STK3/4 Expression Is Regulated in Uterine Endometrial Cells during the Estrous Cycle
Abstract
1. Introduction
2. Material and Methods
2.1. Animal Management
2.2. Estrous Cycle Validation and Uterus Sampling
2.3. Histological and Immunostaining Analysis
2.4. Ovariectomy and Hormone Treatments
2.5. RNA Preparation, RT-PCR, and qRT-PCR
2.6. Knockdown of STK4 Expression in Human Uterine Endometrial Cells
2.7. Western Blot and Statistics
3. Results
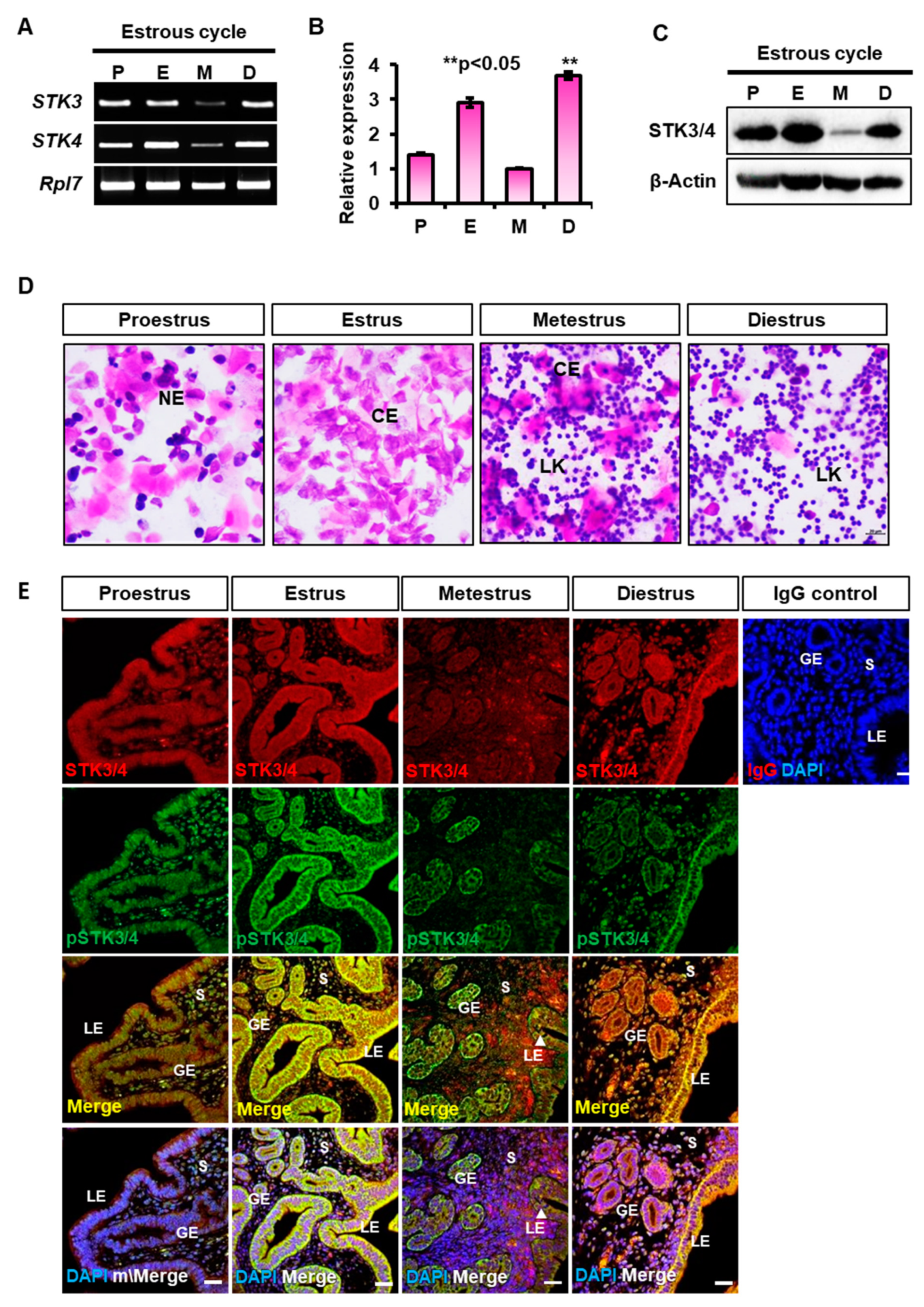
3.1. Expression of STK3 and STK4 in the Mouse Uteri during the Estrous Cycle
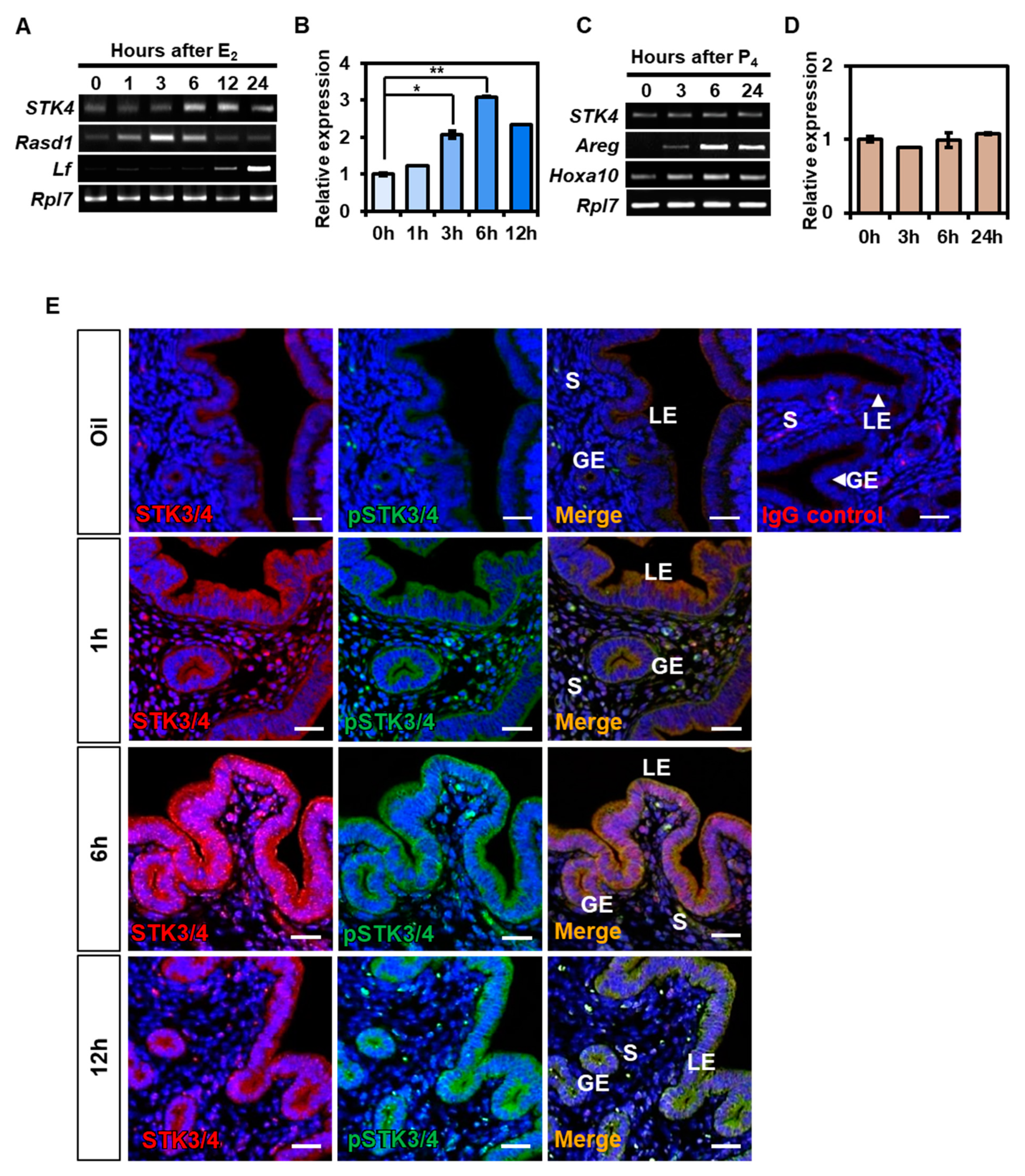
3.2. Hormonal Regulation of STK3/4 Expression in Mouse Uterus
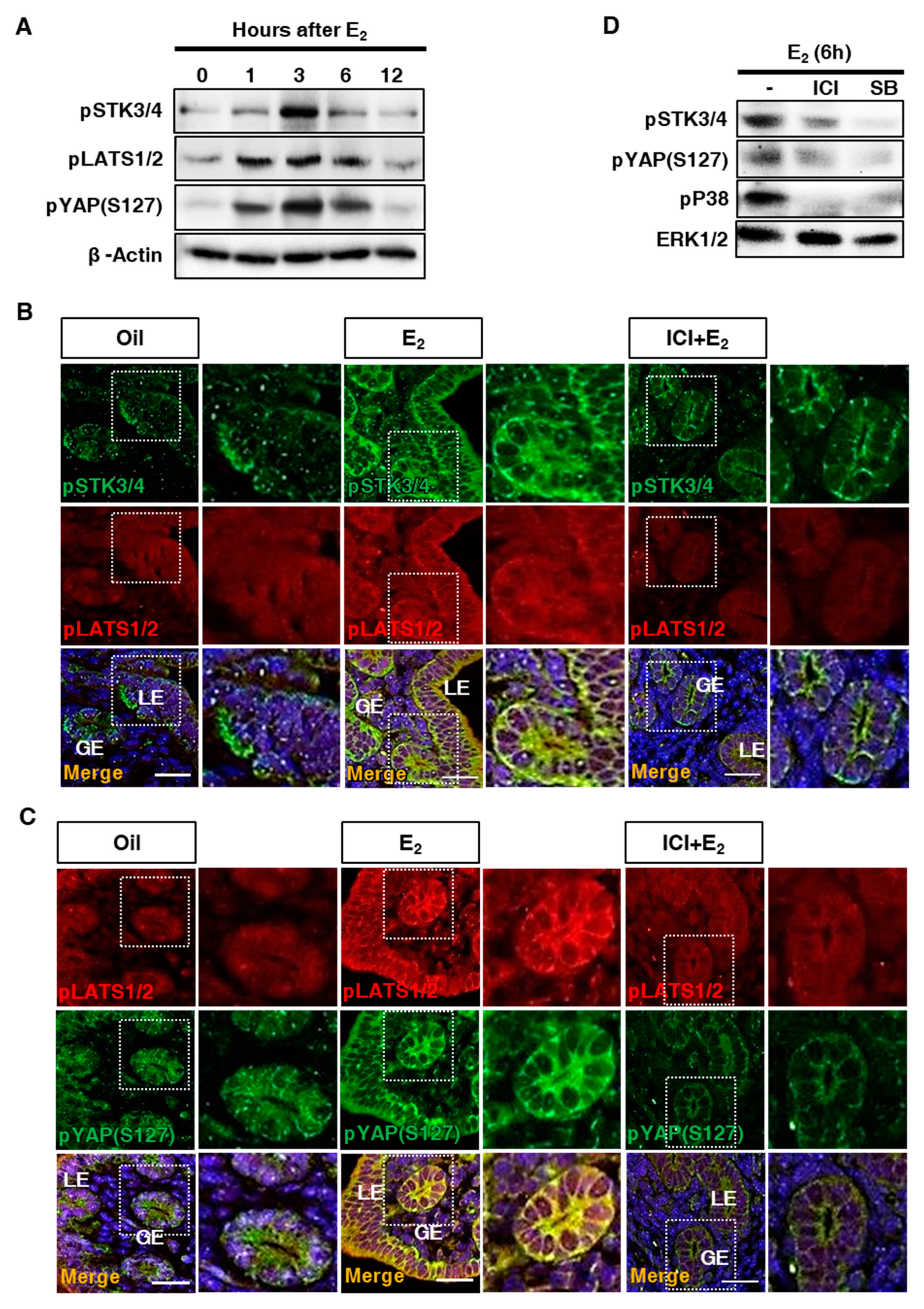
3.3. Estrogen-Dependent Expression of STK3/4 is Mediated via Estrogen Receptor
3.4. Regulation of Downstream Factors in the Hippo Signaling Pathway in the Uterus
3.5. Knockdown of STK3/4 in a Human Endometrial Cell Line, Ishikawa Cell
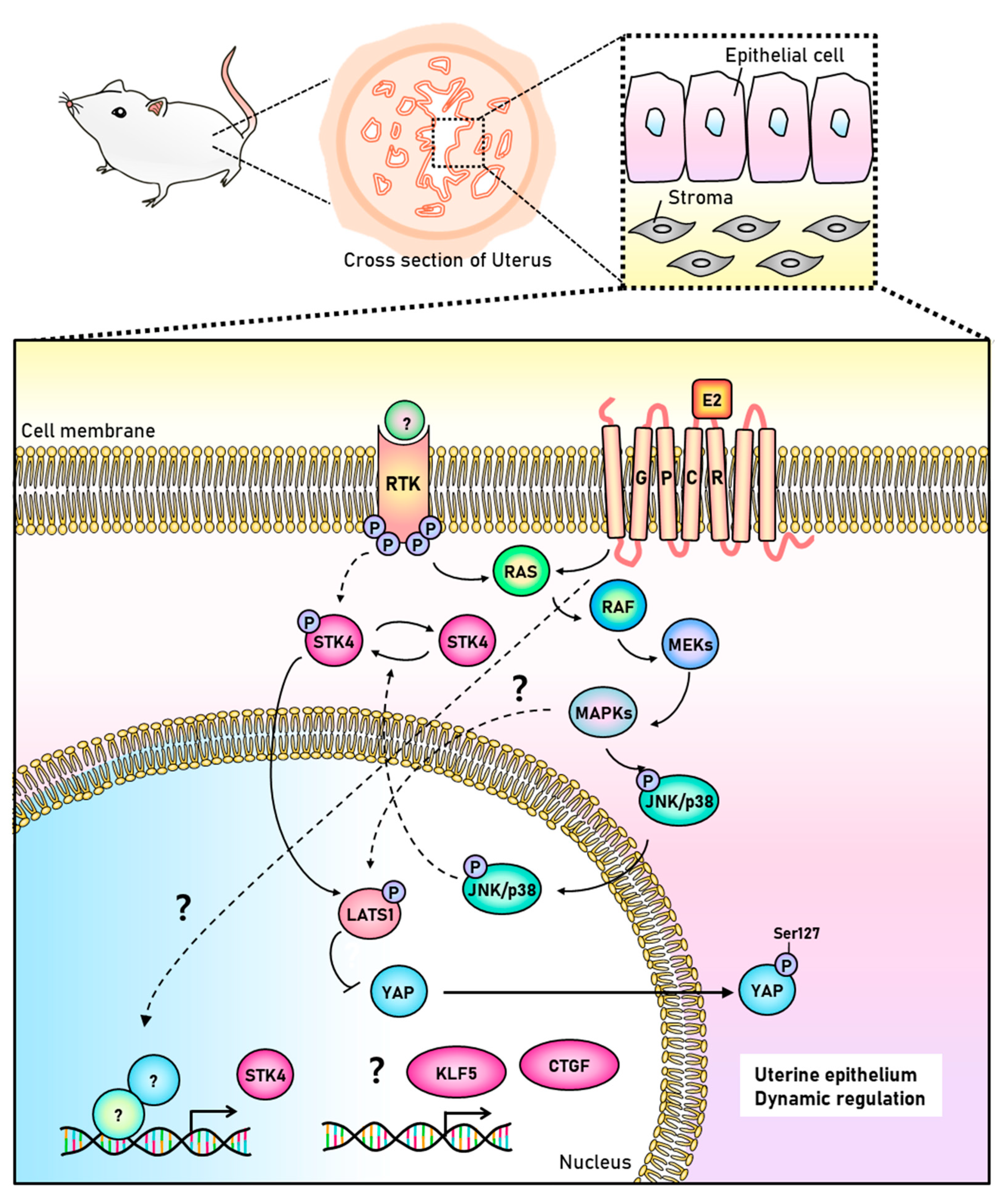
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Bae, S.; Kwon, H.; Yoon, H.; Park, M.; Kim, H.R.; Song, H.; Hong, K.; Choi, Y. Estrogen-dependent expression of sine oculis homeobox 1 in the mouse uterus during the estrous cycle. Biochem. Biophys. Res. Commun. 2016, 472, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-R.; Cho, K.-S.; Kim, E.; Lee, O.-H.; Yoon, H.; Lee, S.; Moon, S.; Park, M.; Hong, K.; Na, Y. Rapid expression of RASD1 is regulated by estrogen receptor-dependent intracellular signaling pathway in the mouse uterus. Mol. Cell. Endocrinol. 