The Host Factor Erlin-1 is Required for Efficient Hepatitis C Virus Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Plasmids, Antibodies and Reagents
2.2. Silencing of Erlin Proteins by siRNA Transfection
2.3. Preparation of Viral Stocks and Infections
2.4. Analysis of HCV Cell Entry Using HCVpp
2.5. In Vitro Transcription and HCV RNA Transfection
2.6. siRNA Transfection Experiments in Acutely HCV Infected Cells
2.7. siRNA Transfection Experiments in Persistently Infected Cells
2.8. Protein Analysis by WB
2.9. RNA Analysis
2.10. Confocal Analysis
2.11. Statistical Analysis
3. Results
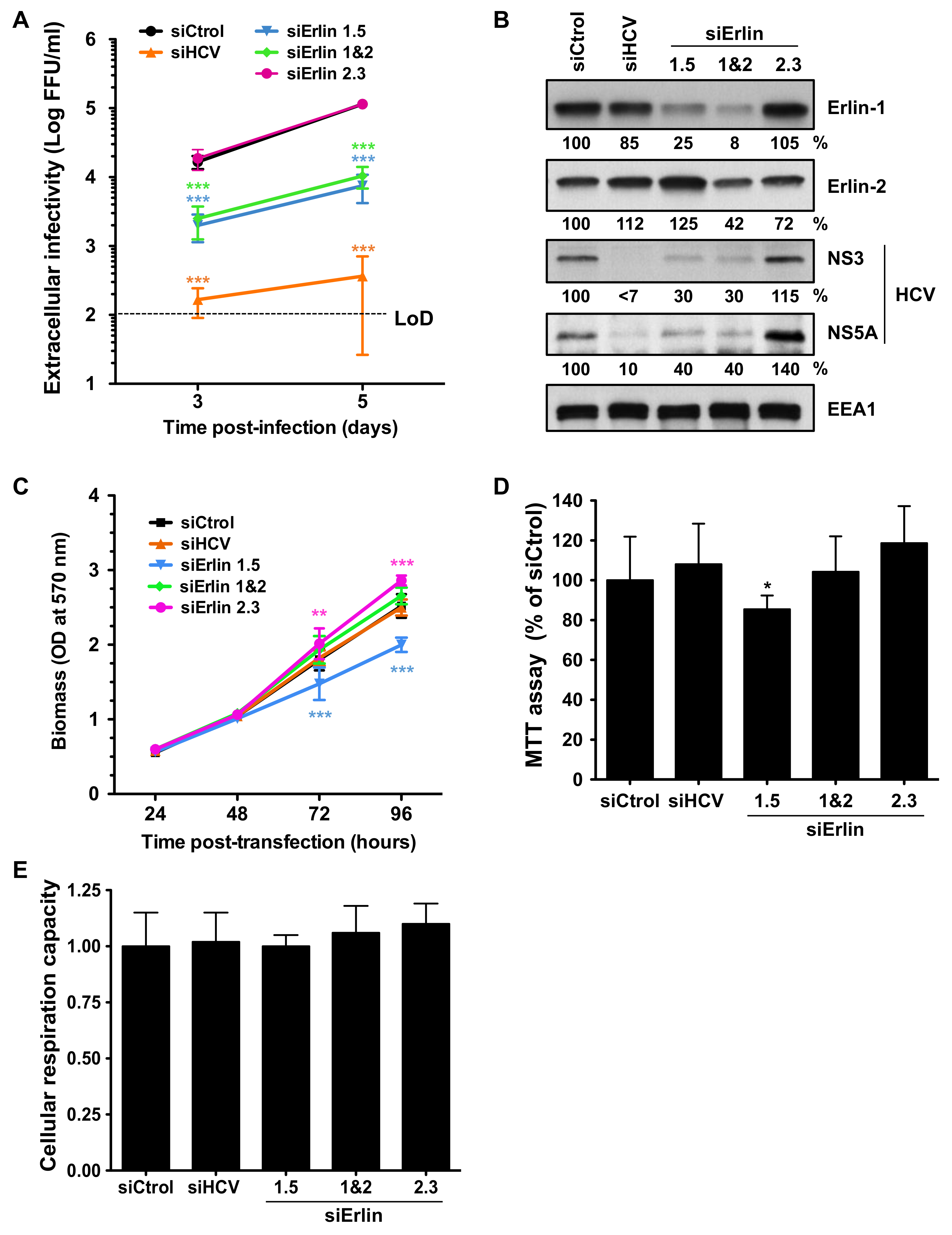
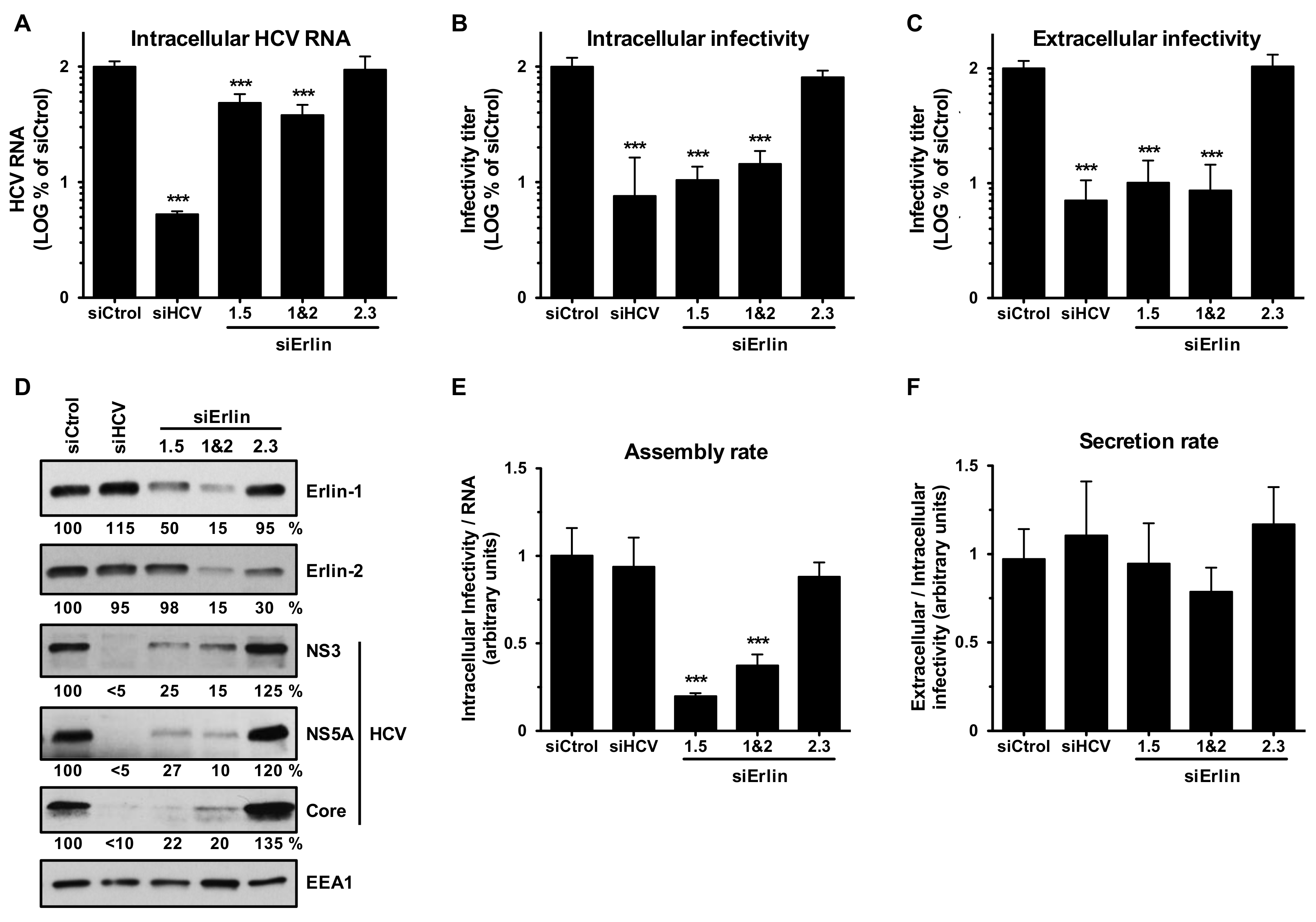
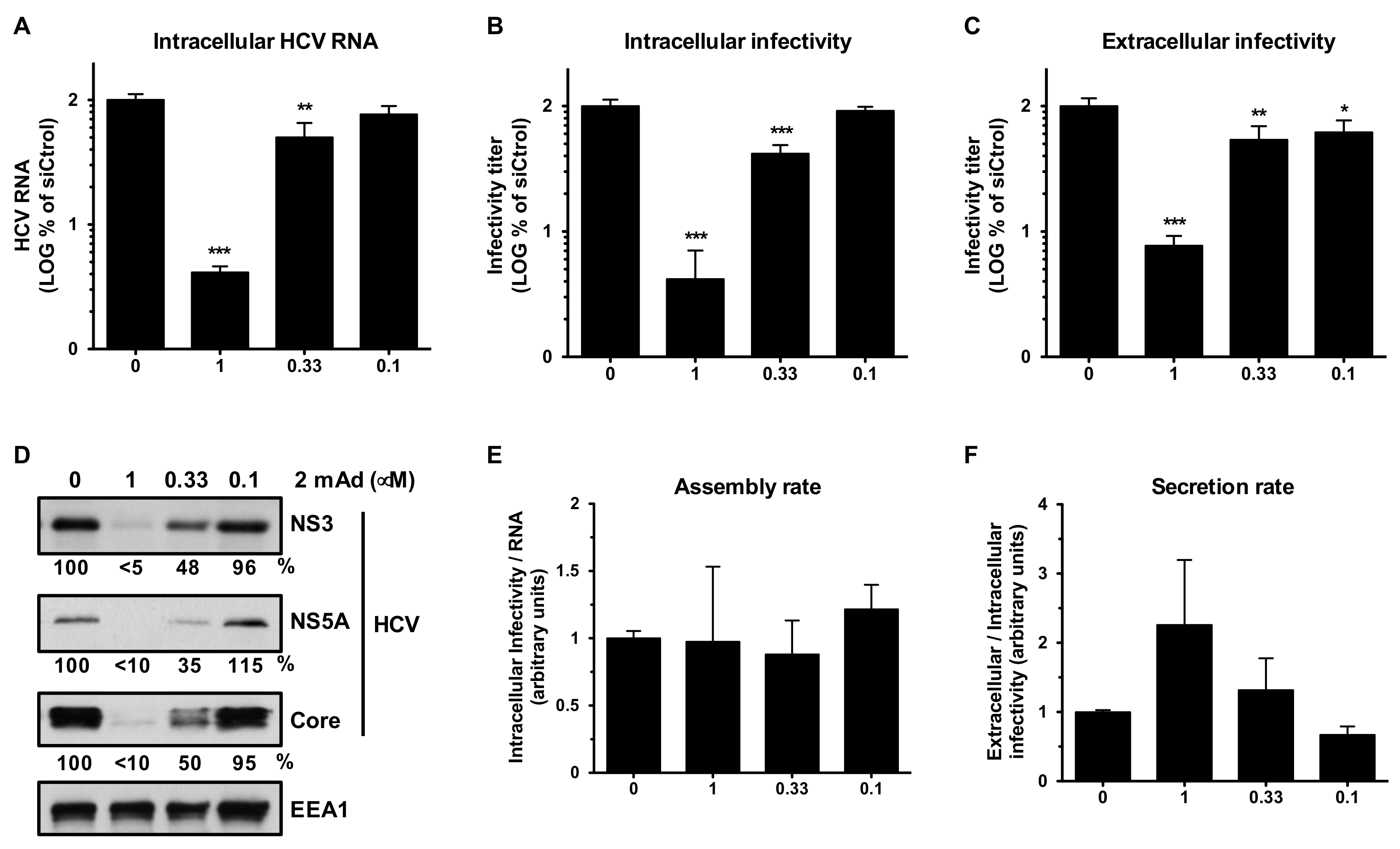
3.1. Erlin-1 Protein Is a Host Factor Required for Efficient HCV Infection
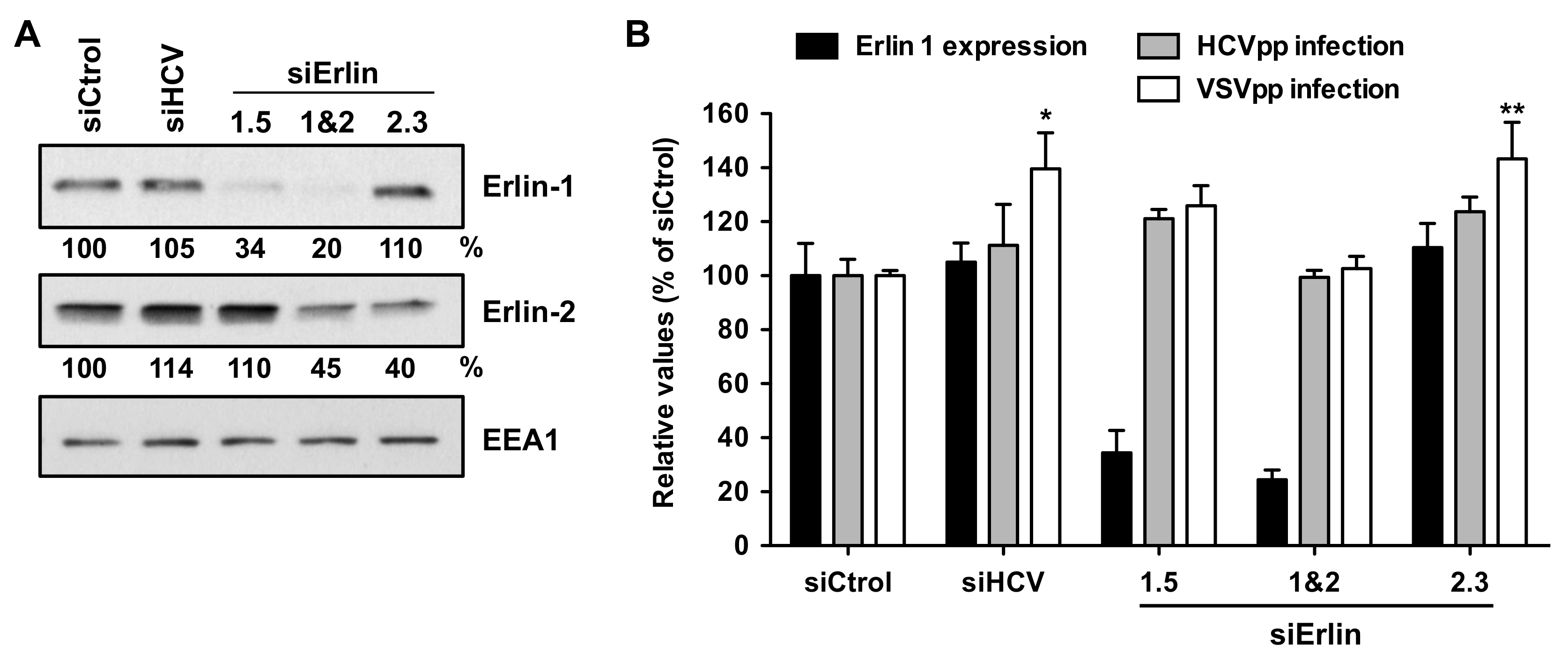
3.2. Erlin-1 Protein Is Not Required for HCV Cell-Entry
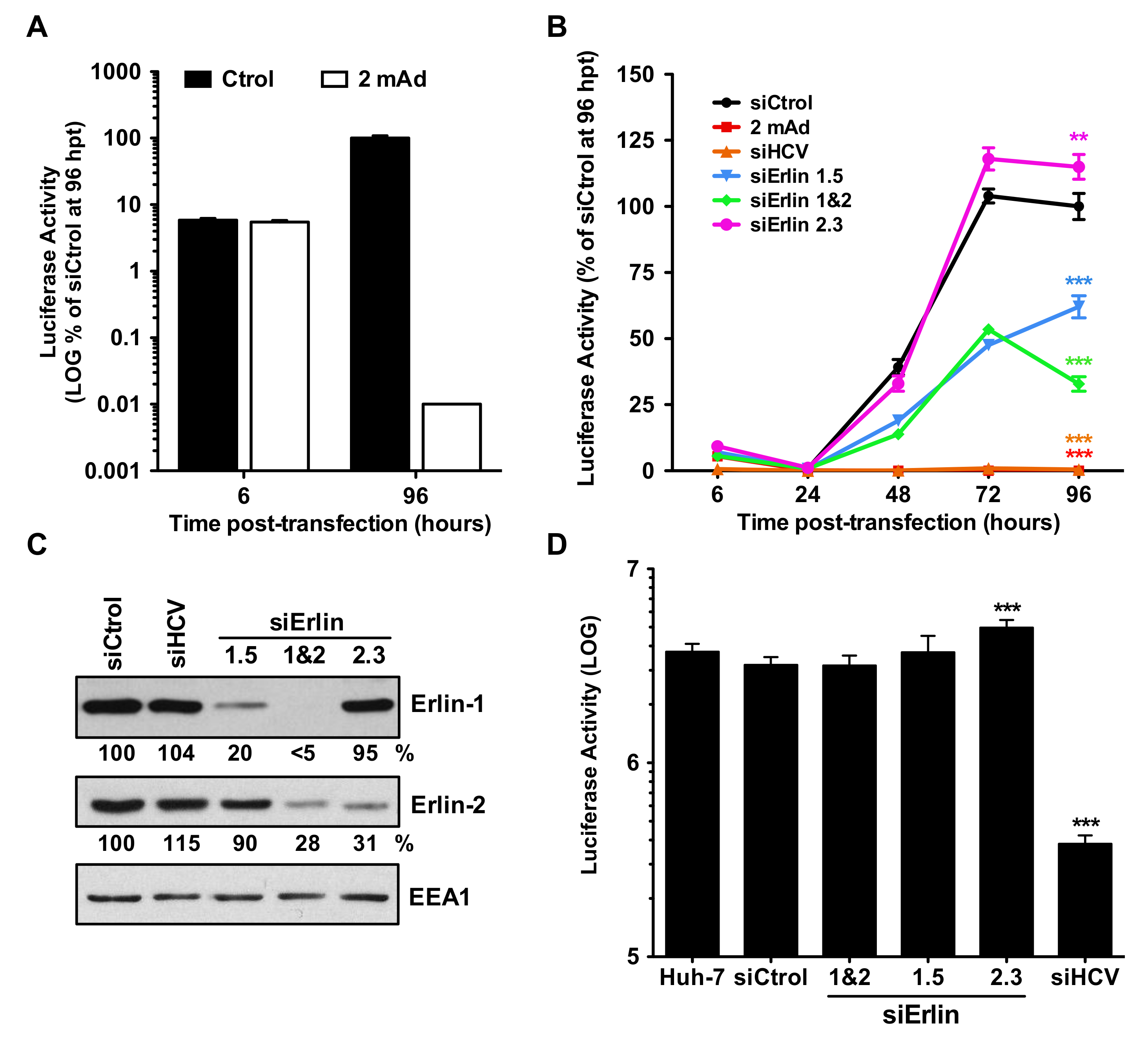
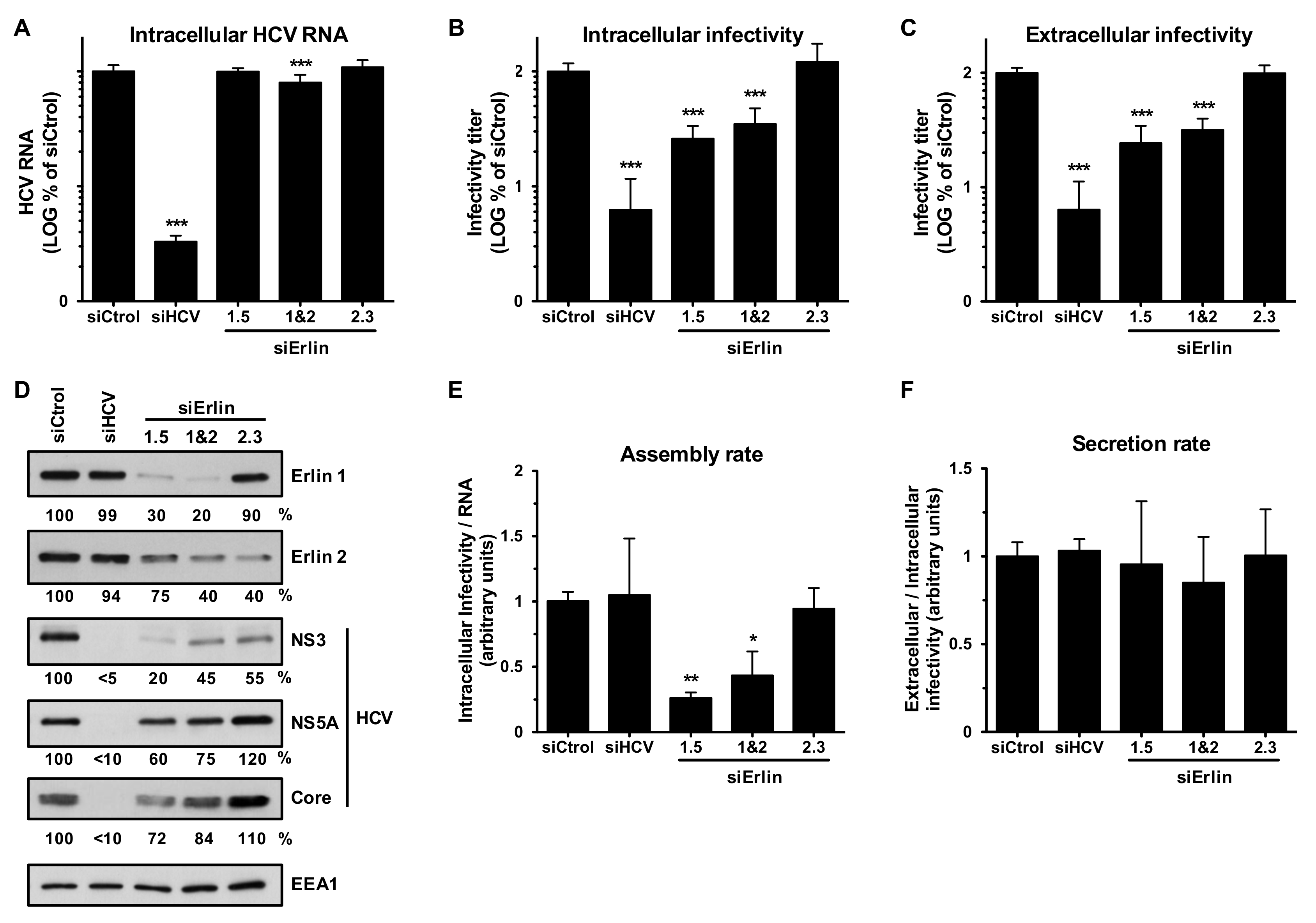
3.3. Erlin-1 Protein Down-Regulation Impairs the Establishment of HCV RNA Replication but Does Not Affect Primary Translation or Maintenance Of Replication
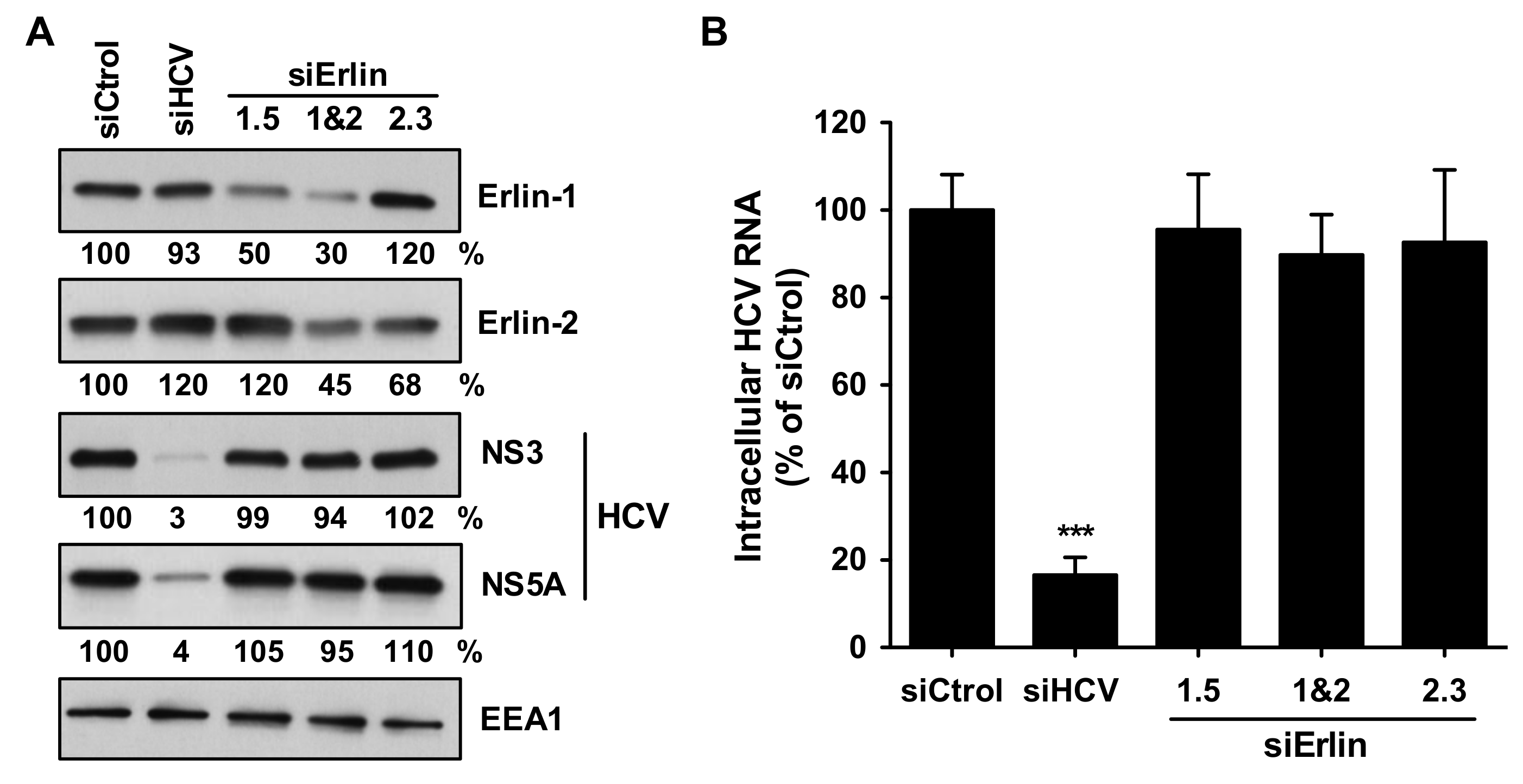
3.4. Erlin-1 Protein Down-Regulation Interferes with HCV Protein and Intracellular Infectious Virus Accumulation
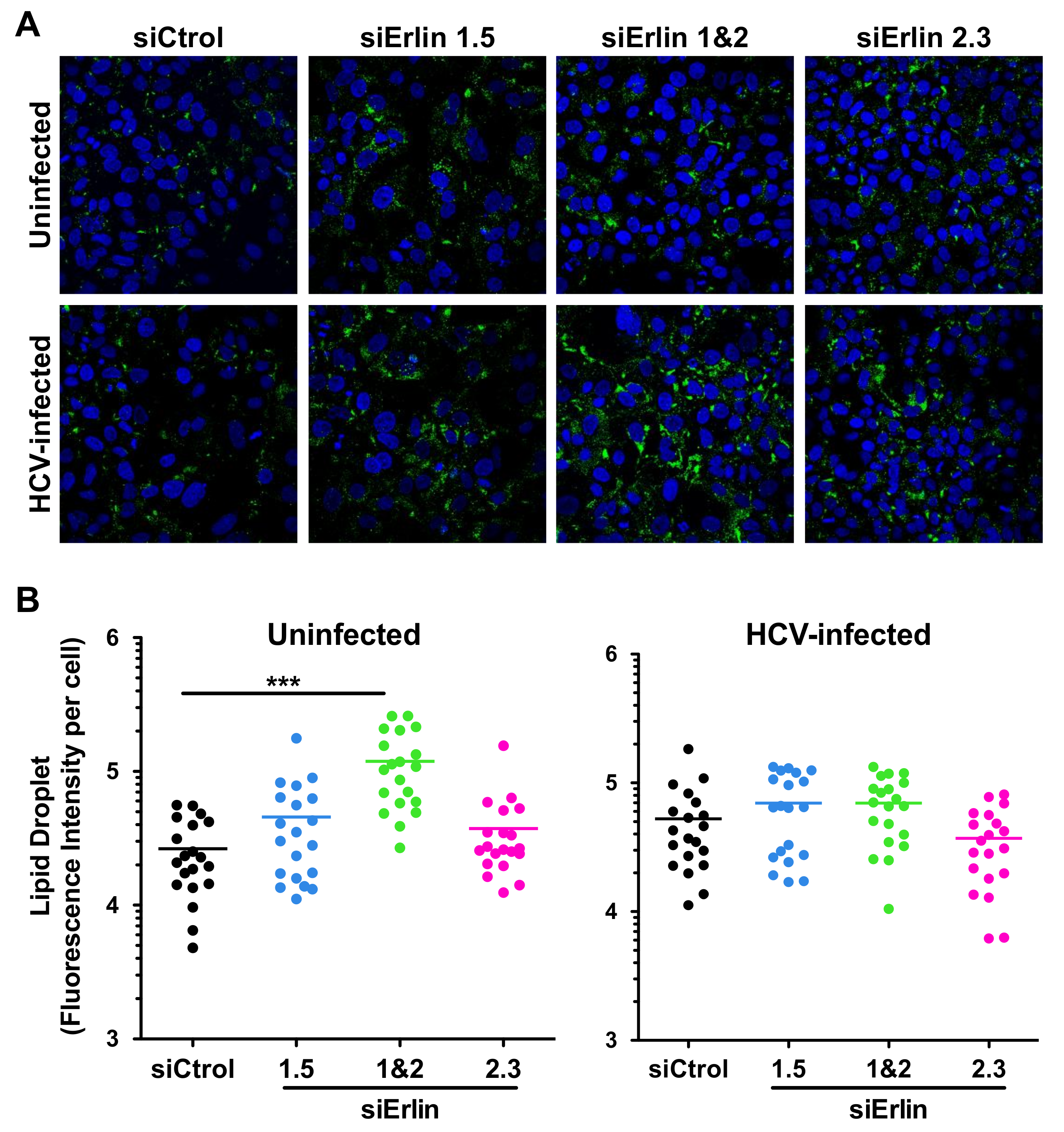
3.5. Erlin-1 Protein Down-Regulation Increases LD Accumulation in Huh-7 Cells
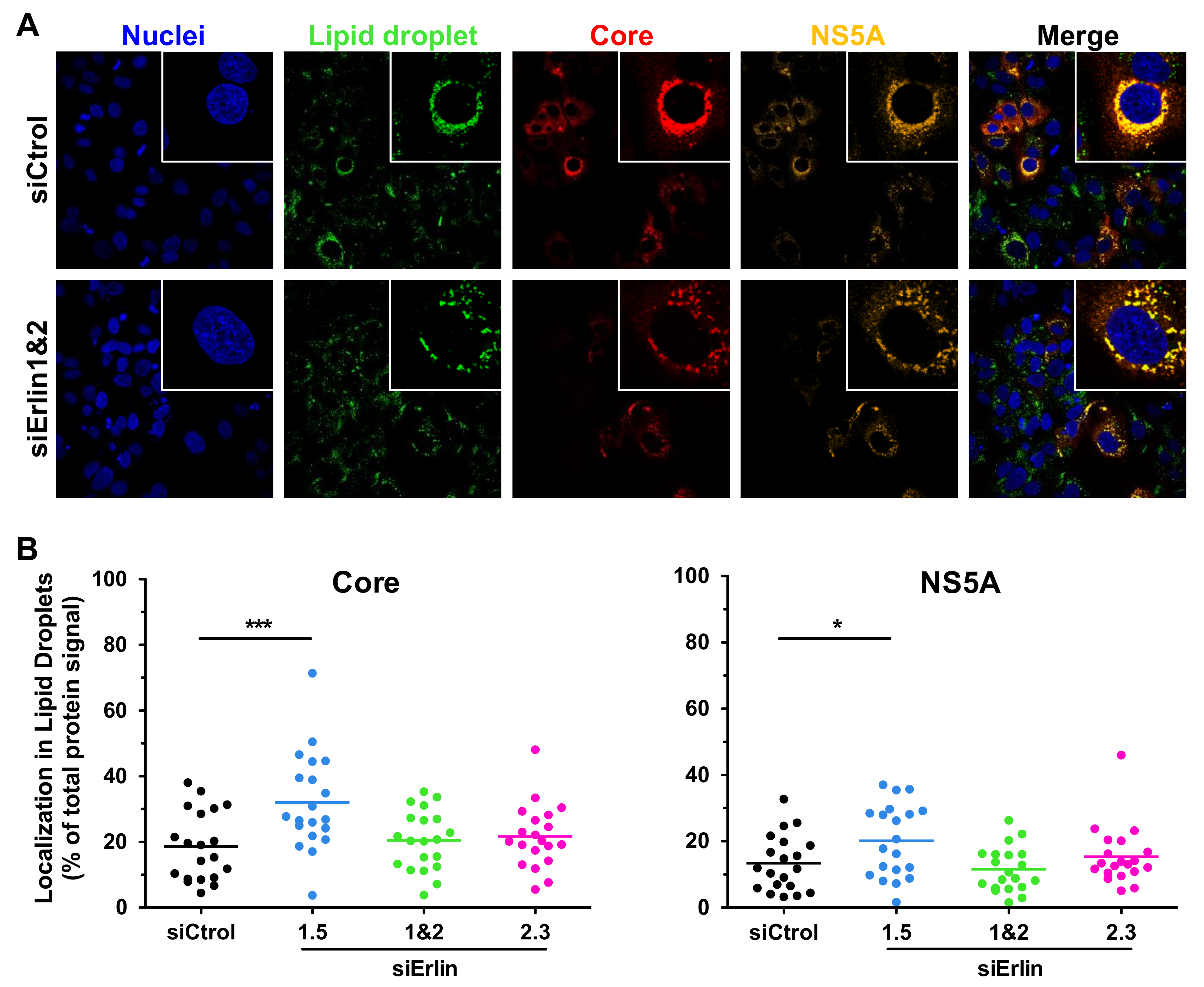
3.6. Erlin-1 Protein Deficiency Does Not Impair HCV Core and NS5A Protein Re-Localization to LDs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maniloff, J. Identification and classification of viruses that have not been propagated. Arch. Virol. 1995, 140, 1515–1520. [Google Scholar] [CrossRef] [PubMed]
