, Rathje CC, Larson EL, Kopania EEK, Good JM, Affara NA, Ellis PJI. Automated Nuclear Cartography Reveals Conserved Sperm Chromosome Territory Localization across 2 Million Years of Mouse Evolution. Genes. 2019; 10(2):109. https://doi.org/10.3390/genes10020109
Chicago/Turabian StyleSkinner, Benjamin Matthew, Joanne Bacon, Claudia Cattoni Rathje, Erica Lee Larson, Emily Emiko Konishi Kopania, Jeffrey Martin Good, Nabeel Ahmed Affara, and Peter James Ivor Ellis. 2019. "Automated Nuclear Cartography Reveals Conserved Sperm Chromosome Territory Localization across 2 Million Years of Mouse Evolution" Genes 10, no. 2: 109. https://doi.org/10.3390/genes10020109
APA StyleSkinner, B. M., Bacon, J., Rathje, C. C., Larson, E. L., Kopania, E. E. K., Good, J. M., Affara, N. A., & Ellis, P. J. I. (2019). Automated Nuclear Cartography Reveals Conserved Sperm Chromosome Territory Localization across 2 Million Years of Mouse Evolution. Genes, 10(2), 109. https://doi.org/10.3390/genes10020109