2017, 446, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Byers, S.L.; Wiles, M.V.; Dunn, S.L.; Taft, R.A. Mouse estrous cycle identification tool and images. PLoS ONE 2012, 7, e35538. [Google Scholar] [CrossRef] [PubMed]
- Champlin, A.K.; Dorr, D.L.; Gates, A.H. Determining the stage of the estrous cycle in the mouse by the appearance of the vagina. Biol. Reprod. 1973, 8, 491–494. [Google Scholar] [CrossRef]
- Yip, K.S.; Suvorov, A.; Connerney, J.; Lodato, N.J.; Waxman, D.J. Changes in mouse uterine transcriptome in estrus and proestrus. Biol. Reprod. 2013, 89, 13. [Google Scholar] [CrossRef]
- Yu, F.X.; Guan, K.L. The Hippo pathway: Regulators and regulations. Genes Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef]
- Strakova, Z.; Reed, J.; Ihnatovych, I. Human transcriptional coactivator with PDZ-binding motif (TAZ) is downregulated during decidualization. Biol. Reprod. 2010, 82, 1112–1118. [Google Scholar] [CrossRef]
- Jeong, J.H.; Park, M.; Park, M.; Lim, E.J.; Kim, H.R.; Song, H.; Park, S.G.; Choi, E.J.; Hong, K.H.; Lee, D.R.; et al. The expression of aminoacyl-tRNA-synthetase-interacting multifunctional protein-1 (Aimp1) is regulated by estrogen in the mouse uterus. Mol. Cell. Endocrinol. 2015, 399, 78–86. [Google Scholar] [CrossRef]
