Chronic In Vivo CRISPR-Cas Genome Editing: Challenges, Long-Term Safety, and Outlook
Abstract
1. Introduction
2. Cas Proteins
2.1. Intrinsic Role of Cas Proteins in Prokaryotes
2.2. Expression of Cas Proteins in Prokaryotes
2.3. Use of Cas Proteins Therapeutically
3. Types of Cas Proteins, Engineering Modifications and Their Delivery
3.1. Cas9
3.2. Cas12
3.3. Cas13
4. Therapeutic Delivery Systems
5. Long-Term Complications of CRISPR Expression
5.1. Double Stranded Breaks
5.2. Off-Target Genomic Editing
5.3. Off-Target Transcriptomic Effects
5.4. Genomic Vector Integration
5.5. Immunogenicity
6. Long-Term Effects of Expression of CRISPR/Cas Systems in In Vitro Models, In Vivo Models, and Humans
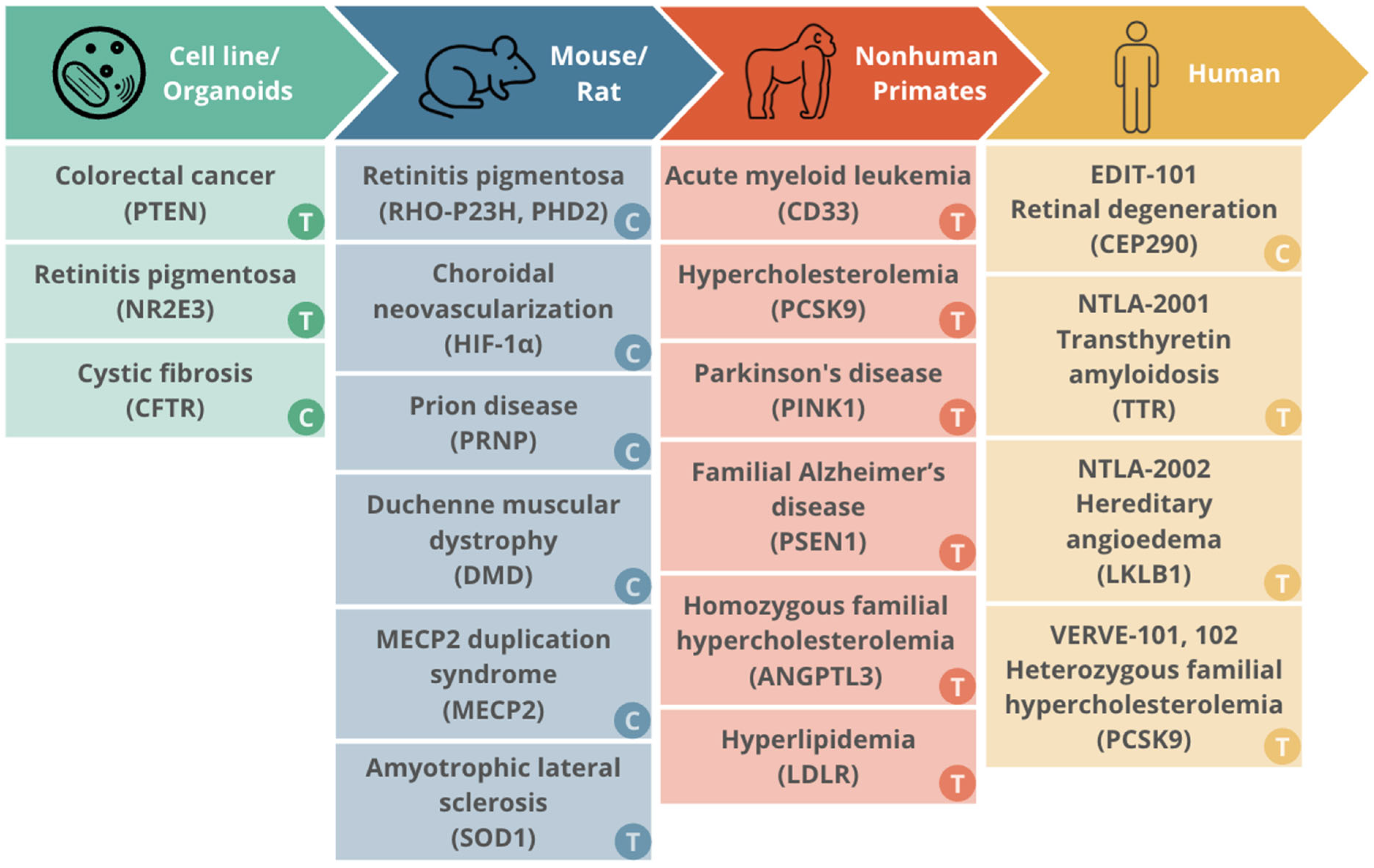
6.1. Cell Line and Organoid Models
6.2. Murine Models
6.3. Nonhuman Primate Models
6.4. Humans
7. Outlook and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CRISPR | clustered regularly interspaced short palindromic repeats |
| Cas | CRISPR-associated |
| crRNA | CRISPR RNAs |
| PAM | protospacer adjacent motif |
| gRNA | guide RNA |
| sgRNA | single-guide RNA |
| tracrRNA | trans-activating CRISPR RNA |
| DSB | double strand break |
| NHEJ | non-homologous end joining |
| HDR | homology-directed repair |
| HEPN | higher eukaryotes and prokaryotes nucleotide-binding |
| ssRNA | single-stranded RNA |
| AAV | adeno-associated virus |
| LNP | lipid nanoparticle |
| RNP | ribonucleoprotein complexes |
| GalNAc | N-acetylgalatosamine |
| Ku | Ku70-Ku80 |
| DNA-PKcs | DNA-dependent protein kinase catalytic subunit |
| MMEJ | microhomology mediated end joining |
| SSA | single stranded annealing |
| indels | insertions and deletions |
| CRISPRi | CRISPR interference |
| eSpCas9 | enhanced specific SpCas9 |
| dCas9 | dead Cas9 |
| CRISPRa | CRISPR activation |
| SpCas9 | Streptococcus pyogenes Cas9 |
| SaCas9 | Staphylococcus aureus Cas9 |
| dCas13 | dead Cas13 |
| m6A | N6-methyladenosine |
| PTEN | phosphatase and tensin homolog deleted on chromosome ten |
| RP | retinitis pigmentosa |
| NR2E3 | nuclear receptor subfamily 2 group E member 3 |
| iPSC | induced pluripotent stem cells |
| CF | cystic fibrosis |
| CFTR | cystic fibrosis transmembrane conductance regulator |
| AAV9 | adeno-associated virus serotype 9 |
| HIF-1α | hypoxia-inducible factor 1α |
| Rho | rhodopsin |
| PRNP | prion protein |
| DMD | Duchenne muscular dystrophy |
| MECP2 | Methyl-CpG-binding protein 2 |
| MDS | MECP2 duplication syndrome |
| ALS | amyotrophic lateral sclerosis |
| SOD1 | superoxide dismutase 1 |
| AML | acute myeloid leukemia |
| CD33 | sialic acid-binding Ig-like lectin 3 |
| CVD | cardiovascular disease |
| LDL-C | low-density lipoprotein cholesterol |
| ABE | adenine base editor |
| PCSK9 | proprotein convertase subtilisin/kexin type 9 |
| PD | Parkinson’s disease |
| PINK1 | PTEN-induced kinase I |
| FAD | familial Alzheimer’s disease |
| PSEN1 | presenilin 1 |
| GalNAc-LNP | GalNAc-conjugated lipid nanoparticle |
| ANGPTL3 | angiopoietin-like 3 |
| LDLR | low density lipoprotein receptor |
| TTR | transthyretin protein |
References
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide Sequence of the Iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia coli, and Identification of the Gene Product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Jansen, R.; Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of Genes That Are Associated with DNA Repeats in Prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Brouns, S.J.J.; Jore, M.M.; Lundgren, M.; Westra, E.R.; Slijkhuis, R.J.H.; Snijders, A.P.L.; Dickman, M.J.; Makarova, K.S.; Koonin, E.V.; van der Oost, J. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science 2008, 321, 960–964. [Google Scholar] [CrossRef]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadán, A.H.; Moineau, S. The CRISPR/Cas Bacterial Immune System Cleaves Bacteriophage and Plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef]
