Cell-Free microRNAs as Potential Oral Cancer Biomarkers: From Diagnosis to Therapy
Abstract
1. Introduction
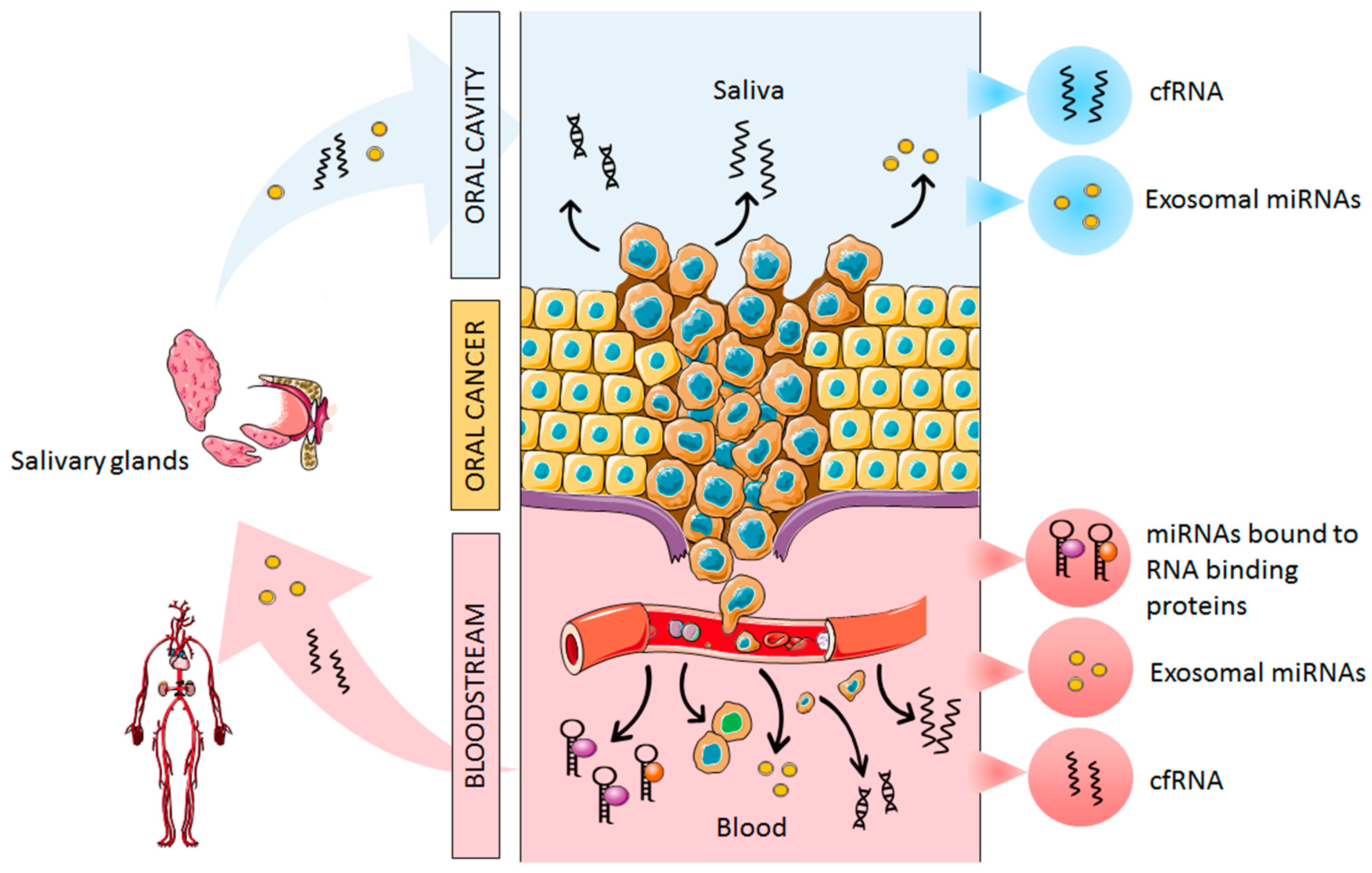
2. Origin of Cell-Free miRNAs in Oral Cancer
3. MiRNAs as Diagnostic Biomarkers
3.1. Plasma and Serum
3.2. Saliva
4. MiRNAs as Prognostic Biomarkers
5. MiRNAs as Therapeutic Targets
6. Conclusions: Future Perspectives and Challenges
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Melchardt, T.; Magnes, T.; Hufnagl, C.; Thorner, A.R.; Ducar, M.; Neureiter, D.; Tränkenschuh, W.; Klieser, E.; Gaggl, A.; Rösch, S.; et al. Clonal evolution and heterogeneity in metastatic head and neck cancer-An analysis of the Austrian study group of medical tumour therapy study group. Eur. J. Cancer 2018, 93, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Perakis, S.; Geigl, J.B.; Speicher, M.R. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. Oncol. 2017, 1, 36. [Google Scholar] [CrossRef] [PubMed]
- Chiu, A.M.; Mitra, M.; Boymoushakian, L.; Coller, H.A. Integrative analysis of the inter-tumoral heterogeneity of triple-negative breast cancer. Sci. Rep. 2018, 8, 11807. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S. Global epidemiology of oral and oropharyngeal cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Güneri, P.; Epstein, J.B. Late stage diagnosis of oral cancer: Components and possible solutions. Oral Oncol. 2014, 50, 1131–1136. [Google Scholar] [CrossRef]
- Gigliotti, J.; Madathil, S.; Makhoul, N. Delays in oral cavity cancer. Int. J. Oral Maxillofac. Surg. 2019, 48, 1131–1137. [Google Scholar] [CrossRef]
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884–11894. [Google Scholar]
- Lafaurie, G.I.; Perdomo, S.J.; Buenahora, M.R.; Amaya, S.; Díaz-Báez, D. Human papilloma virus: An etiological and prognostic factor for oral cancer? J. Investig. Clin. Dent. 2018, 9, e12313. [Google Scholar] [CrossRef]
- Ali, J.; Sabiha, B.; Jan, H.U.; Haider, S.A.; Khan, A.A.; Ali, S.S. Genetic etiology of oral cancer. Oral Oncol. 2017, 70, 23–28. [Google Scholar] [CrossRef]
- Irimie, A.; Ciocan, C.; Gulei, D.; Mehterov, N.; Atanasov, A.; Dudea, D.; Berindan-Neagoe, I. Current insights into oral cancer epigenetics. Int. J. Mol. Sci. 2018, 19, 670. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Vannini, I.; Fanini, F.; Fabbri, M. Emerging roles of microRNAs in cancer. Curr. Opin. Genet. Dev. 2018, 48, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. Mech. Dis. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Schickel, R.; Boyerinas, B.; Park, S.-M.; Peter, M.E. MicroRNAs: Key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008, 27, 5959–5974. [Google Scholar] [CrossRef]
- Barbato, S.; Solaini, G.; Fabbri, M. MicroRNAs in oncogenesis and tumor suppression. Int. Rev. Cell. Mol. Biol. 2017, 333, 229–268. [Google Scholar]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Andersen, G.B.; Tost, J. Circulating miRNAs as biomarker in cancer. Recent Results Cancer Res. 2020, 215, 277–298. [Google Scholar]
- Rapado-González, Ó.; Majem, B.; Muinelo-Romay, L.; Álvarez-Castro, A.; Santamaría, A.; Gil-Moreno, A.; López-López, R.; Suárez-Cunqueiro, M.M. Human salivary microRNAs in cancer. J. Cancer 2018, 9, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Das, S. Profiling cell-free and circulating miRNA: A clinical diagnostic tool for different cancers. Tumor Biol. 2016, 37, 5705–5714. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H. The clinical relevance of circulating, exosomal miRNAs as biomarkers for cancer. Expert Rev. Mol. Diagn. 2015, 15, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Michael, A.; Bajracharya, S.D.; Yuen, P.S.T.; Zhou, H.; Star, R.A.; Illei, G.G.; Alevizos, I. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef]