- Parkening, T.; Collins, T.; Smith, E. Plasma and pituitary concentrations of luteinizing hormone, follicle-stimulating hormone and prolactin in aged, ovariectomized CD-1 and C57BL/6 mice. Exp. Gerontol. 1982, 17, 437–443. [Google Scholar] [CrossRef]
- Mettus, R.V.; Rane, S.G. Characterization of the abnormal pancreatic development, reduced growth and infertility in Cdk4 mutant mice. Oncogene 2003, 22, 8413–8421. [Google Scholar] [CrossRef][Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Galan, J.A.; Avruch, J. MST1/MST2 Protein Kinases: Regulation and Physiologic Roles. Biochemistry 2016, 55, 5507–5519. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.H.; Nousiainen, M.; Chalamalasetty, R.B.; Schafer, A.; Nigg, E.A.; Sillje, H.H. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 2005, 24, 2076–2086. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Li, L.; Lei, Q.; Guan, K.-L. The Hippo–YAP pathway in organ size control and tumorigenesis: An updated version. Genes Dev. 2010, 24, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Cuenda, A.; Rouse, J.; Doza, Y.N.; Meier, R.; Cohen, P.; Gallagher, T.F.; Young, P.R.; Lee, J.C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995, 364, 229–233. [Google Scholar]
- Zhi, X.; Zhao, D.; Zhou, Z.; Liu, R.; Chen, C. YAP promotes breast cell proliferation and survival partially through stabilizing the KLF5 transcription factor. Am. J. Pathol. 2012, 180, 2452–2461. [Google Scholar] [CrossRef]