- Marraffini, L.A.; Sontheimer, E.J. CRISPR Interference Limits Horizontal Gene Transfer in Staphylococci by Targeting DNA. Science 2008, 322, 1843–1845. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, M.; Burmistrz, M. Mechanisms Regulating the CRISPR-Cas Systems. Front. Microbiol. 2023, 14, 1060337. [Google Scholar] [CrossRef] [PubMed]
- Dela Ahator, S.; Liu, Y.; Wang, J.; Zhang, L.-H. The Virulence Factor Regulator and Quorum Sensing Regulate the Type I-F CRISPR-Cas Mediated Horizontal Gene Transfer in Pseudomonas Aeruginosa. Front. Microbiol. 2022, 13, 987656. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Jaenisch, R. Generating Genetically Modified Mice Using CRISPR/Cas-Mediated Genome Engineering. Nat. Protoc. 2014, 9, 1956–1968. [Google Scholar] [CrossRef]
- Patterson, A.G.; Chang, J.T.; Taylor, C.; Fineran, P.C. Regulation of the Type I-F CRISPR-Cas System by CRP-cAMP and GalM Controls Spacer Acquisition and Interference. Nucleic Acids Res. 2015, 43, 6038. [Google Scholar] [CrossRef]
- Simoens, D.; Shravah, V.; Jones, W.K.; Kaja, S. Clinical and Pharmacovigilance Safety Evaluation of LUXTURNA® (Voretigene Neparvovec-Rzyl). Cutan. Ocul. Toxicol. 2025, 44, 361–373. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, Classification and Evolution of CRISPR-Cas Systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Z. CRISPR-Cas Systems: Overview, Innovations and Applications in Human Disease Research and Gene Therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, L.; Jiang, J.; Wu, M.; Lin, P. Applications and Challenges of CRISPR-Cas Gene-Editing to Disease Treatment in Clinics. Precis. Clin. Med. 2021, 4, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered Regularly Interspaced Short Palindrome Repeats (CRISPRs) Have Spacers of Extrachromosomal Origin. Microbiol. Read. Engl. 2005, 151, 2551–2561. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9–crRNA Ribonucleoprotein Complex Mediates Specific DNA Cleavage for Adaptive Immunity in Bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef]
- Zhang, X.-H.; Tee, L.Y.; Wang, X.-G.; Huang, Q.-S.; Yang, S.-H. Off-Target Effects in CRISPR/Cas9-Mediated Genome Engineering. Mol. Ther.-Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, D.; Matsugi, E.; Kishi, K.; Inoue, Y.; Nigorikawa, K.; Nomura, W. SpCas9-HF1 Enhances Accuracy of Cell Cycle-Dependent Genome Editing by Increasing HDR Efficiency, and by Reducing off-Target Effects and Indel Rates. Mol. Ther.-Nucleic Acids 2024, 35, 102124. [Google Scholar] [CrossRef]
- Asmamaw Mengstie, M.; Teshome Azezew, M.; Asmamaw Dejenie, T.; Teshome, A.A.; Tadele Admasu, F.; Behaile Teklemariam, A.; Tilahun Mulu, A.; Mekonnen Agidew, M.; Adugna, D.G.; Geremew, H.; et al. Recent Advancements in Reducing the Off-Target Effect of CRISPR-Cas9 Genome Editing. Biol. Targets Ther. 2024, 18, 21–28. [Google Scholar] [CrossRef]
- Matsumoto, D.; Kishi, K.; Matsugi, E.; Inoue, Y.; Nigorikawa, K.; Nomura, W. Cas9-Geminin and Cdt1-Fused Anti-CRISPR Protein Synergistically Increase Editing Accuracy. FEBS Lett. 2023, 597, 985–994. [Google Scholar] [CrossRef]
- Hossain, K.A.; Nierzwicki, L.; Orozco, M.; Czub, J.; Palermo, G. Flexibility in PAM Recognition Expands DNA Targeting in xCas9. eLife 2025, 13, RP102538. [Google Scholar] [CrossRef]
- CRISPR-Cas9 DNA Base-Editing and Prime-Editing. Available online: https://www.mdpi.com/1422-0067/21/17/6240 (accessed on 21 September 2025).
