NOTCH Receptors and DLK Proteins Enhance Brown Adipogenesis in Mesenchymal C3H10T1/2 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids, Cell Culture, and Transfections
2.2. Quantitative PCR and RT-PCR mRNA Transcription Analysis
2.3. Culture Supernatants and Conditioned Media
2.4. Protein Sample Preparation and Western Blotting
2.5. Luciferase Assays
2.6. Adipogenic Assays and Mitochondrial Biogenesis Analysis
2.7. Lipolytic Potential, Lactate Release to the Extracellular Medium, and Oxygen Consumption Rate (OCR) Assays
2.8. Statistical Analysis
3. Results
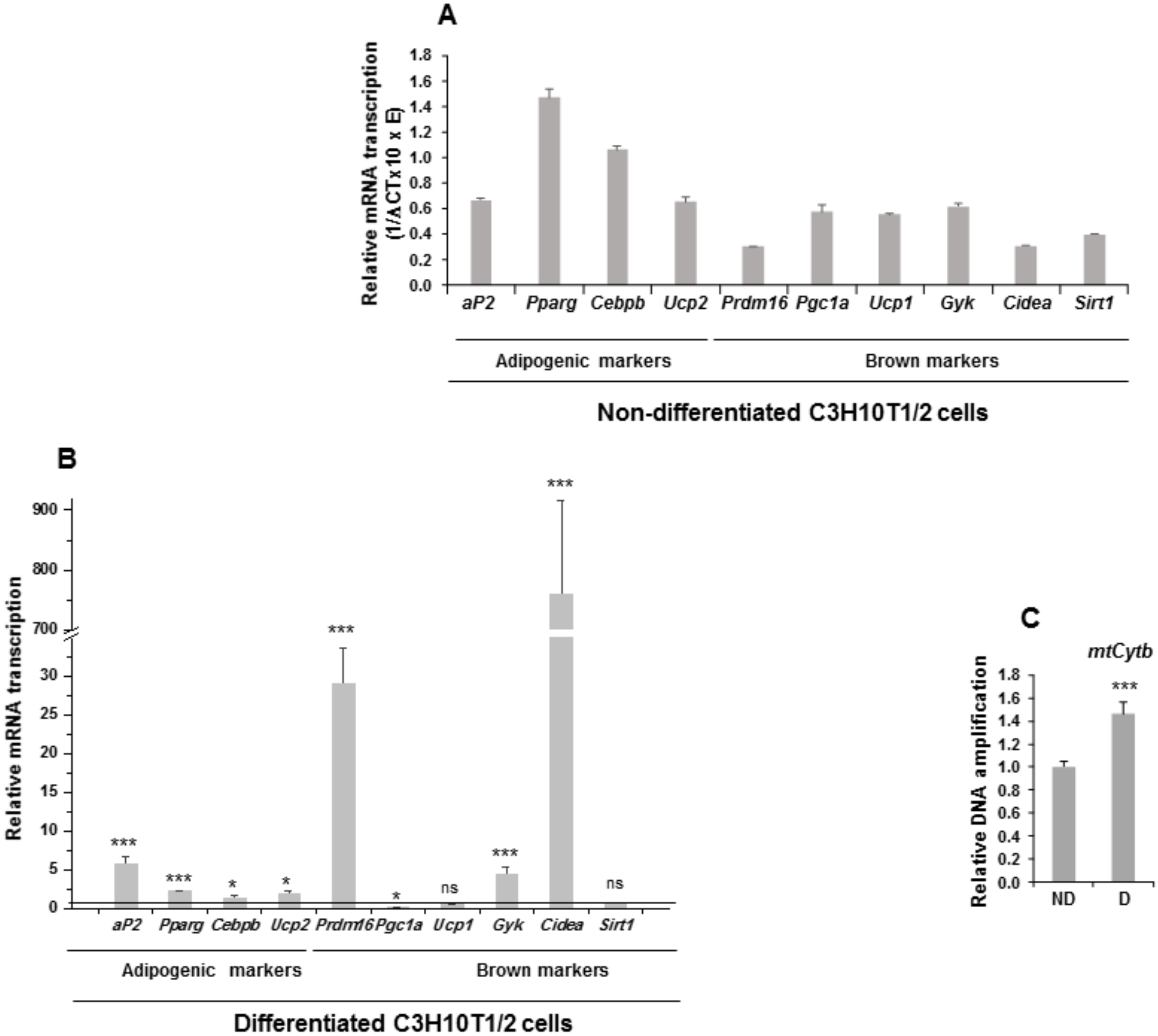
3.1. Comparison of Adipogenesis Levels and Expression Levels of Some of the Notch Family Genes between Multipotent C3H10T1/2 Cells and 3T3-L1 Preadipocytes
3.2. Analysis of the Expression of NOTCH Receptors and DLK Proteins in Multipotent C3H10T1/2 Cells
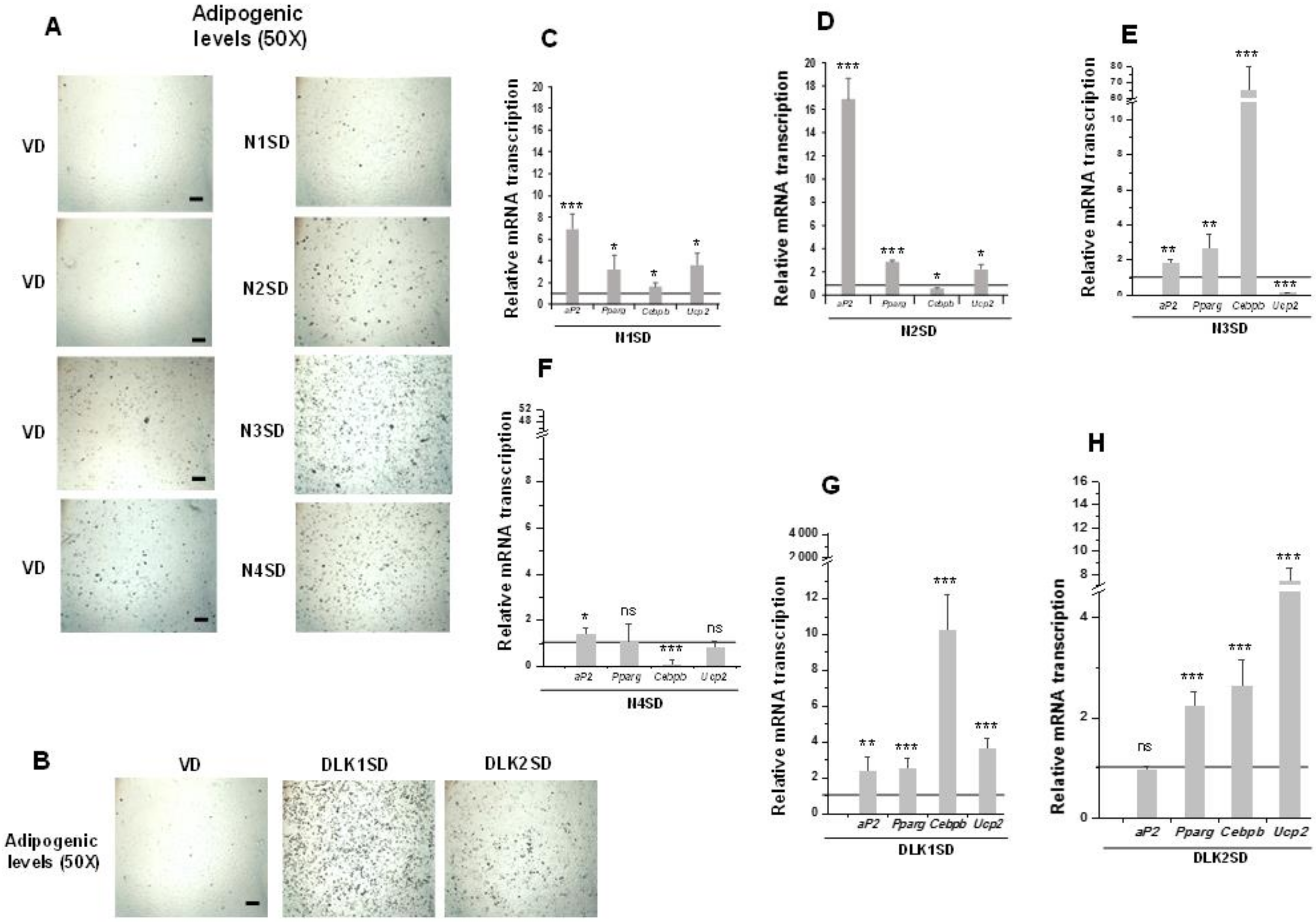
