ZCMM: A Novel Method Using Z-Curve Theory- Based and Position Weight Matrix for Predicting Nucleosome Positioning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Benchmark Datasets for Nucleosomal and Linker Sequences
2.2. Construction of the ZCMM Model for Nucleosomal and Linker Sequences
2.3. ZCMM Method for Identifying Nucleosome Positioning by SVM
2.4. A Set of Metrics for Quantitative Analysis
3. Results
3.1. ZCMM Model Construction and Statistical Analysis
3.2. ZCMM Model Compared with Z-Curve Model and PWM Model
3.3. ZCMM Prediction Results in Different Species
3.4. Performance Compared with Other Methods
3.5. Performance Validation Using Independent Data for S. cerevisiae
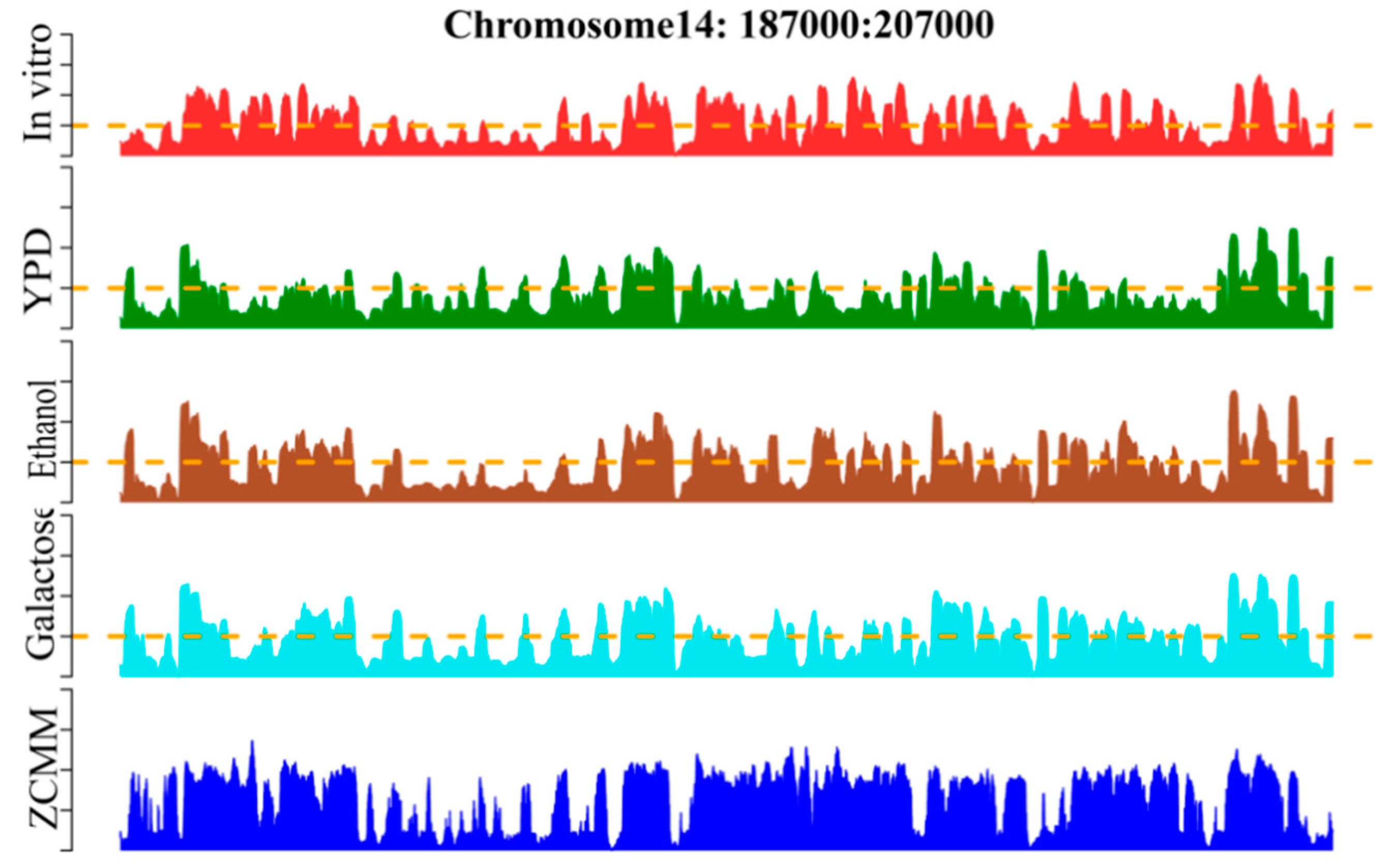
3.6. Occupancy Profile of Predicting Nucleosomal Sequences
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Richmond, T.J.; Davey, C.A. The structure of DNA in the nucleosome core. Nature 2003, 423, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.; Fondufe-Mittendorf, Y.; Chen, L.; Thåström, A.; Field, Y.; Moore, I.K.; Wang, J.P.; Widom, J. A genomic code for nucleosome positioning. Nature 2006, 442, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, R.D.; Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 1999, 98, 285–294. [Google Scholar] [CrossRef]
- Cai, Z.; Heydari, M.; Lin, G. Clustering Binary Oligonucleotide Fingerprint Vectors for DNA Clone Classification Analysis. J. Comb. Optim. 2005, 9, 199–211. [Google Scholar] [CrossRef]
- Taberlay, P.C.; Statham, A.L.; Kelly, T.K.; Clark, S.J.; Jones, P.A. Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome Res. 2014, 24, 1421–1432. [Google Scholar] [CrossRef] [Green Version]
- Struh, K.; Segal, E. Determinants of nucleosome positioning. Nature 2013, 20, 267–273. [Google Scholar]