- Alter, H.J.; Seeff, L.B. Recovery, persistence, and sequelae in hepatitis C virus infection: A perspective on long-term outcome. Semin. Liver Dis. 2000, 20, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Choo, Q.L.; Richman, K.H.; Han, J.H.; Berger, K.; Lee, C.; Dong, C.; Gallegos, C.; Coit, D.; Medina-Selby, R.; Barr, P.J. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 1991, 88, 2451–2455. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Boudet, J.; Simorre, J.-P.; Bartenschlager, R. Kissing-loop interaction in the 3’ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 2005, 79, 380–392. [Google Scholar] [CrossRef]
- Friebe, P.; Lohmann, V.; Krieger, N.; Bartenschlager, R. Sequences in the 5’ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001, 75, 12047–12057. [Google Scholar] [CrossRef]
- Honda, M.; Beard, M.R.; Ping, L.H.; Lemon, S.M. A phylogenetically conserved stem-loop structure at the 5’ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999, 73, 1165–1174. [Google Scholar]
- Penin, F.; Dubuisson, J.; Rey, F.A.; Moradpour, D.; Pawlotsky, J.-M. Structural biology of hepatitis C virus. Hepatology 2004, 39, 5–19. [Google Scholar] [CrossRef]
- Alazard-Dany, N.; Denolly, S.; Boson, B.; Cosset, F.-L. Overview of HCV Life Cycle with a Special Focus on Current and Possible Future Antiviral Targets. Viruses 2019, 11, 30. [Google Scholar] [CrossRef]
- Sharma, N.R.; Mateu, G.; Dreux, M.; Grakoui, A.; Cosset, F.-L.; Melikyan, G.B. Hepatitis C virus is primed by CD81 protein for low pH-dependent fusion. J. Biol. Chem. 2011, 286, 30361–30376. [Google Scholar] [CrossRef]