3.3. Overexpression of NOTCH Receptors or DLK Proteins Enhances the Adipogenic Potential of Multipotent C3H10T1/2 Cells
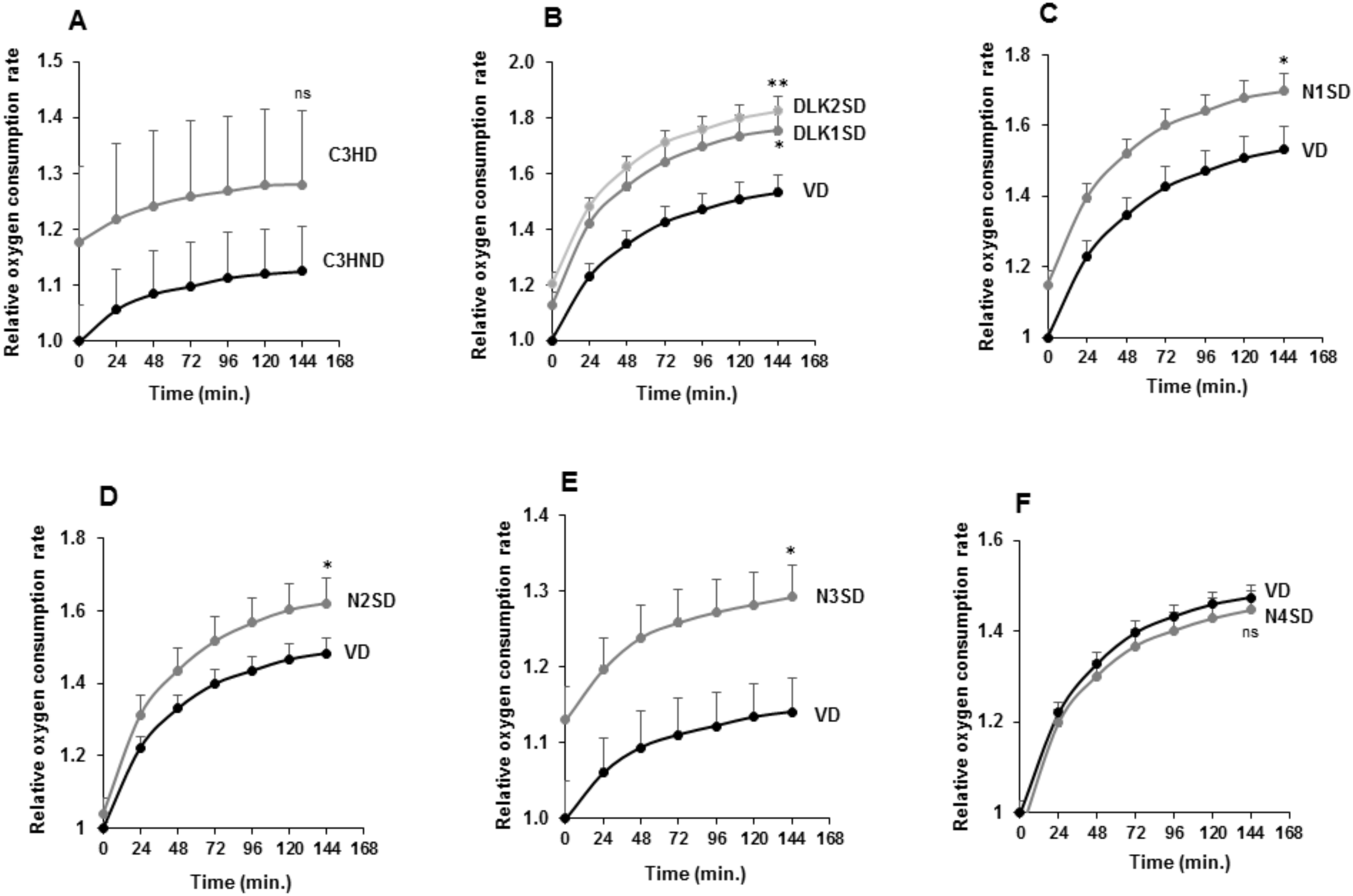
3.4. Overexpression of NOTCH3 Receptor or DLK Proteins Enhances the Brown Adipogenesis of Multipotent C3H10T1/2 Cells
4. Discussion
5. Conclusions
- C3H10T1/2 mesenchymal cells display lower DLK expression levels and higher NOTCH expression, activity, and signaling levels than 3T3-L1 preadipocytes.
- There are complex feedback regulation loops among Notch and Dlk genes in C3H10T1/2 cells.
- Even though Notch1 gene expression is upregulated at the end of the adipogenic process, the expression of the rest of Notch genes and NOTCH receptors’ target genes Hes1 and Hey1 is downregulated.
- The overexpression of each of the four NOTCH receptors, except for NOTCH4, enhances the adipogenic levels of multipotent C3H10T1/2 cells.
- NOTCH activation levels can modulate the adipocyte fate of multipotent C3H10T1/2 cells.