- Wong, T.S.; Liu, X.B.; Wong, B.Y.H.; Ng, R.W.M.; Yuen, A.P.W.; Wei, W.I. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 2008, 14, 2588–2592. [Google Scholar] [CrossRef]
- Santhi, W.S.; Prathibha, R.; Charles, S.; Anurup, K.G.; Reshmi, G.; Ramachandran, S.; Jissa, V.T.; Sebastian, P.; Radhakrishna Pillai, M. Oncogenic microRNAs as biomarkers of oral tumorigenesis and minimal residual disease. Oral Oncol. 2013, 49, 567–575. [Google Scholar] [CrossRef]
- Manikandan, M.; Deva Magendhra Rao, A.K.; Rajkumar, K.S.; Rajaraman, R.; Munirajan, A.K. Altered levels of miR-21, miR-125b-2*, miR-138, miR-155, miR-184, and miR-205 in oral squamous cell carcinoma and association with clinicopathological characteristics. J. Oral Pathol. Med. 2015, 44, 792–800. [Google Scholar] [CrossRef]
- Lin, S.C.; Liu, C.J.; Lin, J.A.; Chiang, W.F.; Hung, P.S.; Chang, K.W. miR-24 up-regulation in oral carcinoma: Positive association from clinical and in vitro analysis. Oral Oncol. 2010, 46, 204–208. [Google Scholar] [CrossRef]
- Zhao, J.; Hu, C.; Chi, J.; Li, J.; Peng, C.; Yun, X.; Li, D.; Yu, Y.; Li, Y.; Gao, M.; et al. miR-24 promotes the proliferation, migration and invasion in human tongue squamous cell carcinoma by targeting FBXW7. Oncol. Rep. 2016, 36, 1143–1149. [Google Scholar] [CrossRef]
- Sochor, M.; Basova, P.; Pesta, M.; Dusilkova, N.; Bartos, J.; Burda, P.; Pospisil, V.; Stopka, T. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 2014, 14, 448. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Tang, J.; Bai, Y.; Lin, H.; You, H.; Jin, H.; Lin, L.; You, P.; Li, J.; Dai, Z.; et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 86. [Google Scholar] [CrossRef] [PubMed]
- Le, H.B.; Zhu, W.Y.; Chen, D.D.; He, J.Y.; Huang, Y.Y.; Liu, X.G.; Zhang, Y.K. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med. Oncol. 2012, 29, 3190–3197. [Google Scholar] [CrossRef] [PubMed]
- Severino, P.; Brüggemann, H.; Andreghetto, F.M.; Camps, C.; Klingbeil, M.d.F.G.; de Pereira, W.O.; Soares, R.M.; Moyses, R.; Wünsch-Filho, V.; Mathor, M.B.; et al. MicroRNA expression profile in head and neck cancer: HOX-cluster embedded microRNA-196a and microRNA-10b dysregulation implicated in cell proliferation. BMC Cancer 2013, 13, 533. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.C.; Chen, Y.J.; Wang, H.M.; Tsai, C.Y.; Chen, W.H.; Huang, Y.C.; Fan, K.H.; Tsai, C.N.; Huang, S.F.; Kang, C.J.; et al. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev. Res. 2012, 5, 665–674. [Google Scholar] [CrossRef]
- Chen, W.; Cai, F.; Zhang, B.; Barekati, Z.; Zhong, X.Y. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: Potential biomarkers. Tumour Biol. 2013, 34, 455–462. [Google Scholar] [CrossRef]
- Ouyang, H.; Gore, J.; Deitz, S.; Korc, M. microRNA-10b enhances pancreatic cancer cell invasion by suppressing TIP30 expression and promoting EGF and TGF-β actions. Oncogene 2014, 33, 4664–4674. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef]
- Hung, P.S.; Liu, C.J.; Chou, C.S.; Kao, S.Y.; Yang, C.C.; Chang, K.W.; Chiu, T.H.; Lin, S.C. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS ONE 2013, 8, e79926. [Google Scholar] [CrossRef]
- Lu, Y.C.; Chang, J.T.C.; Huang, Y.C.; Huang, C.C.; Chen, W.H.; Lee, L.Y.; Huang, B.S.; Chen, Y.J.; Li, H.F.; Cheng, A.J. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin. Biochem. 2015, 48, 115–121. [Google Scholar] [CrossRef]