- Albert, I.; Mavrich, T.N.; Tomsho, L.P.; Qi, J.; Zanton, S.J.; Schuster, S.C.; Pugh, B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 2007, 446, 572–576. [Google Scholar] [CrossRef]
- Yuan, G.C.; Liu, J.S. Genomic sequences is highly predictive of local nucleosome depletion. PLoS Comput. Biol. 2008, 29, 1081–1088. [Google Scholar] [CrossRef]
- González, S.; García, A.; Vázquez, E.; Serrano, R.; Sánchez, M.; Quintales, L.; Antequera, F. Nucleosomal signatures impose nucleosome positioning in coding and noncoding sequences in the genome. Genome Res. 2016, 26, 1532–1543. [Google Scholar] [CrossRef] [Green Version]
- Buckwalter, J.M.; Norouzi, D.; Harutyunyan, A.; Zhurkin, V.B.; Grigoryev, S.A. Regulation of chromatin folding by conformational variations of nucleosome linker DNA. Nucleic Acids Res. 2017, 45, 9372–9387. [Google Scholar] [CrossRef]
- Farman, F.U.; Iqbal, M.; Azam, M.; Saeed, M. Nucleosomes positioning around transcriptional start site of tumor suppressor (Rbl2/p130) gene in breast cancer. Mol. Biol. Rep. 2018, 45, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Murugan, R. Theory of site-specific DNA-protein interactions in the presence of nucleosome roadblocks. Biophys. J. 2018, 114, 2516. [Google Scholar] [CrossRef] [PubMed]
- Nocetti, N.; Whitehouse, I. Nucleosome repositioning underlies dynamic gene expression. Genes Dev. 2016, 30, 660–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, W.K.M.; Pugh, B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nature 2017, 18, 548–562. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Pugh, B.F. Nucleosome positioning and gene regulation: Advances through genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Morozov, A.V. Gene regulation by nucleosome positioning. Cell 2010, 26, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Eaton, M.L.; Kyriaki, G.; Sukhyun, K.; Bell, S.P.; Macalpine, D.M. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010, 24, 748–753. [Google Scholar] [CrossRef] [Green Version]
- Ying, H.; Epps, J.; Williams, R.; Huttley, G. Evidence that localized variation in primate sequence divergence Arises from an influence of nucleosome placement on DNA repair. Mol. Biol. Evol. 2010, 27, 637–649. [Google Scholar] [CrossRef]
- Bevington, S.; Boyes, J. Transcription-coupled eviction of histones H2a/H2b governs V(D)J recombination. EMBO J. 2013, 32, 1381–1392. [Google Scholar] [CrossRef]
- Sabantsev, A.; Levendosky, R.F.; Zhuang, X.; Bowman, G.D.; Deindl, S. Direct observation of coordinated DNA movements on the nucleosome during chromatin remodelling. Nat. Commun. 2019, 10, 1720. [Google Scholar] [CrossRef]
- Lieleg, C.; Krietenstein, N.; Walker, M.; Korber, P. Nucleosome positioning in yeasts: Methods, maps, and mechanisms. Chromosoma. 2015, 124, 131–151. [Google Scholar] [CrossRef]
- Beh, L.Y.; Müller, M.M.; Muir, T.W.; Kaplan, N.; Landweber, L.F. DNA-guided establishment of nucleosome patterns within coding regions of a eukaryotic genome. Genome Res. 2015, 25, 1727–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, H.H.; Meyer, C.A.; Hu, S.S.; Chen, M.W.; Zang, C.; Liu, Y.; Rao, P.K.; Fei, T.; Xu, H.; Long, H.; et al. RefinedDNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods 2013, 11, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Luo, K.; Winter, P.S.; Crawford, G.E.; Iversen, E.S.; Hartemink, A.J. Mapping nucleosome positions using Dnase-seq. Genome Res. 2016, 26, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Bauden, M.; Pamart, D.; Ansari, D.; Herzog, M.; Eccleston, M.; Micallef, J.; Andersson, B.; Andersson, R. Circulating nucleosomes as epigenetic biomarkers in pancreatic cancer. Clin. Epigenet. 