- The overexpression of NOTCH3 receptor or DLK proteins determine a brown adipogenesis fate in multipotent C3H10T1/2 adipocytes.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| aP2 | adipocyte protein 2/fatty acid binding protein 4. |
| APOB | apoliprotein B. |
| BAT | brown adipose tissue. |
| CEBPβ | CCAAT Enhancer Binding Protein Beta. |
| CIDEA | cell death-inducing DNA fragmentation factor, alpha subunit-like effector A. |
| CM | conditioned medium. |
| CSL/RBPJκ | CBF1, Suppressor of Hairless, Lag-1/recombination signal binding protein for immunoglobulin kappa J region. |
| CT | cycle threshold. |
| DAPT | N-[N-(3, 5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester. |
| DLK1 | DELTA-like 1 homolog. |
| DLK2 | DELTA-like 2 homolog. |
| DLK | DELTA-like homolog. |
| DLL4 | DELTA-Like Canonical NOTCH Ligand 4. |
| DNER | DELTA/NOTCH EGF-like Repeat Containing. |
| DOS | DELTA and OSM-11 domain. |
| DSL | DELTA-SERRATE-LAG-2. |
| E | oligonucleotide efficiency. |
| ECAR | extracellular acidification rate. |
| EGFL7 | Epidermal growth factor-like protein 7. |
| GYK | glycerol kinase. |
| HA | influenza hemagglutinin. |
| HES1 | hairy and enhancer of split 1. |
| HEY1 | hairy and enhancer-of-split related with YRPW motif 1. |
| IBMX | 3-Isobutyl-1-methylxanthine. |
| JAG1 | JAGGED canonical NOTCH ligand 1. |
| LP | lipolytic potential. |
| MSCs | mesenchymal stem cells. |
| mtCYTB | mitochondrial cytochrome b. |
| NICD | intracellular active domain of NOTCH receptors. |
| OCR | oxygen consumption rate. |
| P0 | ribosomal protein P0 |
| PGC1α | peroxisome proliferator activated receptor gamma coactivator 1-alpha. |
| PPARγ | peroxisome proliferator activated receptor gamma. |
| PRDM16 | PR domain-containing 16. |
| qPCR/qRT-PCR | quantitative-polymerase chain reaction/quantitative reverse transcription-polymerase chain reaction. |
| SIRT1 | Sirtuin 1. |
| TACE | ADAM metallopeptidase domain 17. |
| UCP1 | mitochondrial uncoupling protein 1. |
| UCP2 | uncoupling protein 2. |
| WAT | white adipose tissue. |
References
- Artavanis-Tsakonas, S.; Rand, M.D.; Lake, R.J. Notch signaling: Cell fate control and signal integration in development. Science 1999, 284, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Bigas, A.; Espinosa, L. The multiple usages of Notch signaling in development, cell differentiation and cancer. Curr. Opin Cell Biol. 2018, 55, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Binshtok, U.; Sprinzak, D. Modeling the Notch Response. Adv. Exp. Med. Biol. 2018, 1066, 79–98. [Google Scholar] [PubMed]
- D’Souza, B.; Meloty-Kapella, L.; Weinmaster, G. Canonical and non-canonical Notch ligands. Curr. Top. Dev. Biol. 2010, 92, 73–129. [Google Scholar] [CrossRef]
- D’Souza, B.; Miyamoto, A.; Weinmaster, G. The many facets of Notch ligands. Oncogene 2008, 27, 5148–5167. [Google Scholar] [CrossRef]
- Kopan, R. Notch: A membrane-bound transcription factor. J. Cell Sci. 2002, 115, 1095–1097. [Google Scholar]
- Laborda, J.; Sausville, E.A.; Hoffman, T.; Notario, V. Dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J. Biol. Chem. 1993, 268, 3817–3820. [Google Scholar]
- Nueda, M.L.; Baladron, V.; Garcia-Ramirez, J.J.; Sanchez-Solana, B.; Ruvira, M.D.; Rivero, S.; Ballesteros, M.A.; Monsalve, E.M.; Diaz-Guerra, M.J.; Ruiz-Hidalgo, M.J.; et al. The novel gene EGFL9/Dlk2, highly homologous to Dlk1, functions as a modulator of adipogenesis. J. Mol. Biol. 2007, 367, 1270–1280. [Google Scholar] [CrossRef]
- Schmidt, M.H.; Bicker, F.; Nikolic, I.; Meister, J.; Babuke, T.; Picuric, S.; Muller-Esterl, W.; Plate, K.H.; Dikic, I. Epidermal growth factor-like domain 7 (EGFL7) modulates Notch signalling and affects neural stem cell renewal. Nat. Cell Biol. 2009, 11, 873–880. [Google Scholar] [CrossRef]
- Eiraku, M.; Hirata, Y.; Takeshima, H.; Hirano, T.; Kengaku, M. Delta/notch-like epidermal growth factor (EGF)-related receptor, a novel EGF-like repeat-containing protein targeted to dendrites of developing and adult central nervous system neurons. J. Biol. Chem. 2002, 277, 25400–25407. [Google Scholar] [CrossRef]
- Shimizu, K.; Chiba, S.; Kumano, K.; Hosoya, N.; Takahashi, T.; Kanda, Y.; Hamada, Y.; Yazaki, Y.; Hirai, H. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J. Biol. Chem. 1999, 274, 32961–32969. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, K.; Chiba, S.; Saito, T.; Kumano, K.; Hirai, H. Physical interaction of Delta1, Jagged1, and Jagged2 with Notch1 and Notch3 receptors. Biochem. Biophys. Res. Commun. 2000, 276, 385–389. [Google Scholar] [CrossRef]
- Miller, A.C.; Lyons, E.L.; Herman, T.G. Cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr. Biol. 2009, 19, 1378–1383. [Google Scholar] [CrossRef]
- Luca, V.C.; Kim, B.C.; Ge, C.; Kakuda, S.; Wu, D.; Roein-Peikar, M.; Haltiwanger, R.S.; Zhu, C.; Ha, T.; Garcia, K.C. Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science 2017, 355, 1320–1324. [Google Scholar] [CrossRef] [PubMed]
- Fiuza, U.M.; Arias, A.M. Cell and molecular biology of Notch. J. Endocrinol. 2007, 194, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Baladron, V.; Ruiz-Hidalgo, M.J.; Nueda, M.L.; Diaz-Guerra, M.J.; Garcia-Ramirez, J.J.; Bonvini, E.; Gubina, E.; Laborda, J. Dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp. Cell Res. 2005, 303, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Nueda, M.L.; Gonzalez-Gomez, M.J.; Rodriguez-Cano, M.M.; Monsalve, E.M.; Diaz-Guerra, M.J.M.; Sanchez-Solana, B.; Laborda, J.; Baladron, V. Author Correction: DLK proteins modulate NOTCH signaling to influence a brown or white 3T3-L1 adipocyte fate. Sci. Rep. 2018, 8, 17784. [Google Scholar] [CrossRef]