- Ma, K.; Xu, Q.; Wang, S.; Zhang, W.; Liu, M.; Liang, S.; Zhu, H.; Xu, N. Nuclear accumulation of Yes-Associated Protein (YAP) maintains the survival of doxorubicin-induced senescent cells by promoting survivin expression. Cancer Lett. 2016, 375, 84–91. [Google Scholar] [CrossRef]
- Di Benedetto, A.; Mottolese, M.; Sperati, F.; Ercolani, C.; Di Lauro, L.; Pizzuti, L.; Vici, P.; Terrenato, I.; Sperduti, I.; Shaaban, A.M.; et al. The Hippo transducers TAZ/YAP and their target CTGF in male breast cancer. Oncotarget 2016, 7, 43188–43198. [Google Scholar] [CrossRef]
- Fitamant, J.; Kottakis, F.; Benhamouche, S.; Tian, H.S.; Chuvin, N.; Parachoniak, C.A.; Nagle, J.M.; Perera, R.M.; Lapouge, M.; Deshpande, V.; et al. YAP Inhibition Restores Hepatocyte Differentiation in Advanced HCC, Leading to Tumor Regression. Cell Rep. 2015, 10, 1692–1707. [Google Scholar] [CrossRef]
- Mo, J.-S.; Meng, Z.; Kim, Y.C.; Park, H.W.; Hansen, C.G.; Kim, S.; Lim, D.-S.; Guan, K.-L. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat. Cell Biol. 2015, 17, 500–510. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Mottier-Pavie, V.; Plouffe, S.W.; Hansen, C.G.; Hong, A.W.; Park, H.W.; Mo, J.-S.; Lu, W.; Lu, S. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat. Commun. 2015, 6, 8357. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Honjo, S.; Jin, J.; Chang, S.S.; Scott, A.W.; Chen, Q.; Kalhor, N.; Correa, A.M.; Hofstetter, W.L.; Albarracin, C.T.; et al. The Hippo Coactivator YAP1 Mediates EGFR Overexpression and Confers Chemoresistance in Esophageal Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 2580–2590. [Google Scholar] [CrossRef] [PubMed]