2015, 7, 106. [Google Scholar] [CrossRef]
- Rodriguez, J.; Lee, L.; Lynch, B.; Tsukiyama, T. Nucleosome occupancy as a novel chromatin parameter for replication origin functions. Genome Res. 2017, 27, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cai, Z.; Wan, X.F.; Hoang, T.; Goebel, R.; Lin, G. Nucleotide Composition String Selection in HIV-1 Subtyping Using Whole Genomes. Bioinformatics 2007, 23, 1744–1752. [Google Scholar] [CrossRef]
- Lin, G.; Cai, Z.; Wu, J.; Wan, X.F.; Xu, L.; Goebel, R. Identifying a Few foot-and-mouth Disease Virus Signature Nucleotide Strings for Computational Genotyping. BMC Bioinform. 2008, 9, 279. [Google Scholar] [CrossRef]
- Zhang, J.; Peng, W.; Wang, L. LeNup: Learning Nucleosome Positioning from DNA Sequences with Improved Convolutional Neural Networks. Bioinformatics 2018, 34, 1–8. [Google Scholar] [CrossRef]
- Yang, K.; Cai, Z.; Li, J.; Lin, G. A Stable Gene Selection in Microarray Data Analysis. BMC Bioinform. 2006, 7, 228. [Google Scholar]
- Cai, Z.; Xu, L.; Shi, Y.; Salavatipour, M.R.; Goebel, R.; Lin, G. Using Gene Clustering to Identify Discriminatory Genes with Higher Classification Accuracy. In Proceedings of the IEEE 6th Symposium on Bioinformatics and Bioengineering (BIBE 2006), Arlington, VA, USA, 16–18 October 2006. [Google Scholar]
