5-aza-2′-Deoxycytidine Induces a RIG-I-Related Innate Immune Response by Modulating Mitochondria Stress in Neuroblastoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Gene Expression Microarray Assay
2.3. Methylation Analysis Using Bisulfite Pyrosequencing
2.4. Gene Knockdown
2.5. MtDNA-Devoid ρ0 Cells
2.6. MtDNA Copy Number
2.7. Western Blotting
2.8. Flow Cytometry Detecting Cell Death and ROS
2.9. Immunofluorescent Imaging
2.10. Statistical Analysis
3. Results
3.1. Dac Preferentially Induces a RIG-I-Related Innate Immune Response and Cell Apoptosis in MYCN Non-Amplified SK-N-AS NB
3.2. mtDNA Plays a Vital Role in Dac-Activated Innate Immune Response and Apoptosis
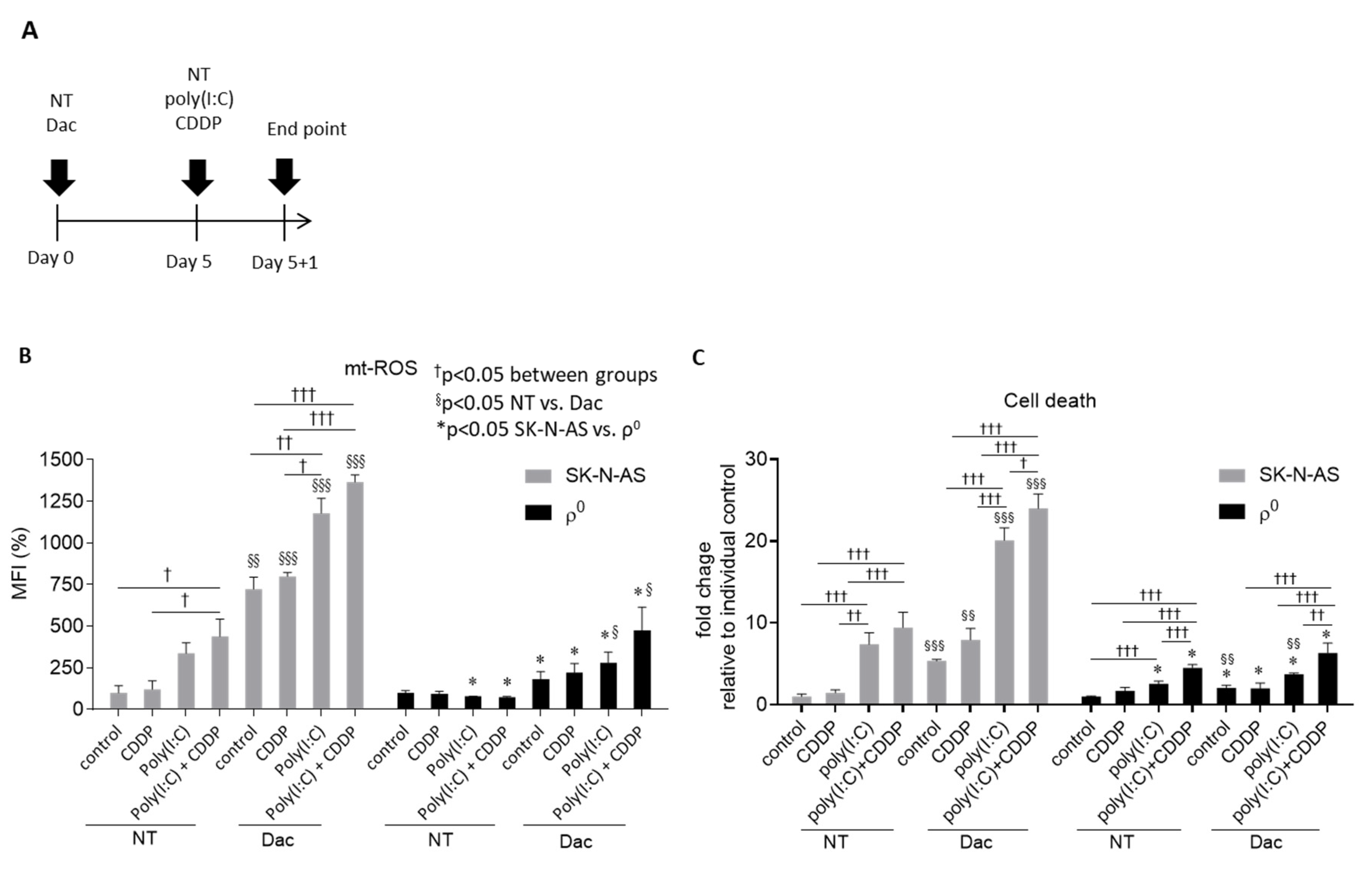
3.3. Dac Potentiates Anti-NB Effect of CDDP and Poly(I:C)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef]
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Mazanek, P.; Dam, V.; Wang, Q.; Zhao, H.; Guo, R.; Jagannathan, J.; Cnaan, A.; Maris, J.M.; Hogarty, M.D. Deregulated Wnt/beta-catenin program in high-risk neuroblastomas without MYCN amplification. Oncogene 2008, 27, 1478–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, J.M.; Yu, J. Promoter hypermethylation of tumour suppressor genes as potential biomarkers in colorectal cancer. Int. J. Mol. Sci. 2015, 16, 2472–2496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philipp, A.B.; Stieber, P.; Nagel, D.; Neumann, J.; Spelsberg, F.; Jung, A.; Lamerz, R.; Herbst, A.; Kolligs, F.T. Prognostic role of methylated free circulating DNA in colorectal cancer. Int. J. Cancer 2012, 131, 2308–2319. [Google Scholar] [CrossRef]
- Herbst, A.; Wallner, M.; Rahmig, K.; Stieber, P.; Crispin, A.; Lamerz, R.; Kolligs, F.T. Methylation of helicase-like transcription factor in serum of patients with colorectal cancer is an independent predictor of disease recurrence. Eur. J. Gastroenterol. Hepatol. 2009, 21, 565–569. [Google Scholar] [CrossRef]
- Wallner, M.; Herbst, A.; Behrens, A.; Crispin, A.; Stieber, P.; Goke, B.; Lamerz, R.; Kolligs, F.T. Methylation of serum DNA is an independent prognostic marker in colorectal cancer. Clin. Cancer Res. 2006, 12, 7347–7352. [Google Scholar] [CrossRef] [Green Version]
- Gomez, S.; Castellano, G.; Mayol, G.; Sunol, M.; Queiros, A.; Bibikova, M.; Nazor, K.L.; Loring, J.F.; Lemos, I.; Rodriguez, E.; et al. DNA methylation fingerprint of neuroblastoma reveals new biological and clinical insights. Epigenomics 2015, 7, 1137–1153. [Google Scholar] [CrossRef] [Green Version]
- Abe, M.; Ohira, M.; Kaneda, A.; Yagi, Y.; Yamamoto, S.; Kitano, Y.; Takato, T.; Nakagawara, A.; Ushijima, T. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005, 65, 828–834. [Google Scholar]