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007, 9, 1089–1097. [Google Scholar] [CrossRef]
- Zayas, M.; Long, G.; Madan, V.; Bartenschlager, R. Coordination of Hepatitis C Virus Assembly by Distinct Regulatory Regions in Nonstructural Protein 5A. PLoS Pathog. 2016, 12, e1005376. [Google Scholar] [CrossRef] [PubMed]
- Popescu, C.-I.; Callens, N.; Trinel, D.; Roingeard, P.; Moradpour, D.; Descamps, V.; Duverlie, G.; Penin, F.; Héliot, L.; Rouille, Y.; et al. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 2011, 7, e1001278. [Google Scholar] [CrossRef] [PubMed]
- Boson, B.; Granio, O.; Bartenschlager, R.; Cosset, F.-L. A concerted action of hepatitis C virus p7 and nonstructural protein 2 regulates core localization at the endoplasmic reticulum and virus assembly. PLoS Pathog. 2011, 7, e1002144. [Google Scholar] [CrossRef] [PubMed]
- Gentzsch, J.; Brohm, C.; Steinmann, E.; Friesland, M.; Menzel, N.; Vieyres, G.; Perin, P.M.; Frentzen, A.; Kaderali, L.; Pietschmann, T. Hepatitis C Virus p7 is Critical for Capsid Assembly and Envelopment. PLoS Pathog. 2013, 9, e1003355. [Google Scholar] [CrossRef]
- Gastaminza, P.; Cheng, G.; Wieland, S.; Zhong, J.; Liao, W.; Chisari, F.V. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J. Virol. 2008, 82, 2120–2129. [Google Scholar] [CrossRef]
- McLauchlan, J.; Lemberg, M.K.; Hope, G.; Martoglio, B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002, 21, 3980–3988. [Google Scholar] [CrossRef]
- Friesland, M.; Mingorance, L.; Chung, J.; Chisari, F.V.; Gastaminza, P. Sigma-1 receptor regulates early steps of viral RNA replication at the onset of hepatitis C virus infection. J. Virol. 2013, 87, 6377–6390. [Google Scholar] [CrossRef]