- Cai, Z.; Goebel, R.; Salavatipour, M.R.; Lin, G. Selecting dissimilar genes for multi-class classification, an application in cancer subtyping. BMC Bioinform. 2007, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Sabaa, H.; Wang, Y.; Goebel, R.; Wang, Z.; Xu, J.; Stothard, P.; Lin, G. Most Parsimonious Haplotype Allele Sharing Determination. BMC Bioinform. 2009, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Allu, P.K.; Dawicki-McKenna, J.M.; VanEeuwen, T.; Slavin, M.; Braitbard, M.; Xu, C.; Kalisman, N.; Murakami, K.; Black, B.E. Structure of the Human Core Centromeric Nucleosome Complex. Curr. Biol. 2019, 29, 2625–2639. [Google Scholar] [CrossRef]
- Chereji, R.V.; Ramachandran, S.; Bryson, T.D.; Henikoff, S. Precise genome-wide mapping of single nucleosomes and linkers in Vivo. Genome Biol. 2018, 19, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.H.; Deng, E.Z.; Xu, L.Q.; Ding, H.; Lin, H.; Chen, W.; Chou, K.C. iNuc-PseKNC: A sequence-based predictor for predicting nucleosome positioning in genomes with pseudo k-tuple nucleotide composition. Bioinformatics 2014, 30, 1522–1529. [Google Scholar] [CrossRef] [PubMed]
- Teif, V.B. Nucleosome positioning: Resources and tools online. Brief. Bioinform. 2016, 17, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Lin, H.; Feng, P.M.; Ding, C.; Zuo, Y.C.; Chou, K.C. iNuc-PhysChem: A Sequence-Based Predictor for Identifying Nucleosomes via Physicochemical Properties. PLoS ONE 2012, 7, e47843. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, B.A.; Alshammari, T.H.; Felton, N.L.; Zhurkin, V.B.; Cui, F. nuMap: A web platform for accurate prediction of nucleosome positioning. Genom. Proteom. Bioinform. 2014, 12, 249–253. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, C.T. Z curves, an intutive tool for visualizing and analyzing the DNA sequences. J. Biomol. Struct. Dyn. 1994, 11, 767–782. [Google Scholar] [CrossRef]