- Bendixen, L.; Jensen, T.I.; Bak, R.O. CRISPR-Cas-Mediated Transcriptional Modulation: The Therapeutic Promises of CRISPRa and CRISPRi. Mol. Ther. 2023, 31, 1920–1937. [Google Scholar] [CrossRef]
- Pattali, R.K.; Ornelas, I.J.; Nguyen, C.D.; Xu, D.; Divekar, N.S.; Nuñez, J.K. CRISPRoff Epigenetic Editing for Programmable Gene Silencing in Human Cells without DNA Breaks. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, J.K.; Chen, J.; Pommier, G.C.; Cogan, J.Z.; Replogle, J.M.; Adriaens, C.; Ramadoss, G.N.; Shi, Q.; Hung, K.L.; Samelson, A.J.; et al. Genome-Wide Programmable Transcriptional Memory by CRISPR-Based Epigenome Editing. Cell 2021, 184, 2503–2519.e17. [Google Scholar] [CrossRef] [PubMed]
- Chey, Y.C.J.; Gierus, L.; Lushington, C.; Arudkumar, J.C.; Geiger, A.B.; Staker, L.G.; Robertson, L.J.; Pfitzner, C.; Kennedy, J.G.; Lee, R.H.B.; et al. Optimal SpCas9- and SaCas9-Mediated Gene Editing by Enhancing gRNA Transcript Levels through Scaffold Poly-T Tract Reduction. BMC Genom. 2025, 26, 138. [Google Scholar] [CrossRef]
- Allen, D.; Rosenberg, M.; Hendel, A. Using Synthetically Engineered Guide RNAs to Enhance CRISPR Genome Editing Systems in Mammalian Cells. Front. Genome Ed. 2021, 2, 617910. [Google Scholar] [CrossRef]
- Strohkendl, I.; Saifuddin, F.A.; Rybarski, J.R.; Finkelstein, I.J.; Russell, R. Kinetic Basis for DNA Target Specificity of CRISPR-Cas12a. Mol. Cell 2018, 71, 816–824.e3. [Google Scholar] [CrossRef] [PubMed]
- Cetin, B.; Erendor, F.; Eksi, Y.E.; Sanlioglu, A.D.; Sanlioglu, S. Advancing CRISPR Genome Editing into Gene Therapy Clinical Trials: Progress and Future Prospects. Expert Rev. Mol. Med. 2025, 27, e16. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wu, Y.; Wang, H.; Liu, H.; Zhou, J.; Chen, J.; Lei, J.; Sun, Z.; Paek, C.; Yin, L. Highly Parallel Profiling of the Activities and Specificities of Cas12a Variants in Human Cells. Nat. Commun. 2025, 16, 3022. [Google Scholar] [CrossRef]
- Xun, G.; Zhu, Z.; Singh, N.; Lu, J.; Jain, P.K.; Zhao, H. Harnessing Noncanonical crRNA for Highly Efficient Genome Editing. Nat. Commun. 2024, 15, 3823. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Macaluso, N.C.; Rakestraw, N.R.; Carman, D.R.; Pizzano, B.L.M.; Hautamaki, R.C.; Rananaware, S.R.; Roberts, I.E.; Jain, P.K. Harnessing Noncanonical crRNAs to Improve Functionality of Cas12a Orthologs. Cell Rep. 2024, 43, 113777. [Google Scholar] [CrossRef]
- Serebriiskii, I.G.; Pavlov, V.; Tricarico, R.; Andrianov, G.; Nicolas, E.; Parker, M.I.; Newberg, J.; Frampton, G.; Meyer, J.E.; Golemis, E.A. Comprehensive Characterization of PTEN Mutational Profile in a Series of 34,129 Colorectal Cancers. Nat. Commun. 2022, 13, 1618. [Google Scholar] [CrossRef]
- Wu, F.; Qiao, X.; Zhao, Y.; Zhang, Z.; Gao, Y.; Shi, L.; Du, H.; Wang, L.; Zhang, Y.-J.; Zhang, Y.; et al. Targeted Mutagenesis in Arabidopsis Thaliana Using CRISPR-Cas12b/C2c1. J. Integr. Plant Biol. 2020, 62, 1653–1658. [Google Scholar] [CrossRef]
- Huang, Z.; Fang, J.; Zhou, M.; Gong, Z.; Xiang, T. CRISPR-Cas13: A New Technology for the Rapid Detection of Pathogenic Microorganisms. Front. Microbiol. 2022, 13, 1011399. [Google Scholar] [CrossRef] [PubMed]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic Acid Detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Bot, J.F.; van der Oost, J.; Geijsen, N. The Double Life of CRISPR–Cas13. Curr. Opin. Biotechnol. 2022, 78, 102789. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Lin, Q.; Jin, S.; Gao, C. The CRISPR-Cas Toolbox and Gene Editing Technologies. Mol. Cell 2022, 82, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cheng, Y.; Yu, J.; Zhu, Y.; Ma, H.; Zhou, Y.; Pu, Z.; Zhu, G.; Yuan, Y.; Zhang, Z.; et al. Compact RNA Editors with Natural Miniature Cas13j Nucleases. Nat. Chem. Biol. 2025, 21, 280–290. [Google Scholar] [CrossRef]
- Tong, H.; Huang, J.; Xiao, Q.; He, B.; Dong, X.; Liu, Y.; Yang, X.; Han, D.; Wang, Z.; Wang, X.; et al. High-Fidelity Cas13 Variants for Targeted RNA Degradation with Minimal Collateral Effects. Nat. Biotechnol. 2023, 41, 108–119. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Franklin, B.; Koob, J.; Kellner, M.J.; Ladha, A.; Joung, J.; Kirchgatterer, P.; Cox, D.B.T.; Zhang, F. A Cytosine Deaminase for Programmable Single-Base RNA Editing. Science 2019, 365, 382–386. [Google Scholar] [CrossRef]
- Tang, H.; Han, S.; Jie, Y.; Jiang, X.; Zhang, Y.; Peng, J.; Wang, F.; Li, X.; Zhou, X.; Jiang, W.; et al. Enhanced or Reversible RNA N6-Methyladenosine Editing by Red/Far-Red Light Induction. Nucleic Acids Res. 2025, 53, gkaf181. [Google Scholar] [CrossRef]