- Browman, D.T.; Hoegg, M.B.; Robbins, S.M. The SPFH domain-containing proteins: More than lipid raft markers. Trends Cell Biol. 2007, 17, 394–402. [Google Scholar] [CrossRef]
- Langhorst, M.F.; Reuter, A.; Stuermer, C.A.O. Scaffolding microdomains and beyond: The function of reggie/flotillin proteins. Cell. Mol. Life Sci. 2005, 62, 2228–2240. [Google Scholar] [CrossRef]
- Browman, D.T.; Resek, M.E.; Zajchowski, L.D.; Robbins, S.M. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 2006, 119, 3149–3160. [Google Scholar] [CrossRef]
- Ikegawa, S.; Isomura, M.; Koshizuka, Y.; Nakamura, Y. Cloning and characterization of a novel gene (C8orf2), a human representative of a novel gene family with homology to C. elegans C42.C1.9. Cytogenet. Cell Genet. 1999, 85, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Hoegg, M.B.; Browman, D.T.; Resek, M.E.; Robbins, S.M. Distinct regions within the erlins are required for oligomerization and association with high molecular weight complexes. J. Biol. Chem. 2009, 284, 7766–7776. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.M.P.; Wormer, D.B.; Wilkens, S.; Wojcikiewicz, R.J.H. An endoplasmic reticulum (ER) membrane complex composed of SPFH1 and SPFH2 mediates the ER-associated degradation of inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 2009, 284, 10433–10445. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.M.P.; Wang, Y.; Kelley, G.G.; Wojcikiewicz, R.J.H. SPFH2 mediates the endoplasmic reticulum-associated degradation of inositol 1,4,5-trisphosphate receptors and other substrates in mammalian cells. J. Biol. Chem. 2007, 282, 20104–20115. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Sguigna, P.V.; DeBose-Boyd, R.A. Membrane-associated ubiquitin ligase complex containing gp78 mediates sterol-accelerated degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 2011, 286, 15022–15031. [Google Scholar] [CrossRef]
- Teranishi, Y.; Hur, J.-Y.; Gu, G.J.; Kihara, T.; Ishikawa, T.; Nishimura, T.; Winblad, B.; Behbahani, H.; Kamali-Moghaddam, M.; Frykman, S.; et al. Erlin-2 is associated with active γ-secretase in brain and affects amyloid β-peptide production. Biochem. Biophys. Res. Commun. 2012, 424, 476–481. [Google Scholar] [CrossRef]
- Huber, M.D.; Vesely, P.W.; Datta, K.; Gerace, L. Erlins restrict SREBP activation in the ER and regulate cellular cholesterol homeostasis. J. Cell Biol. 2013, 203, 427–436. [Google Scholar] [CrossRef]
- Inoue, T.; Tsai, B. Regulated Erlin-dependent release of the B12 transmembrane J-protein promotes ER membrane penetration of a non-enveloped virus. PLoS Pathog. 2017, 13, e1006439. [Google Scholar] [CrossRef]
- Zhong, J.; Gastaminza, P.; Cheng, G.; Kapadia, S.; Kato, T.; Burton, D.R.; Wieland, S.F.; Uprichard, S.L.; Wakita, T.; Chisari, F.V. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 9294–9299. [Google Scholar] [CrossRef]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen Virol. 1977, 36, 59–74. [Google Scholar] [CrossRef]
- Kato, T.; Date, T.; Miyamoto, M.; Furusaka, A.; Tokushige, K.; Mizokami, M.; Wakita, T. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 2003, 125, 1808–1817. [Google Scholar] [CrossRef] [PubMed]