- Lee, W.; Tillo, D.; Bray, N.; Morse, R.H.; Davis, R.W.; Hughes, T.R.; Nislow, C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007, 39, 1235–1244. [Google Scholar] [CrossRef]
- Chen, W.; Feng, P.; Ding, H.; Lin, H.; Chou, K.C. Using deformation energy to analyze nucleosome positioning in genomes. Genomics 2016, 107, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Schones, D.E.; Cui, K.; Cuddapah, S.; Roh, T.Y.; Barski, A.; Wang, Z.; Wei, G.; Zhao, K. Dynamic regulation of nucleosome positioning in the human genome. Cell 2008, 132, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Mavrich, T.N.; Ioshikhes, I.P.; Venters, B.J.; Jiang, C.; Tomsho, L.P.; Qi, J.; Schuster, S.C.; Albert, I.; Pugh, B.F. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008, 18, 1073–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavrich, T.N.; Jiang, C.; Ioshikhes, I.P.; Li, X.; Venters, B.J.; Zanton, S.J.; Tomsho, L.P.; Qi, J.; Glaser, R.L.; Schuster, S.C.; et al. Nucleosome organization in the Drosophila genome. Nature 2008, 453, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M.; Hayat, M. iNuc-STNC: A sequence-based predictor for identification of nucleosome positioning in genomes by extending the concept of SAAC and Chou’s PseAAC. Mol. BioSyst. 2016, 12, 2587–2593. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Pei, Z.; Liu, J.; Qin, S.; Cai, L. Prediction of nucleosome DNA formation potential and nucleosome positioning using increment of diversity combined with quadratic discriminant analysis. Chromosome Res. 2010, 18, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhang, C.T. A Brief Review: The Z-curve Theory and its Application in Genome Analysis. Curr. Genom. 2014, 15, 78–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.Q.; Liu, G.Q.; Zhao, X.J.; Cai, L. An analysis and prediction of nucleosome positioning based on information content. Chromosome Res. 2013, 21, 63–74. [Google Scholar] [CrossRef]