- Nueda, M.L.; Gonzalez-Gomez, M.J.; Rodriguez-Cano, M.M.; Monsalve, E.M.; Diaz-Guerra, M.J.M.; Sanchez-Solana, B.; Laborda, J.; Baladron, V. DLK proteins modulate NOTCH signaling to influence a brown or white 3T3-L1 adipocyte fate. Sci. Rep. 2018, 8, 16923. [Google Scholar] [CrossRef]
- Nueda, M.L.; Naranjo, A.I.; Baladron, V.; Laborda, J. The proteins DLK1 and DLK2 modulate NOTCH1-dependent proliferation and oncogenic potential of human SK-MEL-2 melanoma cells. Biochim. Biophys. Acta 2014, 1843, 2674–2684. [Google Scholar] [CrossRef]
- Nueda, M.L.; Naranjo, A.I.; Baladron, V.; Laborda, J. Different expression levels of DLK1 inversely modulate the oncogenic potential of human MDA-MB-231 breast cancer cells through inhibition of NOTCH1 signaling. FASEB J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Solana, B.; Nueda, M.L.; Ruvira, M.D.; Ruiz-Hidalgo, M.J.; Monsalve, E.M.; Rivero, S.; Garcia-Ramirez, J.J.; Diaz-Guerra, M.J.; Baladron, V.; Laborda, J. The EGF-like proteins DLK1 and DLK2 function as inhibitory non-canonical ligands of NOTCH1 receptor that modulate each other’s activities. Biochim. Biophys. Acta 2011, 1813, 1153–1164. [Google Scholar] [CrossRef]
- Gonzalez, M.J.; Ruiz-Garcia, A.; Monsalve, E.M.; Sanchez-Prieto, R.; Laborda, J.; Diaz-Guerra, M.J.; Ruiz-Hidalgo, M.J. DLK1 is a novel inflammatory inhibitor which interferes with NOTCH1 signaling in TLR-activated murine macrophages. Eur. J. Immunol. 2015, 45, 2615–2627. [Google Scholar] [CrossRef] [PubMed]
- Laborda, J. The role of the epidermal growth factor-like protein dlk in cell differentiation. Histol. Histopathol. 2000, 15, 119–129. [Google Scholar] [PubMed]
- Komatsu, H.; Chao, M.Y.; Larkins-Ford, J.; Corkins, M.E.; Somers, G.A.; Tucey, T.; Dionne, H.M.; White, J.Q.; Wani, K.; Boxem, M.; et al. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 2008, 6, e196. [Google Scholar] [CrossRef]
- Ahfeldt, T.; Schinzel, R.T.; Lee, Y.K.; Hendrickson, D.; Kaplan, A.; Lum, D.H.; Camahort, R.; Xia, F.; Shay, J.; Rhee, E.P.; et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat. Cell Biol. 2012, 14, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Liu, J.; Wu, W.; Xu, Z.; Wang, Y. Roles of Notch Signaling in Adipocyte Progenitor Cells and Mature Adipocytes. J. Cell Physiol. 2017, 232, 1258–1261. [Google Scholar] [CrossRef]
- Nichols, A.M.; Pan, Y.; Herreman, A.; Hadland, B.K.; De Strooper, B.; Kopan, R.; Huppert, S.S. Notch pathway is dispensable for adipocyte specification. Genesis 2004, 40, 40–44. [Google Scholar] [CrossRef]
- Garces, C.; Ruiz-Hidalgo, M.J.; Font de Mora, J.; Park, C.; Miele, L.; Goldstein, J.; Bonvini, E.; Porras, A.; Laborda, J. Notch-1 controls the expression of fatty acid-activated transcription factors and is required for adipogenesis. J. Biol. Chem. 1997, 272, 29729–29734. [Google Scholar] [CrossRef]
- Ba, K.; Yang, X.; Wu, L.; Wei, X.; Fu, N.; Fu, Y.; Cai, X.; Yao, Y.; Ge, Y.; Lin, Y. Jagged-1-mediated activation of notch signalling induces adipogenesis of adipose-derived stem cells. Cell Prolif. 2012, 45, 538–544. [Google Scholar] [CrossRef]
- Lai, P.Y.; Tsai, C.B.; Tseng, M.J. Active form Notch4 promotes the proliferation and differentiation of 3T3-L1 preadipocytes. Biochem. Biophys. Res. Commun. 2012, 430, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.A.; Rao, P.K.; Kadesch, T. Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes. Mol. Cell. Biol. 2004, 24, 3505–3513. [Google Scholar] [CrossRef]
- Urs, S.; Turner, B.; Tang, Y.; Rostama, B.; Small, D.; Liaw, L. Effect of soluble Jagged1-mediated inhibition of Notch signaling on proliferation and differentiation of an adipocyte progenitor cell model. Adipocyte 2012, 1, 46–57. [Google Scholar] [CrossRef]
- Kilian, T.M.; Kloting, N.; Bluher, M.; Beck-Sickinger, A.G. Prenatal notch1 receptor blockade by protein delta homolog 1 (DLK1) modulates adipocyte size in vivo. Int. J. Obes. 2015, 40, 698–705. [Google Scholar] [CrossRef]
- Nueda, M.L.; Baladron, V.; Sanchez-Solana, B.; Ballesteros, M.A.; Laborda, J. The EGF-like protein dlk1 inhibits notch signaling and potentiates adipogenesis of mesenchymal cells. J. Mol. Biol. 2007, 367, 1281–1293. [Google Scholar] [CrossRef]
- Traustadottir, G.A.; Kosmina, R.; Sheikh, S.P.; Jensen, C.H.; Andersen, D.C. Preadipocytes proliferate and differentiate under the guidance of Delta-like 1 homolog (DLK1). Adipocyte 2013, 2, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Traustadottir, G.A.; Lagoni, L.V.; Ankerstjerne, L.B.S.; Bisgaard, H.C.; Jensen, C.H.; Andersen, D.C. The imprinted gene Delta like non-canonical Notch ligand 1 (Dlk1) is conserved in mammals, and serves a growth modulatory role during tissue development and regeneration through Notch dependent and independent mechanisms. Cytokine Growth Factor Rev. 2019, 46, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Sul, H.S.; Smas, C.; Mei, B.; Zhou, L. Function of pref-1 as an inhibitor of adipocyte differentiation. Int. J. Obes. Relat. Metab. Disord. 2000, 24 (Suppl. 4), S15–S19. [Google Scholar] [CrossRef]
- Moon, Y.S.; Smas, C.M.; Lee, K.; Villena, J.A.; Kim, K.H.; Yun, E.J.; Sul, H.S. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol. Cell. Biol. 2002, 22, 5585–5592. [Google Scholar] [CrossRef]
- Lee, K.; Villena, J.A.; Moon, Y.S.; Kim, K.H.; Lee, S.; Kang, C.; Sul, H.S. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J. Clin. Investig. 2003, 111, 453–461. [Google Scholar] [CrossRef]