- Yang, Q.; Tian, Y.; Ostler, K.R.; Chlenski, A.; Guerrero, L.J.; Salwen, H.R.; Godley, L.A.; Cohn, S.L. Epigenetic alterations differ in phenotypically distinct human neuroblastoma cell lines. BMC Cancer 2010, 10, 286. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.C.; Li, H.; Van Neste, L.; Cai, Y.; Robert, C.; Rassool, F.V.; Shin, J.J.; Harbom, K.M.; Beaty, R.; Pappou, E.; et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell 2012, 21, 430–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpinelli, P.; Granata, F.; Augusti-Tocco, G.; Rossi, M.; Bartolucci, S. Antiproliferative effects and DNA hypomethylation by 5-aza-2′-deoxycytidine in human neuroblastoma cell lines. Anticancer Drugs 1993, 4, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Bartolucci, S.; Estenoz, M.; Longo, A.; Santoro, B.; Momparler, R.L.; Rossi, M.; Augusti-Tocco, G. 5-Aza-2′-deoxycytidine as inducer of differentiation and growth inhibition in mouse neuroblastoma cells. Cell Differ. Dev. 1989, 27, 47–55. [Google Scholar] [CrossRef]
- Charlet, J.; Schnekenburger, M.; Brown, K.W.; Diederich, M. DNA demethylation increases sensitivity of neuroblastoma cells to chemotherapeutic drugs. Biochem. Pharmacol. 2012, 83, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Navada, S.C.; Steinmann, J.; Lubbert, M.; Silverman, L.R. Clinical development of demethylating agents in hematology. J. Clin. Investig. 2014, 124, 40–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef] [Green Version]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef] [Green Version]
- Banoth, B.; Cassel, S.L. Mitochondria in innate immune signaling. Transl. Res. 2018, 202, 52–68. [Google Scholar] [CrossRef]
- Wiatrek, D.M.; Candela, M.E.; Sedmik, J.; Oppelt, J.; Keegan, L.P.; O’Connell, M.A. Activation of innate immunity by mitochondrial dsRNA in mouse cells lacking p53 protein. RNA 2019, 25, 713–726. [Google Scholar] [CrossRef]
- Dhir, A.; Dhir, S.; Borowski, L.S.; Jimenez, L.; Teitell, M.; Rotig, A.; Crow, Y.J.; Rice, G.I.; Duffy, D.; Tamby, C.; et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 2018, 560, 238–242. [Google Scholar] [CrossRef]
- Chuang, J.H.; Lin, T.K.; Tai, M.H.; Liou, C.W.; Huang, S.T.; Wu, C.L.; Lin, H.Y.; Wang, P.W. Preferential involvement of mitochondria in Toll-like receptor 3 agonist-induced neuroblastoma cell apoptosis, but not in inhibition of cell growth. Apoptosis 2012, 17, 335–348. [Google Scholar] [CrossRef]
- Ikegaki, N.; Shimada, H.; Fox, A.M.; Regan, P.L.; Jacobs, J.R.; Hicks, S.L.; Rappaport, E.F.; Tang, X.X. Transient treatment with epigenetic modifiers yields stable neuroblastoma stem cells resembling aggressive large-cell neuroblastomas. Proc. Natl. Acad. Sci. USA 2013, 110, 6097–6102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuang, J.H.; Chuang, H.C.; Huang, C.C.; Wu, C.L.; Du, Y.Y.; Kung, M.L.; Chen, C.H.; Chen, S.C.; Tai, M.H. Differential toll-like receptor 3 (TLR3) expression and apoptotic response to TLR3 agonist in human neuroblastoma cells. J. Biomed. Sci. 2011, 18, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castanier, C.; Zemirli, N.; Portier, A.; Garcin, D.; Bidere, N.; Vazquez, A.; Arnoult, D. MAVS ubiquitination by the E3 ligase TRIM25 and degradation by the proteasome is involved in type I interferon production after activation of the antiviral RIG-I-like receptors. BMC Biol. 2012, 10, 44. [Google Scholar] [CrossRef] [Green Version]