- Wu, X.; Liu, H.; Liu, H.; Su, J.; Lv, J.; Cui, Y.; Wang, F.; Zhang, Y. Z curve theory-based analysis of the dynamic nature of nucleosome positioning in Saccharomyces cerevisiae. Gene 2013, 530, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.C.; Liu, Y.J.; Dion, M.F.; Slack, M.D.; Wu, L.F.; Altschuler, S.J.; Rando, O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 2005, 309, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Ioshikhes, I.; Bolshoy, A.; Derenshteyn, K.; Borodovsky, M.; Trifonov, E.N. Nucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequences. J. Mol. Biol. 1996, 262, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, N.; Moore, I.K.; Fondufe-Mittendorf, Y.; Gossett, A.J.; Tillo, D.; Field, Y.; LeProust, E.M.; Hughes, T.R.; Lieb, J.D.; Widom, J.; et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 2009, 458, 362–366. [Google Scholar] [CrossRef] [PubMed]
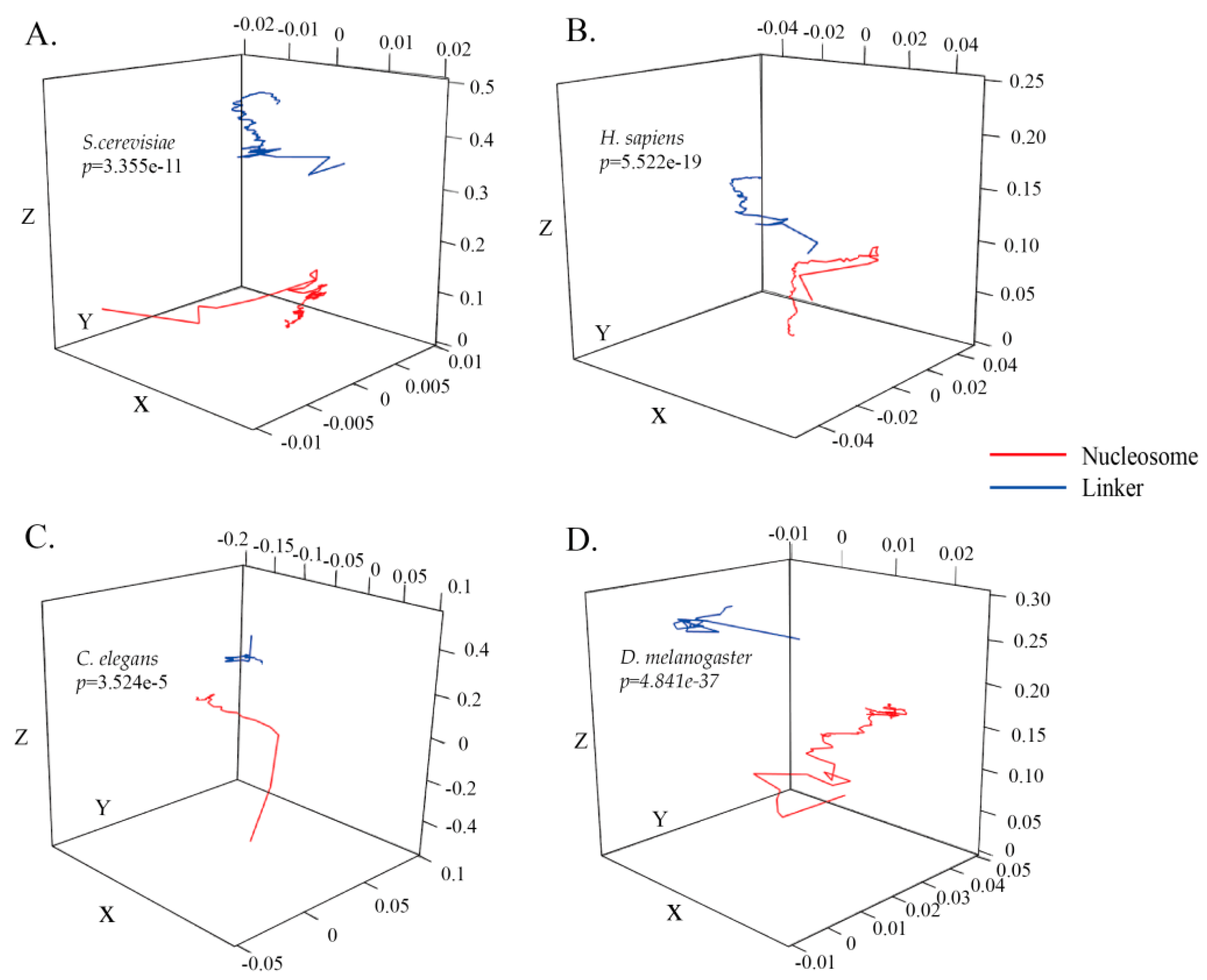
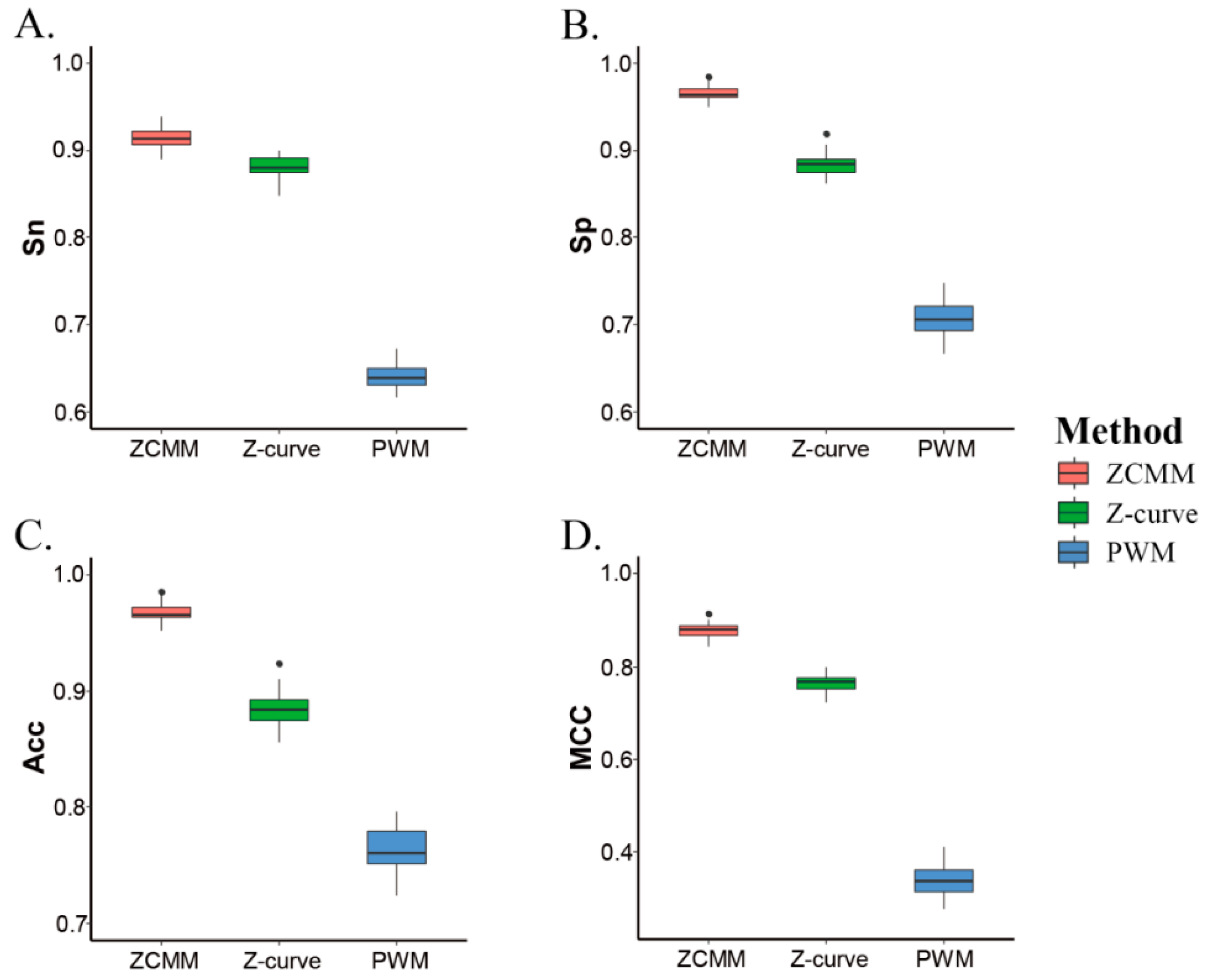
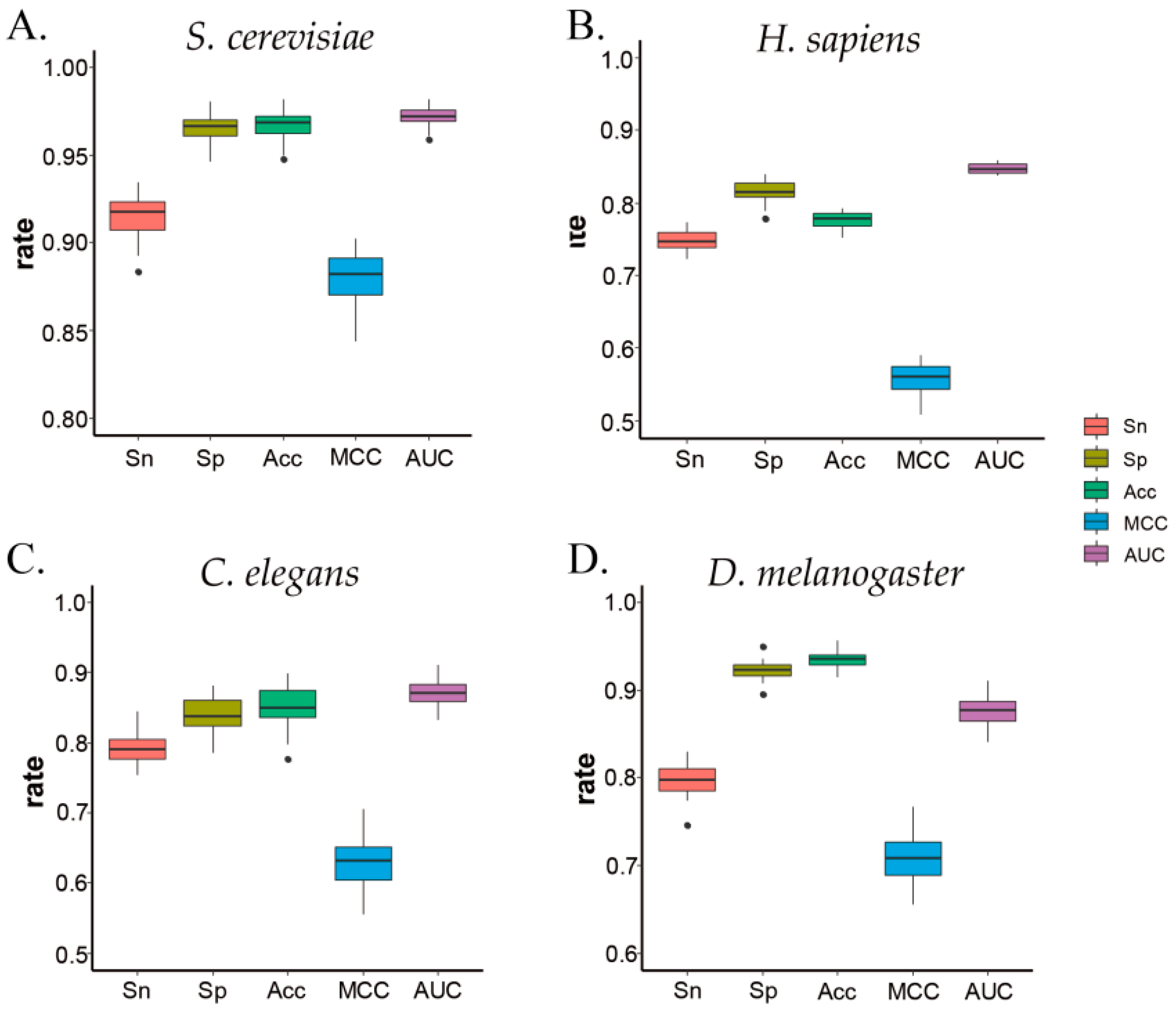
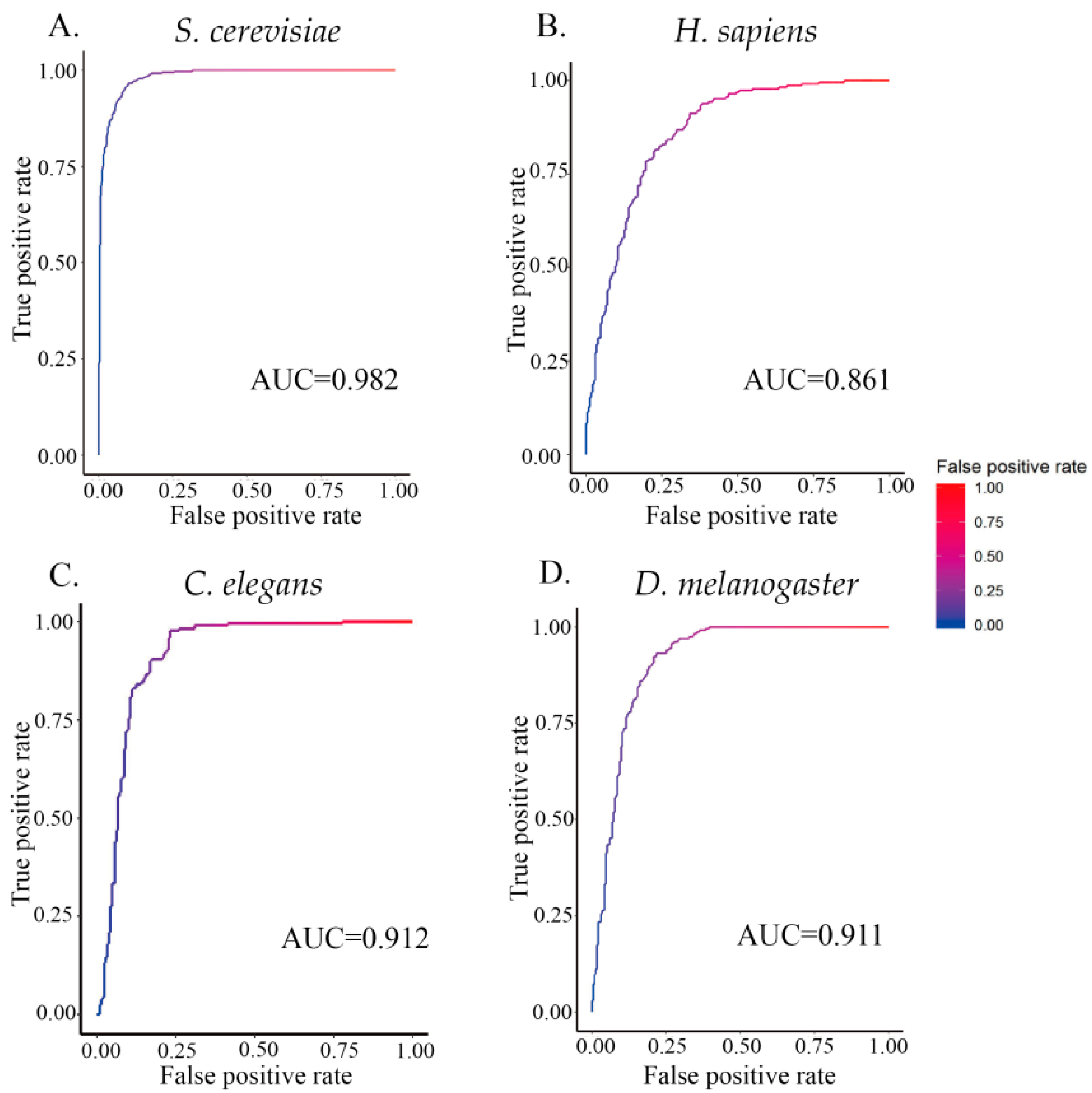
| Species | Sn% | Sp% | Acc% | MCC |
|---|---|---|---|---|
| ZCMM | 91.40 | 96.56 | 96.75 | 0.88 |
| Z-curve | 88.15 | 88.40 | 88.42 | 0.77 |
| PWM | 63.97 | 70.67 | 76.32 | 0.34 |
| Species | Sn% | Sp% | Acc% | MCC | AUC |
|---|---|---|---|---|---|
| S. cerevisiae (S1) | 91.40 | 96.56 | 96.75 | 0.88 | 0.972 |
| H. sapiens | 74.87 | 81.51 | 77.72 | 0.56 | 0.851 |
| C. elegans | 78.80 | 84.10 | 85.34 | 0.62 | 0.872 |
| D. melanogaster | 79.64 | 92.26 | 93.62 | 0.70 | 0.877 |