- Mortensen, S.B.; Jensen, C.H.; Schneider, M.; Thomassen, M.; Kruse, T.A.; Laborda, J.; Sheikh, S.P.; Andersen, D.C. Membrane-tethered delta-like 1 homolog (DLK1) restricts adipose tissue size by inhibiting preadipocyte proliferation. Diabetes 2012, 61, 2814–2822. [Google Scholar] [CrossRef]
- Nueda, M.L.; Garcia-Ramirez, J.J.; Laborda, J.; Baladron, V. Dlk1 specifically interacts with insulin-like growth factor binding protein 1 to modulate adipogenesis of 3T3-L1 cells. J. Mol. Biol. 2008, 379, 428–442. [Google Scholar] [CrossRef]
- Ruiz-Hidalgo, M.J.; Gubina, E.; Tull, L.; Baladron, V.; Laborda, J. Dlk modulates mitogen-activated protein kinase signaling to allow or prevent differentiation. Exp. Cell Res. 2002, 274, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Garces, C.; Ruiz-Hidalgo, M.J.; Bonvini, E.; Goldstein, J.; Laborda, J. Adipocyte differentiation is modulated by secreted delta-like (dlk) variants and requires the expression of membrane-associated dlk. Differentiation 1999, 64, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.H.; Kosmina, R.; Ryden, M.; Baun, C.; Hvidsten, S.; Andersen, M.S.; Christensen, L.L.; Gastaldelli, A.; Marraccini, P.; Arner, P.; et al. The imprinted gene Delta like non-canonical notch ligand 1 (Dlk1) associates with obesity and triggers insulin resistance through inhibition of skeletal muscle glucose uptake. EBioMedicine 2019, 46, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Rivero, S.; Diaz-Guerra, M.J.; Monsalve, E.M.; Laborda, J.; Garcia-Ramirez, J.J. DLK2 is a transcriptional target of KLF4 in the early stages of adipogenesis. J. Mol. Biol. 2012, 417, 36–50. [Google Scholar] [CrossRef]
- Boyer, B.B.; Barnes, B.M.; Lowell, B.B.; Grujic, D. Differential regulation of uncoupling protein gene homologues in multiple tissues of hibernating ground squirrels. Am. J. Physiol. 1998, 275, R1232–R1238. [Google Scholar] [CrossRef]
- Cambon, B.; Reyne, Y.; Nougues, J. In vitro induction of UCP1 mRNA in preadipocytes from rabbit considered as a model of large mammals brown adipose tissue development: Importance of PPARgamma agonists for cells isolated in the postnatal period. Mol. Cell Endocrinol. 1998, 146, 49–58. [Google Scholar] [CrossRef]
- Wolf, G. Brown adipose tissue: The molecular mechanism of its formation. Nutr. Rev. 2009, 67, 167–171. [Google Scholar] [CrossRef]
- Nakagami, H. The mechanism of white and brown adipocyte differentiation. Diabetes Metab. J. 2013, 37, 85–90. [Google Scholar] [CrossRef]
- Carobbio, S.; Rosen, B.; Vidal-Puig, A. Adipogenesis: New insights into brown adipose tissue differentiation. J. Mol. Endocrinol. 2013, 51, T75–T85. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, K.A.; Lidell, M.E.; Orava, J.; Heglind, M.; Westergren, R.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.J.; Enerback, S.; et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009, 360, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Lidell, M.E.; Betz, M.J.; Dahlqvist Leinhard, O.; Heglind, M.; Elander, L.; Slawik, M.; Mussack, T.; Nilsson, D.; Romu, T.; Nuutila, P.; et al. Evidence for two types of brown adipose tissue in humans. Nat. Med. 2013, 19, 631–634. [Google Scholar] [CrossRef]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Pisani, D.F.; Djedaini, M.; Beranger, G.E.; Elabd, C.; Scheideler, M.; Ailhaud, G.; Amri, E.Z. Differentiation of Human Adipose-Derived Stem Cells into “Brite” (Brown-in-White) Adipocytes. Front. Endocrinol. (Lausanne) 2011, 2, 87. [Google Scholar] [CrossRef]
- Wu, J.; Bostrom, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef]
- Ye, L.; Wu, J.; Cohen, P.; Kazak, L.; Khandekar, M.J.; Jedrychowski, M.P.; Zeng, X.; Gygi, S.P.; Spiegelman, B.M. Fat cells directly sense temperature to activate thermogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 12480–12485. [Google Scholar] [CrossRef]
- Cohen, P.; Spiegelman, B.M. Brown and Beige Fat: Molecular Parts of a Thermogenic Machine. Diabetes 2015, 64, 2346–2351. [Google Scholar] [CrossRef]
- Giralt, M.; Villarroya, F. White, brown, beige/brite: Different adipose cells for different functions? Endocrinology 2013, 154, 2992–3000. [Google Scholar] [CrossRef]
- Bouillaud, F. UCP1, UCP2 and UCP3: Are they true uncouplers of respiration? Int. J. Obes. Relat. Metab. Disord. 1999, 23 (Suppl. 6), S19–S23. [Google Scholar] [CrossRef][Green Version]
- Nicholls, D.G. A history of UCP1. Biochem. Soc. Trans. 2001, 29, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Argyropoulos, G.; Harper, M.E. Uncoupling proteins and thermoregulation. J. Appl. Physiol. (1985) 2002, 92, 2187–2198. [Google Scholar] [CrossRef] [PubMed]
- Golozoubova, V.; Cannon, B.; Nedergaard, J. UCP1 is essential for adaptive adrenergic nonshivering thermogenesis. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E350–E357. [Google Scholar] [CrossRef] [PubMed]
- Parker, N.; Crichton, P.G.; Vidal-Puig, A.J.; Brand, M.D. Uncoupling protein-1 (UCP1) contributes to the basal proton conductance of brown adipose tissue mitochondria. J. Bioenerg. Biomembr. 2009, 41, 335–342. [Google Scholar] [CrossRef]
- Ricquier, D. UCP1, the mitochondrial uncoupling protein of brown adipocyte: A personal contribution and a historical perspective. Biochimie 2017, 134, 3–8. [Google Scholar] [CrossRef]
- Wang, W.; Ishibashi, J.; Trefely, S.; Shao, M.; Cowan, A.J.; Sakers, A.; Lim, H.W.; O’Connor, S.; Doan, M.T.; Cohen, P.; et al. A PRDM16-Driven Metabolic Signal from Adipocytes Regulates Precursor Cell Fate. Cell Metab. 2019, 30, 174.e5–189.e5. [Google Scholar] [CrossRef]
- Bargut, T.C.; Aguila, M.B.; Mandarim-de-Lacerda, C.A. Brown adipose tissue: Updates in cellular and molecular biology. Tissue Cell 2016, 48, 452–460. [Google Scholar] [CrossRef]