- Moehle, E.A.; Shen, K.; Dillin, A. Mitochondrial proteostasis in the context of cellular and organismal health and aging. J. Biol. Chem. 2019, 294, 5396–5407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Franco-Iborra, S.; Cuadros, T.; Parent, A.; Romero-Gimenez, J.; Vila, M.; Perier, C. Defective mitochondrial protein import contributes to complex I-induced mitochondrial dysfunction and neurodegeneration in Parkinson's disease. Cell Death Dis. 2018, 9, 1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bomken, S.; Davies, B.; Chong, L.; Cole, M.; Wood, K.M.; McDermott, M.; Tweddle, D.A. Percentage tumor necrosis following chemotherapy in neuroblastoma correlates with MYCN status but not survival. Pediatr. Hematol. Oncol. 2011, 28, 106–114. [Google Scholar] [CrossRef]
- Jubierre, L.; Jimenez, C.; Rovira, E.; Soriano, A.; Sabado, C.; Gros, L.; Llort, A.; Hladun, R.; Roma, J.; Toledo, J.S.; et al. Targeting of epigenetic regulators in neuroblastoma. Exp. Mol. Med. 2018, 50, 51. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Wu, X.; Basu, M.; Dong, C.; Zheng, P.; Liu, Y.; Sandler, A.D. MYCN Amplification Is Associated with Repressed Cellular Immunity in Neuroblastoma: An In Silico Immunological Analysis of TARGET Database. Front. Immunol. 2017, 8, 1473. [Google Scholar] [CrossRef] [Green Version]
- Layer, J.P.; Kronmuller, M.T.; Quast, T.; van den Boorn-Konijnenberg, D.; Effern, M.; Hinze, D.; Althoff, K.; Schramm, A.; Westermann, F.; Peifer, M.; et al. Amplification of N-Myc is associated with a T-cell-poor microenvironment in metastatic neuroblastoma restraining interferon pathway activity and chemokine expression. Oncoimmunology 2017, 6, e1320626. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Zhang, W.; Booth, J.L.; Hutchings, D.C.; Wang, X.; White, V.L.; Youness, H.; Cross, C.D.; Zou, M.H.; Burian, D.; et al. Human primary airway epithelial cells isolated from active smokers have epigenetically impaired antiviral responses. Respir Res. 2016, 17, 111. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Dou, C.; Jia, Y.; Li, Q.; Zheng, X.; Yao, Y.; Liu, Q.; Song, T. RIG-I suppresses the migration and invasion of hepatocellular carcinoma cells by regulating MMP9. Int. J. Oncol. 2015, 46, 1710–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pourcelot, M.; Arnoult, D. Mitochondrial dynamics and the innate antiviral immune response. FEBS J. 2014, 281, 3791–3802. [Google Scholar] [CrossRef] [PubMed]