- Zhan, M.; Ikeda, J.I.; Wada, N.; Hori, Y.; Nojima, S.; Tahara, S.I.; Ueda, Y.; Yoshino, K.; Kimura, T.; Morii, E. Prognostic significance of a component of the Hippo pathway, TAZ, in human uterine endometrioid adenocarcinoma. Oncol. Lett. 2016, 11, 3611–3616. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Song, Y.; Yang, S.; Fu, J.; Feng, X.; Huang, W. YAP mediates human decidualization of the uterine endometrial stromal cells. Placenta 2017, 53, 30–35. [Google Scholar] [CrossRef]
- Dan, I.; Watanabe, N.M.; Kusumi, A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001, 11, 220–230. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, W.; Liu, B.; Deng, H.; Uster, E.; Pan, D. Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev. Cell 2015, 34, 642–655. [Google Scholar] [CrossRef]
- Britschgi, A.; Duss, S.; Kim, S.; Couto, J.P.; Brinkhaus, H.; Koren, S.; De Silva, D.; Mertz, K.D.; Kaup, D.; Varga, Z. The Hippo kinases LATS1 and 2 control human breast cell fate via crosstalk with ERα. Nature 2017, 541, 541–545. [Google Scholar] [CrossRef]
- Hong, K.; Choi, Y. Role of estrogen and RAS signaling in repeated implantation failure. BMB Rep. 2018, 51, 225–229. [Google Scholar] [CrossRef]
- Huang, C.-C.; Orvis, G.D.; Wang, Y.; Behringer, R.R. Stromal-to-epithelial transition during postpartum endometrial regeneration. PLoS ONE 2012, 7, e44285. [Google Scholar] [CrossRef]