- Yang, H.; Patel, D.J. Structures, Mechanisms and Applications of RNA-Centric CRISPR–Cas13. Nat. Chem. Biol. 2024, 20, 673–688. [Google Scholar] [CrossRef]
- Apostolopoulos, A.; Kawamoto, N.; Chow, S.Y.A.; Tsuiji, H.; Ikeuchi, Y.; Shichino, Y.; Iwasaki, S. dCas13-Mediated Translational Repression for Accurate Gene Silencing in Mammalian Cells. Nat. Commun. 2024, 15, 2205. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, Z.; Seun Olajide, J.; Wang, G. CRISPR/Cas-Based Nucleic Acid Detection Strategies: Trends and Challenges. Heliyon 2024, 10, e26179. [Google Scholar] [CrossRef]
- Li, H.; Qiu, Y.; Song, B.; Quan, X.; Zhang, D.; Li, X.; Yang, J.; Liu, X.; Zeng, Z.; Jing, J.; et al. Engineering a Photoactivatable A-to-I RNA Base Editor for Gene Therapy in Vivo. Nat. Biotechnol. 2025, 1–12, Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-H.; Zhan, W.; Gallagher, T.L.; Gao, G. Recombinant Adeno-Associated Virus as a Delivery Platform for Ocular Gene Therapy: A Comprehensive Review. Mol. Ther. 2024, 32, 4185–4207. [Google Scholar] [CrossRef]
- Wang, J.-H.; Gessler, D.J.; Zhan, W.; Gallagher, T.L.; Gao, G. Adeno-Associated Virus as a Delivery Vector for Gene Therapy of Human Diseases. Signal Transduct. Target. Ther. 2024, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Kantor, B. Lentiviral Vectors for Delivery of Gene-Editing Systems Based on CRISPR/Cas: Current State and Perspectives. Viruses 2021, 13, 1288. [Google Scholar] [CrossRef]
- Diakatou, M.; Dubois, G.; Erkilic, N.; Sanjurjo-Soriano, C.; Meunier, I.; Kalatzis, V. Allele-Specific Knockout by CRISPR/Cas to Treat Autosomal Dominant Retinitis Pigmentosa Caused by the G56R Mutation in NR2E3. Int. J. Mol. Sci. 2021, 22, 2607. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Pan, C.; Yong, H.; Wang, F.; Bo, T.; Zhao, Y.; Ma, B.; He, W.; Li, M. Emerging Non-Viral Vectors for Gene Delivery. J. Nanobiotechnol. 2023, 21, 272. [Google Scholar] [CrossRef]
- Jeong, M.; Lee, Y.; Park, J.; Jung, H.; Lee, H. Lipid Nanoparticles (LNPs) for in Vivo RNA Delivery and Their Breakthrough Technology for Future Applications. Adv. Drug Deliv. Rev. 2023, 200, 114990. [Google Scholar] [CrossRef]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Seitzer, J.; O’Connell, D.; Walsh, K.R.; Wood, K.; et al. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 2021, 385, 493–502. [Google Scholar] [CrossRef]
- Geng, G.; Xu, Y.; Hu, Z.; Wang, H.; Chen, X.; Yuan, W.; Shu, Y. Viral and Non-Viral Vectors in Gene Therapy: Current State and Clinical Perspectives. eBioMedicine 2025, 118, 105834. [Google Scholar] [CrossRef]
- Wei, T.; Cheng, Q.; Farbiak, L.; Anderson, D.G.; Langer, R.; Siegwart, D.J. Delivery of Tissue-Targeted Scalpels: Opportunities and Challenges for In Vivo CRISPR/Cas-Based Genome Editing. ACS Nano 2020, 14, 9243–9262. [Google Scholar] [CrossRef]
- Buchlis, G.; Podsakoff, G.M.; Radu, A.; Hawk, S.M.; Flake, A.W.; Mingozzi, F.; High, K.A. Factor IX Expression in Skeletal Muscle of a Severe Hemophilia B Patient 10 Years after AAV-Mediated Gene Transfer. Blood 2012, 119, 3038–3041. [Google Scholar] [CrossRef]
- Xue, C.; Greene, E.C. DNA Repair Pathway Choices in CRISPR-Cas9 Mediated Genome Editing. Trends Genet. TIG 2021, 37, 639–656. [Google Scholar] [CrossRef]
- Hillary, V.E.; Ceasar, S.A. A Review on the Mechanism and Applications of CRISPR/Cas9/Cas12/Cas13/Cas14 Proteins Utilized for Genome Engineering. Mol. Biotechnol. 2023, 65, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Wu, J.; VanDusen, N.J.; Li, Y.; Zheng, Y. CRISPR-Cas9-Mediated Homology-Directed Repair for Precise Gene Editing. Mol. Ther. Nucleic Acids 2024, 35, 102344. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-Homologous DNA End Joining and Alternative Pathways to Double-Strand Break Repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Difilippantonio, M.J.; Zhu, J.; Chen, H.T.; Meffre, E.; Nussenzweig, M.C.; Max, E.E.; Ried, T.; Nussenzweig, A. DNA Repair Protein Ku80 Suppresses Chromosomal Aberrations and Malignant Transformation. Nature 2000, 404, 510–514. [Google Scholar] [CrossRef]
- Stinson, B.M.; Carney, S.M.; Walter, J.C.; Loparo, J.J. Structural Role for DNA Ligase IV in Promoting the Fidelity of Non-Homologous End Joining. Nat. Commun. 2024, 15, 1250. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Watanabe, G.; Gerodimos, C.A.; Ochi, T.; Blundell, T.L.; Jackson, S.P.; Lieber, M.R. Different DNA End Configurations Dictate Which NHEJ Components Are Most Important for Joining Efficiency*. J. Biol. Chem. 2016, 291, 24377–24389. [Google Scholar] [CrossRef]