- Dreux, M.; Gastaminza, P.; Wieland, S.F.; Chisari, F.V. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. USA 2009, 106, 14046–14051. [Google Scholar] [CrossRef] [PubMed]
- Gastaminza, P.; Whitten-Bauer, C.; Chisari, F.V. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. USA 2010, 107, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Gastaminza, P.; Chung, J.; Stamataki, Z.; Isogawa, M.; Cheng, G.; Mckeating, J.A.; Chisari, F.V. Persistent hepatitis C virus infection in vitro: Coevolution of virus and host. J. Virol. 2006, 80, 11082–11093. [Google Scholar] [CrossRef]
- Gastaminza, P.; Kapadia, S.B.; Chisari, F.V. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 2006, 80, 11074–11081. [Google Scholar] [CrossRef]
- Garaigorta, U.; Chisari, F.V. Hepatitis C virus blocks interferon effector function by inducing protein kinase R phosphorylation. Cell Host Microbe 2009, 6, 513–522. [Google Scholar] [CrossRef]
- Bartosch, B.; Dubuisson, J.; Cosset, F.-L. Infectious Hepatitis C Virus Pseudo-particles Containing Functional E1–E2 Envelope Protein Complexes. J. Exp. Med. 2003, 197, 633–642. [Google Scholar] [CrossRef]
- Garaigorta, U.; Heim, M.H.; Boyd, B.; Wieland, S.; Chisari, F.V. Hepatitis C virus (HCV) induces formation of stress granules whose proteins regulate HCV RNA replication and virus assembly and egress. J. Virol. 2012, 86, 11043–11056. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Kräusslich, H.G.; Mizokami, M.; et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005, 11, 791–796. [Google Scholar] [CrossRef]