- Liu, C.J.; Tsai, M.M.; Tu, H.F.; Lui, M.T.; Cheng, H.W.; Lin, S.C. miR-196a Overexpression and miR-196a2 gene polymorphism are prognostic predictors of oral carcinomas. Ann. Surg. Oncol. 2013, 20, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, Y.; Zhang, L.; Weakley, S.M.; Yao, Q. MicroRNA-196: Critical roles and clinical applications in development and cancer. J. Cell. Mol. Med. 2011, 15, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.M.; Wang, C.S.; Tsai, C.Y.; Huang, C.G.; Lee, K.F.; Huang, H.W.; Lin, Y.H.; Chi, H.C.; Kuo, L.M.; Lu, P.H.; et al. Circulating microRNA-196a/b are novel biomarkers associated with metastatic gastric cancer. Eur. J. Cancer 2016, 64, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Slater, E.P.; Strauch, K.; Rospleszcz, S.; Ramaswamy, A.; Esposito, I.; Klöppel, G.; Matthäi, E.; Heeger, K.; Fendrich, V.; Langer, P.; et al. MicroRNA-196a and -196b as potential biomarkers for the early detection of familial pancreatic cancer. Transl. Oncol. 2014, 7, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.A.; Weng, S.L.; Yang, S.F.; Chou, C.H.; Huang, W.C.; Tu, S.J.; Chang, T.H.; Huang, C.N.; Jong, Y.J.; Huang, H.D. A three-microRNA signature as a potential biomarker for the early detection of oral cancer. Int. J. Mol. Sci. 2018, 19, 758. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, F.; Chen, X.; Liu, Z.; Ouyang, Y.; Zhao, W.; Yu, D. Downregulation of miR-221/222 by a microRNA sponge promotes apoptosis in oral squamous cell carcinoma cells through upregulation of PTEN. Oncol. Lett. 2016, 12, 4419–4426. [Google Scholar] [CrossRef]
- Li, Z.; Tao, Y.; Wang, X.; Jiang, P.; Li, J.; Peng, M.; Zhang, X.; Chen, K.; Liu, H.; Zhen, P.; et al. Tumor-secreted exosomal miR-222 promotes tumor progression via regulating p27 expression and re-localization in pancreatic cancer. Cell. Physiol. Biochem. 2018, 51, 610–629. [Google Scholar] [CrossRef]
- Zhang, R.; Pang, B.; Xin, T.; Guo, H.; Xing, Y.; Xu, S.; Feng, B.; Liu, B.; Pang, Q. Plasma miR-221/222 family as novel descriptive and prognostic biomarkers for glioma. Mol. Neurobiol. 2016, 53, 1452–1460. [Google Scholar] [CrossRef]
- Roy, R.; Singh, R.; Chattopadhyay, E.; Ray, A.; De Sarkar, N.; Aich, R.; Paul, R.R.; Pal, M.; Roy, B. MicroRNA and target gene expression based clustering of oral cancer, precancer and normal tissues. Gene 2016, 593, 58–63. [Google Scholar] [CrossRef]
- Lin, J.; Huang, S.; Wu, S.; Ding, J.; Zhao, Y.; Liang, L.; Tian, Q.; Zha, R.; Zhan, R.; He, X. MicroRNA-423 promotes cell growth and regulates G1/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis 2011, 32, 1641–1647. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, A.; Zhang, Z.; Tian, R.; Luo, A.; Li, M.; Zhao, D.; Fu, L.; Fu, L.; Dong, J.T.; et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumor Biol. 2015, 36, 4763–4771. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zeng, X.; Huang, Y.; Chen, S.; Lin, F.; Yang, G.; Yang, N. miR-423-5p serves as a diagnostic indicator and inhibits the proliferation and invasion of ovarian cancer. Exp. Ther. Med. 2018, 15, 4723–4730. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Yu, T.; An, Q.; Cao, X.; Pan, H. MicroRNA-423-5p inhibits colon cancer growth by promoting caspase-dependent apoptosis. Exp. Ther. Med. 2018, 16, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Fayyad-Kazan, H.; Bitar, N.; Najar, M.; Lewalle, P.; Fayyad-Kazan, M.; Badran, R.; Hamade, E.; Daher, A.; Hussein, N.; ElDirani, R.; et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J. Transl. Med. 2013, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yin, J.; Li, T.; Yuan, L.; Wang, D.; He, J.; Du, X.; Lu, J. Upregulated circulating miR-150 is associated with the risk of intrahepatic cholangiocarcinoma. Oncol. Rep. 2015, 33, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Aherne, S.T.; Madden, S.F.; Hughes, D.J.; Pardini, B.; Naccarati, A.; Levy, M.; Vodicka, P.; Neary, P.; Dowling, P.; Clynes, M. Circulating miRNAs miR-34a and miR-150 associated with colorectal cancer progression. BMC Cancer 2015, 15, 329. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, H.; Sho, R.; Takeda, Y.; Zhang, X.; Yoshida, Y.; Narimatsu, H.; Otani, K.; Ishikawa, S.; Fukao, A.; Asao, H.; et al. Circulating miR-223 in oral cancer: Its potential as a novel diagnostic biomarker and therapeutic target. PLoS ONE 2016, 11, e0159693. [Google Scholar] [CrossRef]