- Sfeir, A.; Symington, L.S. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem. Sci. 2015, 40, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, D.M.; Richardson, C.A.; Elliott, B.; Jasin, M. Modeling Oncogenic Translocations: Distinct Roles for Double-Strand Break Repair Pathways in Translocation Formation in Mammalian Cells. DNA Repair 2006, 5, 1065–1074. [Google Scholar] [CrossRef]
- Brunet, E.; Jasin, M. Induction of Chromosomal Translocations with CRISPR-Cas9 and Other Nucleases: Understanding the Repair Mechanisms That Give Rise to Translocations. Adv. Exp. Med. Biol. 2018, 1044, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Dubois, F.; Sidiropoulos, N.; Weischenfeldt, J.; Beroukhim, R. Structural Variations in Cancer and the 3D Genome. Nat. Rev. Cancer 2022, 22, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.M.T.; Samson, C.A.; Rand, A.D.; Sheppard, H.M. Unintended CRISPR-Cas9 Editing Outcomes: A Review of the Detection and Prevalence of Structural Variants Generated by Gene-Editing in Human Cells. Hum. Genet. 2023, 142, 705–720. [Google Scholar] [CrossRef]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-Frequency off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Dey, D.; Chakravarti, R.; Bhattacharjee, O.; Majumder, S.; Chaudhuri, D.; Ahmed, K.T.; Roy, D.; Bhattacharya, B.; Arya, M.; Gautam, A.; et al. A Mechanistic Study on the Tolerance of PAM Distal End Mismatch by SpCas9. J. Biol. Chem. 2024, 300, 107439. [Google Scholar] [CrossRef]
- Kuscu, C.; Arslan, S.; Singh, R.; Thorpe, J.; Adli, M. Genome-Wide Analysis Reveals Characteristics of off-Target Sites Bound by the Cas9 Endonuclease. Nat. Biotechnol. 2014, 32, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Scott, D.A.; Kriz, A.J.; Chiu, A.C.; Hsu, P.D.; Dadon, D.B.; Cheng, A.W.; Trevino, A.E.; Konermann, S.; Chen, S.; et al. Genome-Wide Binding of the CRISPR Endonuclease Cas9 in Mammalian Cells. Nat. Biotechnol. 2014, 32, 670–676. [Google Scholar] [CrossRef]
- Duan, J.; Lu, G.; Xie, Z.; Lou, M.; Luo, J.; Guo, L.; Zhang, Y. Genome-Wide Identification of CRISPR/Cas9 off-Targets in Human Genome. Cell Res. 2014, 24, 1009–1012. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, A.; Zhu, Y.; Groth, T.; Stolfa, G.; Stablewski, A.B.; Singhi, N.; Nemeth, M.; Neelamegham, S. Doxycycline-Dependent Self-Inactivation of CRISPR-Cas9 to Temporally Regulate On- and Off-Target Editing. Mol. Ther. 2020, 28, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, J.; Zhou, R.; Garcia, S.P.; Iyer, S.; Lareau, C.A.; Aryee, M.J.; Joung, J.K. Transcriptome-Wide off-Target RNA Editing Induced by CRISPR-Guided DNA Base Editors. Nature 2019, 569, 433–437. [Google Scholar] [CrossRef]
- Rohatgi, N.; Fortin, J.-P.; Lau, T.; Ying, Y.; Zhang, Y.; Lee, B.L.; Costa, M.R.; Reja, R. Seed Sequences Mediate Off-Target Activity in the CRISPR-Interference System. Cell Genom. 2024, 4, 100693. [Google Scholar] [CrossRef] [PubMed]
- Corsi, G.I.; Gadekar, V.P.; Gorodkin, J.; Seemann, S.E. CRISPRroots: On- and off-Target Assessment of RNA-Seq Data in CRISPR–Cas9 Edited Cells. Nucleic Acids Res. 2022, 50, e20. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, K.S.; Kleinstiver, B.P.; Garcia, S.P.; Zaborowski, M.P.; Volak, A.; Spirig, S.E.; Muller, A.; Sousa, A.A.; Tsai, S.Q.; Bengtsson, N.E.; et al. High Levels of AAV Vector Integration into CRISPR-Induced DNA Breaks. Nat. Commun. 2019, 10, 4439. [Google Scholar] [CrossRef]
- Deyle, D.R.; Russell, D.W. Adeno-Associated Virus Vector Integration. Curr. Opin. Mol. Ther. 2009, 11, 442–447. [Google Scholar]
- Sabatino, D.E.; Bushman, F.D.; Chandler, R.J.; Crystal, R.G.; Davidson, B.L.; Dolmetsch, R.; Eggan, K.C.; Gao, G.; Gil-Farina, I.; Kay, M.A.; et al. Evaluating the State of the Science for Adeno-Associated Virus Integration: An Integrated Perspective. Mol. Ther. 2022, 30, 2646–2663. [Google Scholar] [CrossRef] [PubMed]
- Gil-Farina, I.; Fronza, R.; Kaeppel, C.; Lopez-Franco, E.; Ferreira, V.; D’Avola, D.; Benito, A.; Prieto, J.; Petry, H.; Gonzalez-Aseguinolaza, G.; et al. Recombinant AAV Integration Is Not Associated with Hepatic Genotoxicity in Nonhuman Primates and Patients. Mol. Ther. 2016, 24, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Suchy, F.P.; Karigane, D.; Nakauchi, Y.; Higuchi, M.; Zhang, J.; Pekrun, K.; Hsu, I.; Fan, A.C.; Nishimura, T.; Charlesworth, C.T.; et al. Genome Engineering with Cas9 and AAV Repair Templates Generates Frequent Concatemeric Insertions of Viral Vectors. Nat. Biotechnol. 2025, 43, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Ewaisha, R.; Anderson, K.S. Immunogenicity of CRISPR Therapeutics—Critical Considerations for Clinical Translation. Front. Bioeng. Biotechnol. 2023, 11, 1138596. [Google Scholar] [CrossRef]