- Festuccia, W.T.; Guerra-Sa, R.; Kawashita, N.H.; Garofalo, M.A.; Evangelista, E.A.; Rodrigues, V.; Kettelhut, I.C.; Migliorini, R.H. Expression of glycerokinase in brown adipose tissue is stimulated by the sympathetic nervous system. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 284, R1536–R1541. [Google Scholar] [CrossRef]
- Kawashita, N.H.; Festuccia, W.T.; Brito, M.N.; Moura, M.A.; Brito, S.R.; Garofalo, M.A.; Kettelhut, I.C.; Migliorini, R.H. Glycerokinase activity in brown adipose tissue: A sympathetic regulation? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2002, 282, R1185–R1190. [Google Scholar] [CrossRef]
- Calderon-Dominguez, M.; Sebastian, D.; Fucho, R.; Weber, M.; Mir, J.F.; Garcia-Casarrubios, E.; Obregon, M.J.; Zorzano, A.; Valverde, A.M.; Serra, D.; et al. Carnitine Palmitoyltransferase 1 Increases Lipolysis, UCP1 Protein Expression and Mitochondrial Activity in Brown Adipocytes. PLoS ONE 2016, 11, e0159399. [Google Scholar] [CrossRef]
- Imran, K.M.; Rahman, N.; Yoon, D.; Jeon, M.; Lee, B.T.; Kim, Y.S. Cryptotanshinone promotes commitment to the brown adipocyte lineage and mitochondrial biogenesis in C3H10T1/2 mesenchymal stem cells via AMPK and p38-MAPK signaling. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2017, 1862, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Artsi, H.; Gurt, I.; El-Haj, M.; Muller, R.; Kuhn, G.A.; Ben Shalom, G.; Cohen-Kfir, E.; Abramowitz, E.; Kandel, L.; Safran, O.; et al. Sirt1 Promotes a Thermogenic Gene Program in Bone Marrow Adipocytes: From Mice to (Wo)Men. Front. Endocrinol. (Lausanne) 2019, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Li, X. SIRT1 and energy metabolism. Acta Biochim. Biophys. Sin. 2013, 45, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Bi, P.; Shan, T.; Liu, W.; Yue, F.; Yang, X.; Liang, X.R.; Wang, J.; Li, J.; Carlesso, N.; Liu, X.; et al. Inhibition of Notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nat. Med. 2014, 20, 911–918. [Google Scholar] [CrossRef]
- Gridley, T.; Kajimura, S. Lightening up a notch: Notch regulation of energy metabolism. Nat. Med. 2014, 20, 811–812. [Google Scholar] [CrossRef][Green Version]
- Pasut, A.; Chang, N.C.; Rodriguez, U.G.; Faulkes, S.; Yin, H.; Lacaria, M.; Ming, H.; Rudnicki, M.A. Notch Signaling Rescues Loss of Satellite Cells Lacking Pax7 and Promotes Brown Adipogenic Differentiation. Cell Rep. 2016, 16, 333–343. [Google Scholar] [CrossRef]
- Sparling, D.P.; Yu, J.; Kim, K.; Zhu, C.; Brachs, S.; Birkenfeld, A.L.; Pajvani, U.B. Adipocyte-specific blockade of gamma-secretase, but not inhibition of Notch activity, reduces adipose insulin sensitivity. Mol. Metab. 2016, 5, 113–121. [Google Scholar] [CrossRef]
- Charalambous, M.; Da Rocha, S.T.; Radford, E.J.; Medina-Gomez, G.; Curran, S.; Pinnock, S.B.; Ferron, S.R.; Vidal-Puig, A.; Ferguson-Smith, A.C. DLK1/PREF1 regulates nutrient metabolism and protects from steatosis. Proc. Natl. Acad. Sci. USA 2014, 111, 16088–16093. [Google Scholar] [CrossRef]
- Abdallah, B.M.; Ditzel, N.; Laborda, J.; Karsenty, G.; Kassem, M. DLK1 Regulates Whole-Body Glucose Metabolism: A Negative Feedback Regulation of the Osteocalcin-Insulin Loop. Diabetes 2015, 64, 3069–3080. [Google Scholar] [CrossRef]
- Hermida, C.; Garces, C.; de Oya, M.; Cano, B.; Martinez-Costa, O.H.; Rivero, S.; Garcia-Ramirez, J.J.; Laborda, J.; Aragon, J.J. The serum levels of the EGF-like homeotic protein dlk1 correlate with different metabolic parameters in two hormonally different children populations in Spain. Clin. Endocrinol. 2008, 69, 216–224. [Google Scholar] [CrossRef]
- Wallace, C.; Smyth, D.J.; Maisuria-Armer, M.; Walker, N.M.; Todd, J.A.; Clayton, D.G. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 2009, 42, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Armengol, J.; Villena, J.A.; Hondares, E.; Carmona, M.C.; Sul, H.S.; Iglesias, R.; Giralt, M.; Villarroya, F. Pref-1 in brown adipose tissue: Specific involvement in brown adipocyte differentiation and regulatory role of C/EBPdelta. Biochem. J. 2012, 443, 799–810. [Google Scholar] [CrossRef]
- Rakhshandehroo, M.; Koppen, A.; Kalkhoven, E. Pref-1 preferentially inhibits heat production in brown adipose tissue. Biochem. J. 2012, 443, e3–e5. [Google Scholar] [CrossRef] [PubMed]
- Monsalve, E.; Ruiz-Garcia, A.; Baladron, V.; Ruiz-Hidalgo, M.J.; Sanchez-Solana, B.; Rivero, S.; Garcia-Ramirez, J.J.; Rubio, A.; Laborda, J.; Diaz-Guerra, M.J. Notch1 upregulates LPS-induced macrophage activation by increasing NF-kappaB activity. Eur. J. Immunol. 2009, 39, 2556–2570. [Google Scholar] [CrossRef] [PubMed]
- Laborda, J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res. 1991, 19, 3998. [Google Scholar] [CrossRef]
- Sanchez-Solana, B.; Laborda, J.; Baladron, V. Mouse resistin modulates adipogenesis and glucose uptake in 3T3-L1 preadipocytes through the ROR1 receptor. Mol. Endocrinol. 2012, 26, 110–127. [Google Scholar] [CrossRef]
- Ylikallio, E.; Tyynismaa, H.; Tsutsui, H.; Ide, T.; Suomalainen, A. High mitochondrial DNA copy number has detrimental effects in mice. Hum. Mol. Genet. 2010, 19, 2695–2705. [Google Scholar] [CrossRef]
- Fuke, S.; Kubota-Sakashita, M.; Kasahara, T.; Shigeyoshi, Y.; Kato, T. Regional variation in mitochondrial DNA copy number in mouse brain. Biochim. Biophys. Acta 2011, 1807, 270–274. [Google Scholar] [CrossRef]
- De la Pompa, J.L.; Wakeham, A.; Correia, K.M.; Samper, E.; Brown, S.; Aguilera, R.J.; Nakano, T.; Honjo, T.; Mak, T.W.; Rossant, J.; et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 1997, 124, 1139–1148. [Google Scholar]