- Xu, H.; Yang, Y.; Zhao, H.; Yang, X.; Luo, Y.; Ren, Y.; Liu, W.; Li, N. Serum miR-483-5p: A novel diagnostic and prognostic biomarker for patients with oral squamous cell carcinoma. Tumor Biol. 2016, 37, 447–453. [Google Scholar] [CrossRef]
- Sun, G.; Cao, Y.; Wang, P.; Song, H.; Bie, T.; Li, M.; Huai, D. miR-200b-3p in plasma is a potential diagnostic biomarker in oral squamous cell carcinoma. Biomarkers 2018, 23, 137–141. [Google Scholar] [CrossRef]
- Liu, C.J.; Lin, J.S.; Cheng, H.W.; Hsu, Y.H.; Cheng, C.Y.; Lin, S.C. Plasma miR-187* is a potential biomarker for oral carcinoma. Clin. Oral Investig. 2017, 21, 1131–1138. [Google Scholar] [CrossRef]
- Lu, Z.; He, Q.; Liang, J.; Li, W.; Su, Q.; Chen, Z.; Wan, Q.; Zhou, X.; Cao, L.; Sun, J.; et al. miR-31-5p is a potential circulating biomarker and therapeutic target for oral cancer. Mol. Ther. Nucleic Acids 2019, 16, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Kao, S.Y.; Tu, H.F.; Tsai, M.M.; Chang, K.W.; Lin, S.C. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010, 16, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Srivastava, A.N.; Sharma, R.; Mateen, S.; Shukla, B.; Singh, A.; Chandel, S. Circulating microRNA-21 expression as a novel serum biomarker for oral sub-mucous fibrosis and oral squamous cell carcinoma. Asian Pac. J. Cancer Prev. 2018, 19, 1053–1057. [Google Scholar] [PubMed]
- Ren, W.; Qiang, C.; Gao, L.; Li, S.M.; Zhang, L.M.; Wang, X.L.; Dong, J.W.; Chen, C.; Liu, C.Y.; Zhi, K.Q. Circulating microRNA-21 (MIR-21) and phosphatase and tensin homolog (PTEN) are promising novel biomarkers for detection of oral squamous cell carcinoma. Biomarkers 2014, 19, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, N.; Hanif, M.; Ahmed, A.; Jamal, Q.; Mushtaq, S.; Khan, A.; Saqib, M. Circulating miR-21 as a prognostic and predictive biomarker in oral squamous cell carcinoma. Pakistan J. Med. Sci. 2019, 35, 1408–1412. [Google Scholar] [CrossRef]
- Rabinowits, G.; Bowden, M.; Flores, L.M.; Verselis, S.; Vergara, V.; Jo, V.Y.; Chau, N.; Lorch, J.; Hammerman, P.S.; Thomas, T.; et al. Comparative analysis of microRNA expression among benign and malignant tongue tissue and plasma of patients with tongue cancer. Front. Oncol. 2017, 7, 191. [Google Scholar] [CrossRef]
- Schneider, A.; Victoria, B.; Lopez, Y.N.; Suchorska, W.; Barczak, W.; Sobecka, A.; Golusinski, W.; Masternak, M.M.; Golusinski, P. Tissue and serum microRNA profile of oral squamous cell carcinoma patients. Sci. Rep. 2018, 8, 675. [Google Scholar] [CrossRef]
- Pedersen, N.J.; Jensen, D.H.; Lelkaitis, G.; Kiss, K.; Charabi, B.W.; Ullum, H.; Specht, L.; Schmidt, A.Y.; Nielsen, F.C.; von Buchwald, C. MicroRNA-based classifiers for diagnosis of oral cavity squamous cell carcinoma in tissue and plasma. Oral Oncol. 2018, 83, 46–52. [Google Scholar] [CrossRef]
- Spielmann, N.; Wong, D.T. Saliva: Diagnostics and therapeutic perspectives. Oral Dis. 2011, 17, 345–354. [Google Scholar] [CrossRef]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef]