- Simhadri, V.L.; McGill, J.; McMahon, S.; Wang, J.; Jiang, H.; Sauna, Z.E. Prevalence of Pre-Existing Antibodies to CRISPR-Associated Nuclease Cas9 in the USA Population. Mol. Ther. Methods Clin. Dev. 2018, 10, 105–112. [Google Scholar] [CrossRef]
- Wagner, D.L.; Amini, L.; Wendering, D.J.; Burkhardt, L.-M.; Akyüz, L.; Reinke, P.; Volk, H.-D.; Schmueck-Henneresse, M. High Prevalence of Streptococcus Pyogenes Cas9-Reactive T Cells within the Adult Human Population. Nat. Med. 2019, 25, 242–248. [Google Scholar] [CrossRef]
- Tang, X.-Z.E.; Tan, S.X.; Hoon, S.; Yeo, G.W. Pre-Existing Adaptive Immunity to the RNA-Editing Enzyme Cas13d in Humans. Nat. Med. 2022, 28, 1372–1376. [Google Scholar] [CrossRef]
- Kim, S.; Koo, T.; Jee, H.-G.; Cho, H.-Y.; Lee, G.; Lim, D.-G.; Shin, H.S.; Kim, J.-S. CRISPR RNAs Trigger Innate Immune Responses in Human Cells. Genome Res. 2018, 28, 367–373. [Google Scholar] [CrossRef]
- Keeler, A.M.; Zhan, W.; Ram, S.; Fitzgerald, K.A.; Gao, G. The Curious Case of AAV Immunology. Mol. Ther. 2025, 33, 1946–1965. [Google Scholar] [CrossRef]
- Ertl, H.C.J. Immunogenicity and Toxicity of AAV Gene Therapy. Front. Immunol. 2022, 13, 975803. [Google Scholar] [CrossRef]
- Chand, D.H.; Zaidman, C.; Arya, K.; Millner, R.; Farrar, M.A.; Mackie, F.E.; Goedeker, N.L.; Dharnidharka, V.R.; Dandamudi, R.; Reyna, S.P. Thrombotic Microangiopathy Following Onasemnogene Abeparvovec for Spinal Muscular Atrophy: A Case Series. J. Pediatr. 2021, 231, 265–268. [Google Scholar] [CrossRef]
- Teichler Zallen, D. US Gene Therapy in Crisis. Trends Genet. TIG 2000, 16, 272–275. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. Gene Therapy Community Grapples with Toxicity Issues, as Pipeline Matures. Nat. Rev. Drug Discov. 2021, 20, 804–805. [Google Scholar] [CrossRef]
- Kenjo, E.; Hozumi, H.; Makita, Y.; Iwabuchi, K.A.; Fujimoto, N.; Matsumoto, S.; Kimura, M.; Amano, Y.; Ifuku, M.; Naoe, Y.; et al. Low Immunogenicity of LNP Allows Repeated Administrations of CRISPR-Cas9 mRNA into Skeletal Muscle in Mice. Nat. Commun. 2021, 12, 7101. [Google Scholar] [CrossRef] [PubMed]
- Editas Medicine Nominates EDIT-401, an LDLR-Targeted Medicine, as Lead In Vivo Development Candidate | Editas Medicine. Available online: https://ir.editasmedicine.com/news-releases/news-release-details/editas-medicine-nominates-edit-401-ldlr-targeted-medicine-lead (accessed on 21 September 2025).
- Tashiro, H.; Sauer, T.; Shum, T.; Parikh, K.; Mamonkin, M.; Omer, B.; Rouce, R.H.; Lulla, P.; Rooney, C.M.; Gottschalk, S.; et al. Treatment of Acute Myeloid Leukemia with T Cells Expressing Chimeric Antigen Receptors Directed to C-Type Lectin-like Molecule 1. Mol. Ther. 2017, 25, 2202–2213. [Google Scholar] [CrossRef]
- Pelcovits, A.; Niroula, R. Acute Myeloid Leukemia: A Review. Rhode Isl. Med. J. 2020, 103, 38–40. [Google Scholar]
- Cora, D.; Novo, M.; Al-Soufi, W.; Sánchez, L.; Arana, Á.J. CRISPR/Cas9 Delivery Systems to Enhance Gene Editing Efficiency. Int. J. Mol. Sci. 2025, 26, 4420. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.-H.; Liu, S.; Xiao, J.; Zhou, Y.-X.; Dong, L.-W.; Li, Y.-F.; Zhang, Y.-Q.; Li, W.-H.; Wang, J.-Q.; Wang, Y.; et al. New Loss-of-Function Mutations in PCSK9 Reduce Plasma LDL Cholesterol. Arterioscler. Thromb. Vasc. Biol. 2023, 43, 1219–1233. [Google Scholar] [CrossRef]
- Wang, L.; Breton, C.; Warzecha, C.C.; Bell, P.; Yan, H.; He, Z.; White, J.; Zhu, Y.; Li, M.; Buza, E.L.; et al. Long-Term Stable Reduction of Low-Density Lipoprotein in Nonhuman Primates Following in Vivo Genome Editing of PCSK9. Mol. Ther. 2021, 29, 2019–2029. [Google Scholar] [CrossRef]
- Tanner, C.M.; Ostrem, J.L. Parkinson’s Disease. N. Engl. J. Med. 2024, 391, 442–452. [Google Scholar] [CrossRef]
- Petit, D.; Fernández, S.G.; Zoltowska, K.M.; Enzlein, T.; Ryan, N.S.; O’Connor, A.; Szaruga, M.; Hill, E.; Vandenberghe, R.; Fox, N.C.; et al. Aβ Profiles Generated by Alzheimer’s Disease Causing PSEN1 Variants Determine the Pathogenicity of the Mutation and Predict Age at Disease Onset. Mol. Psychiatry 2022, 27, 2821–2832. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.M.; Tanzi, R.E. Monogenic Determinants of Familial Alzheimer’s Disease: Presenilin-1 Mutations. Cell. Mol. Life Sci. CMLS 1998, 54, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Skoufou-Papoutsaki, N.; Adler, S.; D’Santos, P.; Mannion, L.; Mehmed, S.; Kemp, R.; Smith, A.; Perrone, F.; Nayak, K.; Russell, A.; et al. Efficient Genetic Editing of Human Intestinal Organoids Using Ribonucleoprotein-Based CRISPR. Dis. Models Mech. 2023, 16, dmm050279. [Google Scholar] [CrossRef]