| Species | Method | Sn% | Sp% | Acc% | MCC | AUC |
|---|---|---|---|---|---|---|
| H. sapiens | ZCMM | 74.87 | 81.51 | 77.72 | 0.56 | 0.861 |
| Wu’s method | 65.82 | 64.12 | 64.74 | 0.30 | ||
| iNuc-PseKNC | 86.27 | 87.86 | 84.70 | 0.73 | 0.925 | |
| C. elegans | ZCMM | 78.80 | 84.10 | 85.34 | 0.62 | 0.912 |
| Wu’s method | 89.90 | 71.64 | 76.05 | 0.62 | ||
| iNuc-PseKNC | 86.90 | 90.30 | 83.55 | 0.74 | 0.935 | |
| D. melanogaster | ZCMM | 79.64 | 92.26 | 93.62 | 0.70 | 0.911 |
| Wu’s method | 79.17 | 41.07 | 57.34 | 0.22 | ||
| iNuc-PseKNC | 79.97 | 78.31 | 81.65 | 0.60 | 0.874 |
| Dataset | Method | Sn% | Sp% | Acc% | MCC |
|---|---|---|---|---|---|
| S1 | ZCMM | 91.40 | 96.56 | 96.75 | 0.88 |
| Wu’s method | 85.73 | 86.62 | 86.52 | 0.72 | |
| S2 | ZCMM | 96.72 | 96.54 | 94.10 | 0.88 |
| Wu’s method | 86.20 | 84.89 | 85.57 | 0.71 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Xu, Z.; Li, J. ZCMM: A Novel Method Using Z-Curve Theory- Based and Position Weight Matrix for Predicting Nucleosome Positioning. Genes 2019, 10, 765. https://doi.org/10.3390/genes10100765
Cui Y, Xu Z, Li J. ZCMM: A Novel Method Using Z-Curve Theory- Based and Position Weight Matrix for Predicting Nucleosome Positioning. Genes. 2019; 10(10):765. https://doi.org/10.3390/genes10100765
Chicago/Turabian StyleCui, Ying, Zelong Xu, and Jianzhong Li. 2019. "ZCMM: A Novel Method Using Z-Curve Theory- Based and Position Weight Matrix for Predicting Nucleosome Positioning" Genes 10, no. 10: 765. https://doi.org/10.3390/genes10100765
APA StyleCui, Y., Xu, Z., & Li, J. (2019). ZCMM: A Novel Method Using Z-Curve Theory- Based and Position Weight Matrix for Predicting Nucleosome Positioning. Genes, 10(10), 765. https://doi.org/10.3390/genes10100765