- Lindenbach, B.D.; Evans, M.J.; Syder, A.J.; Wolk, B.; Tellinghuisen, T.L.; Liu, C.C.; Maruyama, T.; Hynes, R.O.; Burton, D.R.; Mckeating, J.A.; et al. Complete Replication of Hepatitis C Virus in Cell Culture. Science 2005, 309, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Alvisi, G.; Madan, V.; Bartenschlager, R. Hepatitis c virus and host cell lipids: An intimate connection. RNA Biol. 2014, 8, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.-Y.; Zhang, L.-L. Host restriction factors for hepatitis C virus. World J. Gastroenterol. 2016, 22, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Asabe, S.; Wieland, S.; Garaigorta, U.; Gastaminza, P.; Isogawa, M.; Chisari, F.V. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc. Natl. Acad. Sci. USA 2010, 107, 7431–7436. [Google Scholar] [CrossRef]
- Dreux, M.; Garaigorta, U.; Boyd, B.; Décembre, E.; Chung, J.; Whitten-Bauer, C.; Wieland, S.; Chisari, F.V. Short-Range Exosomal Transfer of Viral RNA from Infected Cells to Plasmacytoid Dendritic Cells Triggers Innate Immunity. Cell Host Microbe 2012, 12, 558–570. [Google Scholar] [CrossRef]
- Padmanabhan, P.; Garaigorta, U.; Dixit, N.M. Emergent properties of the interferon-signalling network may underlie the success of hepatitis C treatment. Nat. Commun. 2014, 5, 3872. [Google Scholar] [CrossRef]
- Egger, D.; Wolk, B.; Gosert, R.; Bianchi, L.; Blum, H.E.; Moradpour, D.; Bienz, K. Expression of Hepatitis C Virus Proteins Induces Distinct Membrane Alterations Including a Candidate Viral Replication Complex. J. Virol. 2002, 76, 5974–5984. [Google Scholar] [CrossRef]