- Byun, J.S.; Hong, S.H.; Choi, J.K.; Jung, J.K.; Lee, H.J. Diagnostic profiling of salivary exosomal microRNAs in oral lichen planus patients. Oral Dis. 2015, 21, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Rapado-González, Ó.; Majem, B.; Álvarez-Castro, A.; Díaz-Peña, R.; Abalo, A.; Suárez-Cabrera, L.; Gil-Moreno, A.; Santamaría, A.; López-López, R.; Muinelo-Romay, L.; et al. A novel saliva-based miRNA signature for colorectal cancer diagnosis. J. Clin. Med. 2019, 8, 2029. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.T. Salivary Diagnostics: Amazing as it might seem, doctors can detect and monitor diseases using molecules found in a sample of spit. Am. Sci. 2008, 96, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [PubMed]
- Arunkumar, G.; Deva Magendhra Rao, A.; Manikandan, M.; Prasanna Srinivasa Rao, H.; Subbiah, S.; Ilangovan, R.; Murugan, A.; Munirajan, A. Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma. Oncol. Lett. 2017, 15, 649–657. [Google Scholar] [CrossRef]
- Hsing, E.W.; Shiah, S.G.; Peng, H.Y.; Chen, Y.W.; Chuu, C.P.; Hsiao, J.R.; Lyu, P.C.; Chang, J.Y. TNF-α-induced miR-450a mediates TMEM182 expression to promote oral squamous cell carcinoma motility. PLoS ONE 2019, 14, e0213463. [Google Scholar] [CrossRef]
- Peng, S.Y.; Tu, H.F.; Yang, C.C.; Wu, C.H.; Liu, C.J.; Chang, K.W.; Lin, S.C. miR-134 targets PDCD7 to reduce E-cadherin expression and enhance oral cancer progression. Int. J. Cancer 2018, 143, 2892–2904. [Google Scholar] [CrossRef]
- Lu, W.C.; Kao, S.Y.; Yang, C.C.; Tu, H.F.; Wu, C.H.; Chang, K.W.; Lin, S.C. EGF up-regulates miR-31 through the C/EBPβ signal cascade in oral carcinoma. PLoS ONE 2014, 9, e108049. [Google Scholar] [CrossRef]
- Liu, C.J.; Lin, S.C.; Yang, C.C.; Cheng, H.W.; Chang, K.W. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck 2012, 34, 219–224. [Google Scholar] [CrossRef]
- Hung, K.F.; Liu, C.J.; Chiu, P.C.; Lin, J.S.; Chang, K.W.; Shih, W.Y.; Kao, S.Y.; Tu, H.F. MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder. Oral Oncol. 2016, 53, 42–47. [Google Scholar] [CrossRef]
- Zahran, F.; Ghalwash, D.; Shaker, O.; Al-Johani, K.; Scully, C. Salivary microRNAs in oral cancer. Oral Dis. 2015, 21, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Trachtenberg, A.J.; Kuo, W.P.; Cheng, Y.S. Genomewide study of salivary microRNAs for detection of oral cancer. J. Dent. Res. 2014, 93, 86S–93S. [Google Scholar] [CrossRef]
- Liu, B.; Chen, W.; Cao, G.; Dong, Z.; Xu, J.; Luo, T.; Zhang, S. MicroRNA-27b inhibits cell proliferation in oral squamous cell carcinoma by targeting FZD7 and Wnt signaling pathway. Arch. Oral Biol. 2017, 83, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Duz, M.B.; Karatas, O.F.; Guzel, E.; Turgut, N.F.; Yilmaz, M.; Creighton, C.J.; Ozen, M. Identification of miR-139-5p as a saliva biomarker for tongue squamous cell carcinoma: A pilot study. Cell. Oncol. 2016, 39, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yu, T.; Cabay, R.J.; Jin, Y.; Mahjabeen, I.; Luan, X.; Huang, L.; Dai, Y.; Zhou, X. miR-486-3p, miR-139-5p, and miR-21 as biomarkers for the detection of oral tongue squamous cell carcinoma. Biomark. Cancer 2017, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, T.; Wong, D.T. Saliva-exosomics in cancer: Molecular characterization of cancer-derived exosomes in saliva. Enzymes 2017, 42, 125–151. [Google Scholar]
- Gai, C.; Camussi, F.; Broccoletti, R.; Gambino, A.; Cabras, M.; Molinaro, L.; Carossa, S.; Camussi, G.; Arduino, P.G. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. BMC Cancer 2018, 18, 439. [Google Scholar] [CrossRef]
- Yap, T.; Seers, C.; Koo, K.; Cheng, L.; Vella, L.J.; Hill, A.F.; Reynolds, E.; Nastri, A.; Cirillo, N.; McCullough, M. Non-invasive screening of a microRNA-based dysregulation signature in oral cancer and oral potentially malignant disorders. Oral Oncol. 2019, 96, 113–120. [Google Scholar] [CrossRef]