- Suchting, S.; Freitas, C.; Eichmann, A. Angiogenesis under Delta-Notch couple control. Med. Sci. (Paris) 2007, 23, 347–348. [Google Scholar] [CrossRef]
- Kaneta, M.; Osawa, M.; Sudo, K.; Nakauchi, H.; Farr, A.G.; Takahama, Y. A role for pref-1 and HES-1 in thymocyte development. J. Immunol. 2000, 164, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sul, H.S. Ectodomain shedding of preadipocyte factor 1 (Pref-1) by tumor necrosis factor alpha converting enzyme (TACE) and inhibition of adipocyte differentiation. Mol. Cell. Biol. 2006, 26, 5421–5435. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, B.M.; Beck-Nielsen, H.; Gaster, M. FA1 Induces Pro-Inflammatory and Anti-Adipogenic Pathways/Markers in Human Myotubes Established from Lean, Obese, and Type 2 Diabetic Subjects but Not Insulin Resistance. Front. Endocrinol. (Lausanne) 2013, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, B.M.; Ding, M.; Jensen, C.H.; Ditzel, N.; Flyvbjerg, A.; Jensen, T.G.; Dagnaes-Hansen, F.; Gasser, J.A.; Kassem, M. Dlk1/FA1 is a novel endocrine regulator of bone and fat mass and its serum level is modulated by growth hormone. Endocrinology 2007, 148, 3111–3121. [Google Scholar] [CrossRef]
- Smas, C.M.; Chen, L.; Sul, H.S. Cleavage of membrane-associated pref-1 generates a soluble inhibitor of adipocyte differentiation. Mol. Cell. Biol. 1997, 17, 977–988. [Google Scholar] [CrossRef]
- Falix, F.A.; Aronson, D.C.; Lamers, W.H.; Gaemers, I.C. Possible roles of DLK1 in the Notch pathway during development and disease. Biochim. Biophys. Acta 2012, 1822, 988–995. [Google Scholar] [CrossRef]
- Garcia-Gallastegui, P.; Ibarretxe, G.; Garcia-Ramirez, J.J.; Baladron, V.; Aurrekoetxea, M.; Nueda, M.L.; Naranjo, A.I.; Santaolalla, F.; Sanchez-del Rey, A.; Laborda, J.; et al. DLK1 regulates branching morphogenesis and parasympathetic innervation of salivary glands through inhibition of NOTCH signalling. Biol. Cell 2014, 106, 237–253. [Google Scholar] [CrossRef]
- Puertas-Avendano, R.A.; Gonzalez-Gomez, M.J.; Ruvira, M.D.; Ruiz-Hidalgo, M.J.; Morales-Delgado, N.; Laborda, J.; Diaz, C.; Bello, A.R. Role of the non-canonical notch ligand delta-like protein 1 in hormone-producing cells of the adult male mouse pituitary. J. Neuroendocrinol. 2011, 23, 849–859. [Google Scholar] [CrossRef]
- Bray, S.J.; Takada, S.; Harrison, E.; Shen, S.C.; Ferguson-Smith, A.C. The atypical mammalian ligand Delta-like homologue 1 (Dlk1) can regulate Notch signalling in Drosophila. BMC Dev. Biol. 2008, 8, 11. [Google Scholar] [CrossRef]
- Garcia-Gallastegui, P.; Luzuriaga, J.; Aurrekoetxea, M.; Baladron, V.; Ruiz-Hidalgo, M.J.; Garcia-Ramirez, J.J.; Laborda, J.; Unda, F.; Ibarretxe, G. Reduced salivary gland size and increased presence of epithelial progenitor cells in DLK1-deficient mice. Cell Tissue Res. 2015, 364, 513–525. [Google Scholar] [CrossRef]
- Traustadottir, G.A.; Jensen, C.H.; Thomassen, M.; Beck, H.C.; Mortensen, S.B.; Laborda, J.; Baladron, V.; Sheikh, S.P.; Andersen, D.C. Evidence of non-canonical NOTCH signaling: Delta-like 1 homolog (DLK1) directly interacts with the NOTCH1 receptor in mammals. Cell. Signal. 2016, 28, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.; Higueras, M.A.; Gonzalez-Rajal, A.; Alfranca, A.; Fierro-Fernandez, M.; Garcia-Fernandez, R.A.; Ruiz-Hidalgo, M.J.; Monsalve, M.; Rodriguez-Pascual, F.; Redondo, J.M.; et al. The non-canonical NOTCH ligand DLK1 exhibits a novel vascular role as a strong inhibitor of angiogenesis. Cardiovasc. Res. 2012, 93, 232–241. [Google Scholar] [CrossRef]
- Andersen, D.C.; Laborda, J.; Baladron, V.; Kassem, M.; Sheikh, S.P.; Jensen, C.H. Dual role of delta-like 1 homolog (DLK1) in skeletal muscle development and adult muscle regeneration. Development 2013, 140, 3743–3753. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, L.; Smas, C.; Sul, H.S. Pref-1 interacts with fibronectin to inhibit adipocyte differentiation. Mol. Cell. Biol. 2010, 30, 3480–3492. [Google Scholar] [CrossRef] [PubMed]
- Ohno, N.; Izawa, A.; Hattori, M.; Kageyama, R.; Sudo, T. Dlk inhibits stem cell factor-induced colony formation of murine hematopoietic progenitors: Hes-1-independent effect. Stem Cells 2001, 19, 71–79. [Google Scholar] [CrossRef]
- Ferron, S.R.; Charalambous, M.; Radford, E.; McEwen, K.; Wildner, H.; Hind, E.; Morante-Redolat, J.M.; Laborda, J.; Guillemot, F.; Bauer, S.R.; et al. Postnatal loss of Dlk1 imprinting in stem cells and niche astrocytes regulates neurogenesis. Nature 2011, 475, 381–385. [Google Scholar] [CrossRef]
- Qi, X.; Chen, Z.; Liu, D.; Cen, J.; Gu, M. Expression of Dlk1 gene in myelodysplastic syndrome determined by microarray, and its effects on leukemia cells. Int. J. Mol. Med. 2008, 22, 61–68. [Google Scholar] [CrossRef][Green Version]
- Li, L.; Tan, J.; Zhang, Y.; Han, N.; Di, X.; Xiao, T.; Cheng, S.; Gao, Y.; Liu, Y. DLK1 promotes lung cancer cell invasion through upregulation of MMP9 expression depending on Notch signaling. PLoS ONE 2014, 9, e91509. [Google Scholar] [CrossRef]
- Shamis, Y.; Cullen, D.E.; Liu, L.; Yang, G.; Ng, S.F.; Xiao, L.; Bell, F.T.; Ray, C.; Takikawa, S.; Moskowitz, I.P.; et al. Maternal and zygotic Zfp57 modulate NOTCH signaling in cardiac development. Proc. Natl. Acad. Sci. USA 2015, 112, E2020–E2029. [Google Scholar] [CrossRef]
- Traustadottir, G.A.; Jensen, C.H.; Garcia Ramirez, J.J.; Beck, H.C.; Sheikh, S.P.; Andersen, D.C. The non-canonical NOTCH1 ligand Delta-like 1 homolog (DLK1) self interacts in mammals. Int. J. Biol. Macromol. 2017, 97, 460–467. [Google Scholar] [CrossRef]