- Mingorance, L.; Castro, V.; Ávila-Pérez, G.; Calvo, G.; Rodriguez, M.J.; Carrascosa, J.L.; Pérez-del-Pulgar, S.; Forns, X.; Gastaminza, P. Host phosphatidic acid phosphatase lipin1 is rate limiting for functional hepatitis C virus replicase complex formation. PLoS Pathog. 2018, 14, e1007284. [Google Scholar] [CrossRef]
- Herker, E.; Harris, C.; Hernandez, C.; Carpentier, A.; Kaehlcke, K.; Rosenberg, A.R.; Farese, R.V.; Ott, M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat. Med. 2010, 16, 1295–1298. [Google Scholar] [CrossRef]
- Neveu, G.; Barouch-Bentov, R.; Ziv-Av, A.; Gerber, D.; Jacob, Y.; Einav, S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012, 8, e1002845. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whitten-Bauer, C.; Chung, J.; Gómez-Moreno, A.; Gomollón-Zueco, P.; Huber, M.D.; Gerace, L.; Garaigorta, U. The Host Factor Erlin-1 is Required for Efficient Hepatitis C Virus Infection. Cells 2019, 8, 1555. https://doi.org/10.3390/cells8121555
Whitten-Bauer C, Chung J, Gómez-Moreno A, Gomollón-Zueco P, Huber MD, Gerace L, Garaigorta U. The Host Factor Erlin-1 is Required for Efficient Hepatitis C Virus Infection. Cells. 2019; 8(12):1555. https://doi.org/10.3390/cells8121555
Chicago/Turabian StyleWhitten-Bauer, Christina, Josan Chung, Andoni Gómez-Moreno, Pilar Gomollón-Zueco, Michael D. Huber, Larry Gerace, and Urtzi Garaigorta. 2019. "The Host Factor Erlin-1 is Required for Efficient Hepatitis C Virus Infection" Cells 8, no. 12: 1555. https://doi.org/10.3390/cells8121555
APA StyleWhitten-Bauer, C., Chung, J., Gómez-Moreno, A., Gomollón-Zueco, P., Huber, M. D., Gerace, L., & Garaigorta, U. (2019). The Host Factor Erlin-1 is Required for Efficient Hepatitis C Virus Infection. Cells, 8(12), 1555. https://doi.org/10.3390/cells8121555