| Genes | Accession No. | Primer Sequence | Annealing Temperature (°C) | Product Size (bp) |
|---|---|---|---|---|
| Stk4 | NM_021420.3 | (1) Fwd; 5′-CAGCCCGAGGAAGTGTTTGA-3′ (2) Rev; 5′-CACGGGCACTTGCTTGATTG-3′ | 60 | 117 |
| Stk3 | NM_019635.2 | (1) Fwd; 5′-TGGTGAAGAGTCCTGAGCAG-3′ (2) Rev; 5′-CCACGCTTTCTGAACTCGTC-3′ | 60 | 228 |
| Rpl7 | NM_011291.5 | (1) Fwd; 5′-TCAATGGAGTAAGCCCAAAG-3′ (2) Rev; 5′-GAAGAGACCGAGCAATCAAG-3′ | 60 | 246 |
| Lf | NM_008522.3 | (1) Fwd; 5′-AGGAAAGCCCCCCTACAAAC-3′ (2) Rev; 5′-GGAACACAGCTCTTTGAGAAGAAC-3′ | 58 | 141 |
| Rasd1 | NM_009026 | (1) Fwd; 5′-GATGTGCCCAAGCGACTCT-3′ (2) Rev; 5′-TGAGGAAGCGCGACACAAT-3′ | 60 | 110 |
| Areg | NM_009704.4 | (1) Fwd; 5′-GGTCTTAGGCTCAGGCCATTA-3′ (2) Rev; 5′-CGCTTATGGTGGAAACCTCTC-3′ | 60 | 137 |
| Hoxa10 | NM_008263.3 | (1) Fwd; 5′-CCTGCCGCGAACTCCTTTT-3′ (2) Rev; 5′-GGCGCTTCATTACGCTTGC-3′ | 60 | 203 |
| Gapdh | BC092294 | (1) Fwd; 5′-AGGTCGGTGTGAACGGATTTG-3′ (2) Rev; 5′-TGTAGACCATGTAGTTGAGGTCA-3′ | 60 | 123 |
| STK4 | NM_006282.3 | (1) Fwd; 5′-CACCCATTTGTCAGGAGTGC-3′ (2) Rev; 5′-GAACCATCGTGCCAGAATCC-3′ | 65 | 163 |
| VEFGA | NM_001171623.1 | (1) Fwd; 5′-GGCCAGCACATAGGAGAGAT-3′ (2) Rev; 5′-ACGCTCCAGGACTTATACCG-3′ | 65 | 155 |
| KLF5 | NM_001730.4 | (1) Fwd; 5′-ACCCTGCCAGTTAACTCACA-3′ (2) Rev; 5′-ACCAGGGTAATCGCAGTAGT-3′ | 65 | 102 |
| BIRC5 | NM_001012271.1 | (1) Fwd; 5′-AGGACCACCGCATCTCTACAT-3′ (2) Rev; 5′-AAGTCTGGCTCGTTCTCAGTG-3′ | 60 | 118 |