- Bulcaen, M.; Kortleven, P.; Liu, R.B.; Maule, G.; Dreano, E.; Kelly, M.; Ensinck, M.M.; Thierie, S.; Smits, M.; Ciciani, M.; et al. Prime Editing Functionally Corrects Cystic Fibrosis-Causing CFTR Mutations in Human Organoids and Airway Epithelial Cells. Cell Rep. Med. 2024, 5, 101544. [Google Scholar] [CrossRef] [PubMed]
- Shahin, S.; Xu, H.; Lu, B.; Mercado, A.; Jones, M.K.; Bakondi, B.; Wang, S. AAV-CRISPR/Cas9 Gene Editing Preserves Long-Term Vision in the P23H Rat Model of Autosomal Dominant Retinitis Pigmentosa. Pharmaceutics 2022, 14, 824. [Google Scholar] [CrossRef]
- Wu, W.-H.; Tsai, Y.-T.; Huang, I.-W.; Cheng, C.-H.; Hsu, C.-W.; Cui, X.; Ryu, J.; Quinn, P.M.J.; Caruso, S.M.; Lin, C.-S.; et al. CRISPR Genome Surgery in a Novel Humanized Model for Autosomal Dominant Retinitis Pigmentosa. Mol. Ther. 2022, 30, 1407–1420. [Google Scholar] [CrossRef]
- Liu, X.; Qiao, J.; Jia, R.; Zhang, F.; Meng, X.; Li, Y.; Yang, L. Allele-Specific Gene-Editing Approach for Vision Loss Restoration in RHO-Associated Retinitis Pigmentosa. eLife 2023, 12, e84065. [Google Scholar] [CrossRef]
- Yu, W.; Mookherjee, S.; Chaitankar, V.; Hiriyanna, S.; Kim, J.-W.; Brooks, M.; Ataeijannati, Y.; Sun, X.; Dong, L.; Li, T.; et al. Nrl Knockdown by AAV-Delivered CRISPR/Cas9 Prevents Retinal Degeneration in Mice. Nat. Commun. 2017, 8, 14716. [Google Scholar] [CrossRef]
- Du, S.W.; Newby, G.A.; Salom, D.; Gao, F.; Menezes, C.R.; Suh, S.; Choi, E.H.; Chen, P.Z.; Liu, D.R.; Palczewski, K. In Vivo Photoreceptor Base Editing Ameliorates Rhodopsin-E150K Autosomal-Recessive Retinitis Pigmentosa in Mice. Proc. Natl. Acad. Sci. USA 2024, 121, e2416827121. [Google Scholar] [CrossRef]
- Nolan, N.D.; Cui, X.; Robbings, B.M.; Demirkol, A.; Pandey, K.; Wu, W.-H.; Hu, H.F.; Jenny, L.A.; Lin, C.-S.; Hass, D.T.; et al. CRISPR Editing of Anti-Anemia Drug Target Rescues Independent Preclinical Models of Retinitis Pigmentosa. Cell Rep. Med. 2024, 5, 101459. [Google Scholar] [CrossRef]
- Jo, D.H.; Koo, T.; Cho, C.S.; Kim, J.H.; Kim, J.-S.; Kim, J.H. Long-Term Effects of In Vivo Genome Editing in the Mouse Retina Using Campylobacter jejuni Cas9 Expressed via Adeno-Associated Virus. Mol. Ther. 2019, 27, 130–136. [Google Scholar] [CrossRef]
- An, M.; Davis, J.R.; Levy, J.M.; Serack, F.E.; Harvey, J.W.; Brauer, P.P.; Pirtle, C.P.; Berríos, K.N.; Newby, G.A.; Yeh, W.-H.; et al. In Vivo Base Editing Extends Lifespan of a Humanized Mouse Model of Prion Disease. Nat. Med. 2025, 31, 1319–1328. [Google Scholar] [CrossRef]
- Li, H.L.; Fujimoto, N.; Sasakawa, N.; Shirai, S.; Ohkame, T.; Sakuma, T.; Tanaka, M.; Amano, N.; Watanabe, A.; Sakurai, H.; et al. Precise Correction of the Dystrophin Gene in Duchenne Muscular Dystrophy Patient Induced Pluripotent Stem Cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 2015, 4, 143–154. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, C.; Li, H.; Wang, P.; Gao, Y.; Mokadam, N.A.; Ma, J.; Arnold, W.D.; Han, R. Efficient Precise in Vivo Base Editing in Adult Dystrophic Mice. Nat. Commun. 2021, 12, 3719. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Wu, X.; Yao, Y.; Duan, M.; Wang, X.; Li, G.; Guo, A.; Wu, M.; Liu, Y.; Zheng, J.; et al. An RNA Editing Strategy Rescues Gene Duplication in a Mouse Model of MECP2 Duplication Syndrome and Nonhuman Primates. Nat. Neurosci. 2025, 28, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, M.; Su, B. Application of the Genome Editing Tool CRISPR/Cas9 in Non-Human Primates. Zool. Res. 2016, 37, 214–219. [Google Scholar] [CrossRef]
- Petty, N.E.; Radtke, S.; Fields, E.; Humbert, O.; Llewellyn, M.J.; Laszlo, G.S.; Zhu, H.; Jerome, K.R.; Walter, R.B.; Kiem, H.-P. Efficient Long-Term Multilineage Engraftment of CD33-Edited Hematopoietic Stem/Progenitor Cells in Nonhuman Primates. Mol. Ther. Methods Clin. Dev. 2023, 31, 101121. [Google Scholar] [CrossRef]
- Kim, M.Y.; Yu, K.-R.; Kenderian, S.S.; Ruella, M.; Chen, S.; Shin, T.-H.; Aljanahi, A.A.; Schreeder, D.; Klichinsky, M.; Shestova, O.; et al. Genetic Inactivation of CD33 in Hematopoietic Stem Cells to Enable CAR T Cell Immunotherapy for Acute Myeloid Leukemia. Cell 2018, 173, 1439–1453.e19. [Google Scholar] [CrossRef]
- Tu, T.; Song, Z.; Liu, X.; Wang, S.; He, X.; Xi, H.; Wang, J.; Yan, T.; Chen, H.; Zhang, Z.; et al. A Precise and Efficient Adenine Base Editor. Mol. Ther. 2022, 30, 2933–2941. [Google Scholar] [CrossRef]
- Musunuru, K.; Chadwick, A.C.; Mizoguchi, T.; Garcia, S.P.; DeNizio, J.E.; Reiss, C.W.; Wang, K.; Iyer, S.; Dutta, C.; Clendaniel, V.; et al. In Vivo CRISPR Base Editing of PCSK9 Durably Lowers Cholesterol in Primates. Nature 2021, 593, 429–434. [Google Scholar] [CrossRef]