- Baladron, V.; Ruiz-Hidalgo, M.J.; Gubina, E.; Bonvini, E.; Laborda, J. Specific regions of the extracellular domain of dlk, an EGF-like homeotic protein involved in differentiation, participate in intramolecular interactions. Front. Biosci. 2001, 6, A25–A32. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, X.; Wu, Y.; Jing, W.; Cai, X.; Tang, W.; Liu, L.; Liu, Y.; Grottkau, B.E.; Lin, Y. Gamma-secretase inhibitor induces adipogenesis of adipose-derived stem cells by regulation of Notch and PPAR-gamma. Cell Prolif. 2010, 43, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Chartoumpekis, D.V.; Palliyaguru, D.L.; Wakabayashi, N.; Khoo, N.K.; Schoiswohl, G.; O’Doherty, R.M.; Kensler, T.W. Notch intracellular domain overexpression in adipocytes confers lipodystrophy in mice. Mol. Metab. 2015, 4, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Jeong, Y.H.; Kim, H.M.; Park, H.Y.; Yoon, D.; Kim, D.H.; Saeki, S.; Moon, S.J.; Kang, M.J. Presenilin enhancer-2 (PSENEN), a component of the gamma-secretase complex, is involved in adipocyte differentiation. Domest. Anim. Endocrinol. 2009, 37, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.T.; Cheng, H.T.; Chang, L.W.; Ohtsuka, T.; Kageyama, R.; Stormo, G.D.; Kopan, R. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 2006, 281, 5106–5119. [Google Scholar] [CrossRef] [PubMed]
- Beatus, P.; Lundkvist, J.; Oberg, C.; Lendahl, U. The notch 3 intracellular domain represses notch 1-mediated activation through Hairy/Enhancer of split (HES) promoters. Development 1999, 126, 3925–3935. [Google Scholar]
- Beatus, P.; Lundkvist, J.; Oberg, C.; Pedersen, K.; Lendahl, U. The origin of the ankyrin repeat region in Notch intracellular domains is critical for regulation of HES promoter activity. Mech. Dev. 2001, 104, 3–20. [Google Scholar] [CrossRef]
- Sprinzak, D.; Lakhanpal, A.; LeBon, L.; Garcia-Ojalvo, J.; Elowitz, M.B. Mutual inactivation of Notch receptors and ligands facilitates developmental patterning. PLoS Comput. Biol. 2011, 7, e1002069. [Google Scholar] [CrossRef]
- Sprinzak, D.; Lakhanpal, A.; Lebon, L.; Santat, L.A.; Fontes, M.E.; Anderson, G.A.; Garcia-Ojalvo, J.; Elowitz, M.B. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 2010, 465, 86–90. [Google Scholar] [CrossRef]
- Semenova, D.; Bogdanova, M.; Kostina, A.; Golovkin, A.; Kostareva, A.; Malashicheva, A. Dose-dependent mechanism of Notch action in promoting osteogenic differentiation of mesenchymal stem cells. Cell Tissue Res. 2020, 379, 169–179. [Google Scholar] [CrossRef]
- Seymour, P.A.; Collin, C.A.; Egeskov-Madsen, A.R.; Jorgensen, M.C.; Shimojo, H.; Imayoshi, I.; de Lichtenberg, K.H.; Kopan, R.; Kageyama, R.; Serup, P. Jag1 Modulates an Oscillatory Dll1-Notch-Hes1 Signaling Module to Coordinate Growth and Fate of Pancreatic Progenitors. Dev. Cell 2020, 52, 731–747. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D. Notch Signaling-Induced Oscillatory Gene Expression May Drive Neurogenesis in the Developing Retina. Front. Mol. Neurosci. 2019, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Kageyama, R.; Niwa, Y.; Shimojo, H.; Kobayashi, T.; Ohtsuka, T. Ultradian oscillations in Notch signaling regulate dynamic biological events. Curr. Top. Dev. Biol. 2010, 92, 311–331. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Mizuno, H.; Imayoshi, I.; Furusawa, C.; Shirahige, K.; Kageyama, R. The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 2009, 23, 1870–1875. [Google Scholar] [CrossRef]
- Peschechera, A.; Eckel, J. “Browning” of adipose tissue—Regulation and therapeutic perspectives. Arch. Physiol. Biochem. 2013, 119, 151–160. [Google Scholar] [CrossRef]
- Fruhbeck, G.; Becerril, S.; Sainz, N.; Garrastachu, P.; Garcia-Velloso, M.J. BAT: A new target for human obesity? Trends Pharmacol. Sci. 2009, 30, 387–396. [Google Scholar] [CrossRef]





| Protein | Dilution of Primary and Secondary Antibodies | Company |
|---|---|---|
| NOTCH1 | Rabbit anti-NOTCH1 C20R (1:1000) | Santa Cruz Biotechnology |
| NOTCH2 | Goat anti-NOTCH2 M20 (1:500) | Santa Cruz Biotechnology |
| NOTCH3 | Rabbit anti-NOTCH3 (1:1000) | Abcam |
| NOTCH4 | Rabbit anti-NOTCH4 (1:1000) | Upstate Millipore |
| HA | Mouse anti-HA 16B12 (1:5000) | Covance |
| DLK1 (DELTA-like 1 homolog) | Rabbit anti-DLK1 (1:1000) | Nueda et al., 2008 |
| DLK2 (DELTA-like 2 homolog) | Rabbit polyclonal anti-mouse DLK2-C-terminal (1:500) | Abcam |
| α-Tubulin | Mouse anti-alpha-Tubulin (1:5000) | Sigma |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Cano, M.-M.; González-Gómez, M.-J.; Sánchez-Solana, B.; Monsalve, E.-M.; Díaz-Guerra, M.-J.M.; Laborda, J.; Nueda, M.-L.; Baladrón, V. NOTCH Receptors and DLK Proteins Enhance Brown Adipogenesis in Mesenchymal C3H10T1/2 Cells. Cells 2020, 9, 2032. https://doi.org/10.3390/cells9092032
Rodríguez-Cano M-M, González-Gómez M-J, Sánchez-Solana B, Monsalve E-M, Díaz-Guerra M-JM, Laborda J, Nueda M-L, Baladrón V. NOTCH Receptors and DLK Proteins Enhance Brown Adipogenesis in Mesenchymal C3H10T1/2 Cells. Cells. 2020; 9(9):2032. https://doi.org/10.3390/cells9092032
Chicago/Turabian StyleRodríguez-Cano, María-Milagros, María-Julia González-Gómez, Beatriz Sánchez-Solana, Eva-María Monsalve, María-José M. Díaz-Guerra, Jorge Laborda, María-Luisa Nueda, and Victoriano Baladrón. 2020. "NOTCH Receptors and DLK Proteins Enhance Brown Adipogenesis in Mesenchymal C3H10T1/2 Cells" Cells 9, no. 9: 2032. https://doi.org/10.3390/cells9092032
APA StyleRodríguez-Cano, M.-M., González-Gómez, M.-J., Sánchez-Solana, B., Monsalve, E.-M., Díaz-Guerra, M.-J. M., Laborda, J., Nueda, M.-L., & Baladrón, V. (2020). NOTCH Receptors and DLK Proteins Enhance Brown Adipogenesis in Mesenchymal C3H10T1/2 Cells. Cells, 9(9), 2032. https://doi.org/10.3390/cells9092032