- Sazanov, A.A.; Kiselyova, E.V.; Zakharenko, A.A.; Romanov, M.N.; Zaraysky, M.I. Plasma and saliva miR-21 expression in colorectal cancer patients. J. Appl. Genet. 2017, 58, 231–237. [Google Scholar] [CrossRef]
- Xie, Z.J.; Chen, G.; Zhang, X.C.; Li, D.F.; Huang, J.; Li, Z.J. Saliva supernatant miR-21: A novel potential biomarker for esophageal cancer detection. Asian Pac. J. Cancer Prev. 2012, 13, 6145–6149. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, G.; Zhang, X.; Li, D.; Huang, J.; Yang, C.; Zhang, P.; Qin, Y.; Duan, Y.; Gong, B.; et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS ONE 2013, 8, e57502. [Google Scholar] [CrossRef] [PubMed]
- Humeau, M.; Vignolle-Vidoni, A.; Sicard, F.; Martins, F.; Bournet, B.; Buscail, L.; Torrisani, J.; Cordelier, P. Salivary microRNA in pancreatic cancer patients. PLoS ONE 2015, 10, e0130996. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, J.S.; Attumi, T.; Opekun, A.R.; Abraham, B.; Hou, J.; Shelby, H.; Graham, D.Y.; Streckfus, C.; Klein, J.R. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Mehdipour, M.; Shahidi, M.; Manifar, S.; Jafari, S.; Mashhadi, F.M.; Barati, M.; Mortazavi, H.; Shirkhoda, M.; Farzanegan, A.; Rankohi, Z.E. Diagnostic and prognostic relevance of salivary microRNA-21, -125a, -31 and -200a levels in patients with oral lichen planus - a short report. Cell. Oncol. 2018, 41, 329–334. [Google Scholar] [CrossRef]
- Stasio, D.D.; Mosca, L.; Lucchese, A.; Cave, D.D.; Kawasaki, H.; Lombardi, A.; Porcelli, M.; Caraglia, M. Salivary mir-27b expression in oral lichen planus patients: A series of cases and a narrative review of litterature. Curr. Top. Med. Chem. 2019, 21. [Google Scholar] [CrossRef]
- Jakob, M.; Mattes, L.M.; Küffer, S.; Unger, K.; Hess, J.; Bertlich, M.; Haubner, F.; Ihler, F.; Canis, M.; Weiss, B.G.; et al. MicroRNA expression patterns in oral squamous cell carcinoma: hsa-mir-99b-3p and hsa-mir-100-5p as novel prognostic markers for oral cancer. Head Neck 2019, 41, 3499–3515. [Google Scholar] [CrossRef]
- Sun, L.; Liu, L.; Fu, H.; Wang, Q.; Shi, Y. Association of decreased expression of serum miR-9 with poor prognosis of oral squamous cell carcinoma patients. Med. Sci. Monit. 2016, 22, 289–294. [Google Scholar] [CrossRef]
- Yan, Y.; Wang, X.; Venø, M.T.; Bakholdt, V.; Sørensen, J.A.; Krogdahl, A.; Sun, Z.; Gao, S.; Kjems, J. Circulating miRNAs as biomarkers for oral squamous cell carcinoma recurrence in operated patients. Oncotarget 2017, 8, 8206–8214. [Google Scholar] [CrossRef]
- Chen, L.; Hu, J.; Pan, L.; Yin, X.; Wang, Q.; Chen, H. Diagnostic and prognostic value of serum miR-99a expression in oral squamous cell carcinoma. Cancer Biomark. 2018, 23, 333–339. [Google Scholar] [CrossRef]
- Shi, J.; Bao, X.; Liu, Z.; Zhang, Z.; Chen, W.; Xu, Q. Serum miR-626 and miR-5100 are promising prognosis predictors for oral squamous cell carcinoma. Theranostics 2019, 9, 920–931. [Google Scholar] [CrossRef]
- Nakashima, H.; Yoshida, R.; Hirosue, A.; Kawahara, K.; Sakata, J.; Arita, H.; Yamamoto, T.; Toya, R.; Murakami, R.; Hiraki, A.; et al. Circulating miRNA-1290 as a potential biomarker for response to chemoradiotherapy and prognosis of patients with advanced oral squamous cell carcinoma: A single-center retrospective study. Tumor Biol. 2019, 41, 101042831982685. [Google Scholar] [CrossRef]
- Mollaei, H.; Safaralizadeh, R.; Rostami, Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019, 234, 12369–12384. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Weiler, J.; Hunziker, J.; Hall, J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease? Gene Ther. 2006, 13, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. MicroRNA sponges: Progress and possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef]