- Agod, Z.; Fekete, T.; Budai, M.M.; Varga, A.; Szabo, A.; Moon, H.; Boldogh, I.; Biro, T.; Lanyi, A.; Bacsi, A.; et al. Regulation of type I interferon responses by mitochondria-derived reactive oxygen species in plasmacytoid dendritic cells. Redox Biol. 2017, 13, 633–645. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sanchez, E.; Marin, J.J.; Perez, M.J. The expression of genes involved in hepatocellular carcinoma chemoresistance is affected by mitochondrial genome depletion. Mol. Pharm. 2014, 11, 1856–1868. [Google Scholar] [CrossRef]
- Montopoli, M.; Bellanda, M.; Lonardoni, F.; Ragazzi, E.; Dorigo, P.; Froldi, G.; Mammi, S.; Caparrotta, L. “Metabolic reprogramming” in ovarian cancer cells resistant to cisplatin. Curr. Cancer Drug Targets 2011, 11, 226–235. [Google Scholar] [CrossRef]
- Qian, W.; Nishikawa, M.; Haque, A.M.; Hirose, M.; Mashimo, M.; Sato, E.; Inoue, M. Mitochondrial density determines the cellular sensitivity to cisplatin-induced cell death. Am. J. Physiol. Cell Physiol. 2005, 289, 1466–1475. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Choi, B.; Cheon, H.; Pak, Y.K.; Kulawiec, M.; Singh, K.K.; Lee, M.S. Cellular aging of mitochondrial DNA-depleted cells. Biochem. Biophys. Res. Commun. 2004, 325, 1399–1405. [Google Scholar] [CrossRef]
- Tajeddine, N.; Galluzzi, L.; Kepp, O.; Hangen, E.; Morselli, E.; Senovilla, L.; Araujo, N.; Pinna, G.; Larochette, N.; Zamzami, N.; et al. Hierarchical involvement of Bak, VDAC1 and Bax in cisplatin-induced cell death. Oncogene 2008, 27, 4221–4232. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Schumaker, L.M.; Egorin, M.J.; Zuhowski, E.G.; Guo, Z.; Cullen, K.J. Cisplatin preferentially binds mitochondrial DNA and voltage-dependent anion channel protein in the mitochondrial membrane of head and neck squamous cell carcinoma: Possible role in apoptosis. Clin. Cancer Res. 2006, 12, 5817–5825. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, H.-Y.; Chuang, J.-H.; Wang, P.-W.; Lin, T.-K.; Wu, M.-T.; Hsu, W.-M.; Chuang, H.-C. 5-aza-2′-Deoxycytidine Induces a RIG-I-Related Innate Immune Response by Modulating Mitochondria Stress in Neuroblastoma. Cells 2020, 9, 1920. https://doi.org/10.3390/cells9091920
Lin H-Y, Chuang J-H, Wang P-W, Lin T-K, Wu M-T, Hsu W-M, Chuang H-C. 5-aza-2′-Deoxycytidine Induces a RIG-I-Related Innate Immune Response by Modulating Mitochondria Stress in Neuroblastoma. Cells. 2020; 9(9):1920. https://doi.org/10.3390/cells9091920
Chicago/Turabian StyleLin, Hung-Yu, Jiin-Haur Chuang, Pei-Wen Wang, Tsu-Kung Lin, Min-Tsui Wu, Wen-Ming Hsu, and Hui-Ching Chuang. 2020. "5-aza-2′-Deoxycytidine Induces a RIG-I-Related Innate Immune Response by Modulating Mitochondria Stress in Neuroblastoma" Cells 9, no. 9: 1920. https://doi.org/10.3390/cells9091920
APA StyleLin, H.-Y., Chuang, J.-H., Wang, P.-W., Lin, T.-K., Wu, M.-T., Hsu, W.-M., & Chuang, H.-C. (2020). 5-aza-2′-Deoxycytidine Induces a RIG-I-Related Innate Immune Response by Modulating Mitochondria Stress in Neuroblastoma. Cells, 9(9), 1920. https://doi.org/10.3390/cells9091920