| CTGF | NM_001901.2 | (1) Fwd; 5′-TCTTCGGTGGTACGGTGTAC-3′ (2) Rev; 5′-TGTCTTCCAGTCGGTAAGCC-3′ | 63 | 249 |
| E2F | NM_005225.2 | (1) Fwd; 5′-CTTCGTAGCATTGCAGACCC-3′ (2) Rev; 5′-AAAACATCGATCGGGCCTTG-3′ | 65 | 137 |
| CTNNB1 | NM_001904.3 | (1) Fwd; 5′-CTTACACCCACCATCCCACT-3′ (2) Rev; 5′-CCTCCACAAATTGCTGCTGT-3′ | 65 | 197 |
| GAPDH | AY340484.1 | (1) Fwd; 5′-ATGGGGAAGGTGAAGGTCG-3′ (2) Rev; 5′-GGGTCATTGATGGCAACAAT-3′ | 60 | 107 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, S.; Lee, O.-H.; Lee, S.; Lee, J.; Park, H.; Park, M.; Chang, E.M.; Park, K.-H.; Choi, Y. STK3/4 Expression Is Regulated in Uterine Endometrial Cells during the Estrous Cycle. Cells 2019, 8, 1643. https://doi.org/10.3390/cells8121643
Moon S, Lee O-H, Lee S, Lee J, Park H, Park M, Chang EM, Park K-H, Choi Y. STK3/4 Expression Is Regulated in Uterine Endometrial Cells during the Estrous Cycle. Cells. 2019; 8(12):1643. https://doi.org/10.3390/cells8121643
Chicago/Turabian StyleMoon, Sohyeon, Ok-Hee Lee, Sujin Lee, Jihyun Lee, Haeun Park, Miseon Park, Eun Mi Chang, Keun-Hong Park, and Youngsok Choi. 2019. "STK3/4 Expression Is Regulated in Uterine Endometrial Cells during the Estrous Cycle" Cells 8, no. 12: 1643. https://doi.org/10.3390/cells8121643
APA StyleMoon, S., Lee, O.-H., Lee, S., Lee, J., Park, H., Park, M., Chang, E. M., Park, K.-H., & Choi, Y. (2019). STK3/4 Expression Is Regulated in Uterine Endometrial Cells during the Estrous Cycle. Cells, 8(12), 1643. https://doi.org/10.3390/cells8121643