- Choi, W.Y.; Giraldez, A.J.; Schier, A.F. Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR-430. Science 2007, 318, 271–274. [Google Scholar] [CrossRef]
- Gumireddy, K.; Young, D.D.; Xiong, X.; Hogenesch, J.B.; Huang, Q.; Deiters, A. Small-molecule inhibitors of microRNA miR-21 function. Angew. Chemie Int. Ed. 2008, 47, 7482–7484. [Google Scholar] [CrossRef]
| Study | Preanalytical Variables | RNA Extraction | Study Cohort | Technique | Molecular Profile (oral cancer vs. HC) | Sensitivity/Specificity (%) | AUC |
|---|---|---|---|---|---|---|---|
| Park et al., 2009 [74] | Cell-free saliva Preservation: SUPERase·In (Ambion) | Volume: 400 μL Kit: mirVana miRNA Isolation (Ambion) | 50 OSCC 50 HC | RT-preamp-qPCR | miR-200a (↓) miR-125a (↓) miR-200a + miR-125a (↓) | ND | 0.65 0.62 0.66 |
| Liu et al., 2012 [79] | Cell-free saliva Storage: –80 °C | Volume: 600 µL Kit: mirVana PARIS Isolation (Ambion) | 45 OSCC 10 OVL 24 HC | TaqMan qRT-PCR | miR-31 (↑) | 80/68 | 0.82 |
| Momen-Heravi et al., 2014 [83] | Cell-free saliva Centrifugation: 2600 × g 15 min at 4 °C Preservation: 5 µL of SUPERase·In (Ambion) per milliliter of supernatant Storage: −80 °C | Volume: 440 µL Kit: RNeasy (Qiagen) | 9 OSCC 8 OSCC-R 8 LP 9 HC | TaqMan qRT-PCR | miR-27b (↑) miR-136 (↓) | 85.71/100 88.89/100 | 0.964 0.968 |
| Zahran et al., 2015 [81] | Cell-free saliva Centrifugation: (i) 2500 × g 10 min at 4 °C, (ii) 10,000 × g 1 min | Volume: 200 µL Kit: miRNeasy serum/plasma extraction (Qiagen) | 20 OSCC 40 OPMD 20 RAS 20 HC | SYBR Green qRT-PCR | miR-21 (↑) miR-145 (↓) miR-184 (↑) | NS | NS |
| Duz et al., 2016 [84] | Cell-free saliva Centrifugation: 2600 × g 15 min at 4 °C Storage: −80 ◦C | Volume: NS Kit: mirVana PARIS (Ambion) | 25 TSCC 25 HC | TaqMan qRT-PCR | miR-139-5p (↓) | 73.9/85 | 0.805 |
| Gai et al., 2018 [87] | Cell-free saliva Centrifugation: 2600 × g 15 min at 4 °C Storage: −80 ◦C | Volume: 250 µL Kit: mirVana Isolation (Thermo Fisher Scientific) | 21 OSCC 11 HC | SYBR Green qRT-PCR | miR-512-3p (↑) miR-412-3p (↑) | ND | 0.847 0.871 |
| Yap et al. 2019 [88] | Cell-free oral swirls Centrifugation: 4000 × g 4 min at 4 °C Storage: −20 °C | Volume: NS Kit: mirVana Isolation (Life Technologies) | 53 OSCC 54 NMA 9 HNE 74 OPMD | qRT-PCR | miR-24-3p + miR-21-5p + miR-99a-5p + let-7c-5p + miR-100-5p (↑) | NS | 0.867 |
| Study | Preanalytical Variables | RNA Extraction | Study Cohort | Technique | Molecular Profile (oral cancer vs. HC) | Sensitivity/Specificity (%) | AUC | Clinical Application |
|---|---|---|---|---|---|---|---|---|
| Wong et al., 2008 [26] | Plasma | Volume: NS Kit: mirVana miRNA Isolation (Ambion) | 30 TSCC 38 HC | TaqMan qRT-PCR | miR-184 (↑) | ND | ND | Diagnosis |
| Lin et al., 2010 [29] | Plasma | Volume: NS Kit: NS | 33 OSCC 10 HC | qRT-PCR | miR-24 (↑) | 70/92 | 0.82 | Diagnosis |
| Liu et al., 2010 [62] | Plasma | Volume: 600 µL Kit: mirVana PARIS Isolation (Ambion) | 43 OSCC 21 HC | TaqMan qRT-PCR | miR-31 (↑) | ND | 0.82 | Diagnosis |
| Lu et al., 2012 [35] | Plasma | Volume: 200 µL Kit: miRNeasy Mini (Qiagen) | 54 OSCC 7 OLK 36 HC | TaqMan qRT-PCR | miR-10b (↑) | 94.4/80 | 0.932 | Diagnosis |
| Hung et al., 2013 [39] | Plasma | Volume: NS Kit: NS | 51 OSCC 12 HC | TaqMan qRT-PCR | miR-146a (↑) | 79/92 | 0.86 | Diagnosis |
| Liu et al., 2013 [41] | Plasma | Volume: NS Kit: mirVana PARIS (Ambion) | 65 OSCC 24 HC | TaqMan qRT-PCR | miR-196a (↑) | ND | 0.75 | Diagnosis and prognosis |
| Lu et al., 2015 [40] | Plasma | Volume: 200 µL Kit: miRNeasy mini (Qiagen) | 90 OC 16 OPMD 53 HC | TaqMan qRT-PCR | miR-196a (↑) miR-196b (↑) miR-196a + miR-196b (↑) | 66.7/96.2 97.8/81.1 87.8/92.5 | 0.864 0.960 0.963 | Diagnosis |
| Xu et al., 2016 [58] | Serum Storage: −80 °C | Volume: NS Kit: miRNeasy RNA Isolation (Qiagen) | 101 OSCC 103 HC | SYBR Green qRT-PCR | miR-483-5p (↑) | 85.3/74.6 | 0.85 | Diagnosis and prognosis |
| Sun et al., 2016 [97] | Serum Centrifugation: 3500 × g 5 min Storage: −80 °C | Volume: NS Kit: miRNeasy (Qiagen) | 104 OSCC 30 OLK 40 HC | SYBR Green qRT-PCR | miR-9 (↓) | ND | ND | Diagnosis and prognosis |
| Tachibana et al., 2016 [57] | Plasma Storage: −80 °C | Volume: 100 μL Kit: miRNeasy (Qiagen) | 31 GSCC 31 HC | qRT-PCR | miR-223 (↑) | 67.7/61.3 | 0.703 | Diagnosis |
| Liu et al., 2017 [60] | Plasma | Volume: NS Kit: mirVana PARIS Isolation (Ambion) | 63 OSCC 26 HC | qRT-PCR | miR-187-5p (↑) | ND | 0.73 | Diagnosis and prognosis |
| Chang et al., 2018 [45] | Plasma Centrifugation: 3000 × g 10 min Storage: −80 °C | Volume: 200 µL Kit: miRNeasy Serum/Plasma (Qiagen) | 70 HC 66 OLK 114 OSCC | SYBR Green qRT-PCR | miR-150-5p (↑) miR-423-5p (↑) miR-150-5p + miR-423-5p (↑) | 60.55/77.14 58.72/72.86 70.91/72.86 | 0.702 0.677 0.749 | Diagnosis |
| Sun et al., 2018 [59] | Plasma Storage: −80 °C | Volume: 200 μL Kit: miRNeasy Mini (Qiagen) | 80 OSCC 80 HC | TaqMan qRT-PCR | miR-200b-3p (↑) | 90/88.75 | 0.917 | Diagnosis and prognosis |
| Chen et al., 2018 [99] | Serum Centrifugation: 3500 × g 5 min Storage: −80 °C | Volume: NS Kit: TRIzol Reagent | 121 OSCC 55 HC | TaqMan qRT-PCR | miR-99a (↓) | 80.2/83.6 | 0.911 | Diagnosis and prognosis |
| Pedersen et al., 2018 [68] | Plasma | Volume: NS Kit: miRCURY RNA isolation (Exiqon) | 55 OSCC 15 HC | TaqMan qRT-PCR | miR-30a-5p (↑) miR-370-3p (↑) miR-144-5p (↑) miR-769-5p (↑) miR-30a-5p + miR-769-5p (↑) | ND | 0.97 ND ND 0.94 1 | Diagnosis |
| Mahmood et al., 2019 [65] | Plasma | Volume: NS Kit: Favorgen Nucleic acid extraction | 100 OSCC 100 HC | SYBR Green qRT-PCR | miR-21 (↑) | 91/54 | 0.829 | Diagnosis |
| Nakashima et al., 2019 [101] | Plasma | Volume: NS Kit: miRNeasy Serum/Plasma (Qiagen) | 55 OSCC 10 HC | SYBR Green qRT-PCR | miR-1290 (↓) | ND | ND | Diagnosis, prognosis and predictive resistance to therapy |
| Lu et al., 2019 [61] | Serum Centrifugation: (i) 4000 rpm 10 min 4 °C, (ii) 12,000 rpm 15 min 4 °CStorage: −80 °C | Volume: NS Kit: miRcute miRNA Isolation (Tiangen Biotech) | 82 OSCC 53 HC | SYBR Green qRT-PCR | miR-31-5p (↑) | 69.8/52.4 | 0.661 | Diagnosis and therapeutic target |
| Shi et al., 2019 [100] | Serum | Volume: 200 µL Kit: miRNeasy Mini Kit (Qiagen) | 218 OSCC 90 HC | TaqMan qRT-PCR | miR-626 (↑) miR-5100 (↑) | 76.8/77.3 | 0.771 | Prognosis |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rapado-González, Ó.; López-López, R.; López-Cedrún, J.L.; Triana-Martínez, G.; Muinelo-Romay, L.; Suárez-Cunqueiro, M.M. Cell-Free microRNAs as Potential Oral Cancer Biomarkers: From Diagnosis to Therapy. Cells 2019, 8, 1653. https://doi.org/10.3390/cells8121653
Rapado-González Ó, López-López R, López-Cedrún JL, Triana-Martínez G, Muinelo-Romay L, Suárez-Cunqueiro MM. Cell-Free microRNAs as Potential Oral Cancer Biomarkers: From Diagnosis to Therapy. Cells. 2019; 8(12):1653. https://doi.org/10.3390/cells8121653
Chicago/Turabian StyleRapado-González, Óscar, Rafael López-López, José Luis López-Cedrún, Gabriel Triana-Martínez, Laura Muinelo-Romay, and María Mercedes Suárez-Cunqueiro. 2019. "Cell-Free microRNAs as Potential Oral Cancer Biomarkers: From Diagnosis to Therapy" Cells 8, no. 12: 1653. https://doi.org/10.3390/cells8121653
APA StyleRapado-González, Ó., López-López, R., López-Cedrún, J. L., Triana-Martínez, G., Muinelo-Romay, L., & Suárez-Cunqueiro, M. M. (2019). Cell-Free microRNAs as Potential Oral Cancer Biomarkers: From Diagnosis to Therapy. Cells, 8(12), 1653. https://doi.org/10.3390/cells8121653