- Chen, Z.-Z.; Wang, J.-Y.; Kang, Y.; Yang, Q.-Y.; Gu, X.-Y.; Zhi, D.-L.; Yan, L.; Long, C.-Z.; Shen, B.; Niu, Y.-Y. PINK1 Gene Mutation by Pair Truncated sgRNA/Cas9-D10A in Cynomolgus Monkeys. Zool. Res. 2021, 42, 469–477. [Google Scholar] [CrossRef]
- Li, M.; Guan, M.; Lin, J.; Zhu, K.; Zhu, J.; Guo, M.; Li, Y.; Chen, Y.; Chen, Y.; Zou, Y.; et al. Early Blood Immune Molecular Alterations in Cynomolgus Monkeys with a PSEN1 Mutation Causing Familial Alzheimer’s Disease. Alzheimers Dement. 2024, 20, 5492–5510. [Google Scholar] [CrossRef]
- Lee, R.; Mazzola, A.; Denizio, J.; Mizoguchi, T.; Clendaniel, V.; Garrity, R.; Cox, N.; Glass, Z.; Chamarthi, H.; Flannigan, S.; et al. An Investigational in Vivo Base Editing Medicine Targeting ANGPTL3, VERVE-201, Achieves Precise and Durable Liver Editing in Nonclinical Studies. Atherosclerosis 2024, 395, 118496. [Google Scholar] [CrossRef]
- Streilein, J.W. Unraveling Immune Privilege. Science 1995, 270, 1158. [Google Scholar] [CrossRef]
- Irigoyen, C.; Amenabar Alonso, A.; Sanchez-Molina, J.; Rodríguez-Hidalgo, M.; Lara-López, A.; Ruiz-Ederra, J. Subretinal Injection Techniques for Retinal Disease: A Review. J. Clin. Med. 2022, 11, 4717. [Google Scholar] [CrossRef] [PubMed]
- Toral, M.A.; Charlesworth, C.T.; Ng, B.; Chemudupati, T.; Homma, S.; Nakauchi, H.; Bassuk, A.G.; Porteus, M.H.; Mahajan, V.B. Investigation of Cas9 Antibodies in the Human Eye. Nat. Commun. 2022, 13, 1053. [Google Scholar] [CrossRef] [PubMed]
- Safety and Efficacy of MCO-010 Optogenetic Therapy in Patients with Stargardt Disease in USA (STARLIGHT): An Open-Label Multi-Center Ph2 Trial—PMC. Available online: https://pmc.ncbi.nlm.nih.gov/articles/PMC12362400/ (accessed on 1 November 2025).
- Editas Medicine, Inc. Open-Label, Single Ascending Dose Study to Evaluate the Safety, Tolerability, and Efficacy of EDIT-101 in Adult and Pediatric Participants with Leber Congenital Amaurosis Type 10 (LCA10), with Centrosomal Protein 290 (CEP290)-Related Retinal Degeneration Caused by a Compound Heterozygous or Homozygous Mutation Involving c.2991+1655A>G in Intron 26 (IVS26) of the CEP290 Gene (*LCA10-IVS26*); Editas Medicine, Inc.: Cambridge, MA, USA, 2022. Available online: https://clinicaltrials.gov/study/NCT03872479 (accessed on 1 November 2025).
- Pierce, E.A.; Aleman, T.S.; Jayasundera, K.T.; Ashimatey, B.S.; Kim, K.; Rashid, A.; Jaskolka, M.C.; Myers, R.L.; Lam, B.L.; Bailey, S.T.; et al. Gene Editing for CEP290-Associated Retinal Degeneration. N. Engl. J. Med. 2024, 390, 1972–1984. [Google Scholar] [CrossRef]
- Maeder, M.L.; Stefanidakis, M.; Wilson, C.J.; Baral, R.; Barrera, L.A.; Bounoutas, G.S.; Bumcrot, D.; Chao, H.; Ciulla, D.M.; DaSilva, J.A.; et al. Development of a Gene-Editing Approach to Restore Vision Loss in Leber Congenital Amaurosis Type 10. Nat. Med. 2019, 25, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikova, M.; Lima de Carvalho, J.R.; Oh, J.K.; Soucy, M.; Demirkol, A.; Kim, A.H.; Tsang, S.H.; Breazzano, M.P. Phenotypic Variability of Retinal Disease Among a Cohort of Patients with Variants in the CLN Genes. Investig. Ophthalmol. Vis. Sci. 2023, 64, 23. [Google Scholar] [CrossRef]
- Wei, A.; Yin, D.; Zhai, Z.; Ling, S.; Le, H.; Tian, L.; Xu, J.; Paludan, S.R.; Cai, Y.; Hong, J. In Vivo CRISPR Gene Editing in Patients with Herpes Stromal Keratitis. medRxiv 2023. medRxiv:2023.02.21.23285822. [Google Scholar] [CrossRef]
- Fontana, M.; Solomon, S.D.; Kachadourian, J.; Walsh, L.; Rocha, R.; Lebwohl, D.; Smith, D.; Täubel, J.; Gane, E.J.; Pilebro, B.; et al. CRISPR-Cas9 Gene Editing with Nexiguran Ziclumeran for ATTR Cardiomyopathy. N. Engl. J. Med. 2024, 391, 2231–2241. [Google Scholar] [CrossRef]
- Intellia Therapeutics Announces First Quarter 2025 Financial Results and Highlights Recent Company Progress—Intellia Therapeutics. Available online: https://ir.intelliatx.com/news-releases/news-release-details/intellia-therapeutics-announces-first-quarter-2025-financial (accessed on 28 September 2025).
- Verve Therapeutics, Inc. Open-Label, Phase 1b, Single-Ascending Dose and Optional Re Dosing Study to Evaluate the Safety of VERVE-101 Administered to Patients with Heterozygous Familial Hypercholesterolemia, Atherosclerotic Cardiovascular Disease, and Uncontrolled Hypercholesterolemia; Verve Therapeutics, Inc.: Boston, MA, USA, 2025. Available online: https://clinicaltrials.gov/study/NCT05398029 (accessed on 22 December 2025).
- Verve Therapeutics. Verve Therapeutics Announces Updates on Its PCSK9 Program; Verve Therapeutics: Boston, MA, USA, 2024. [Google Scholar]
- Vafai, S.; Karsten, V.; Jensen, C.; Falzone, R.; Lister, T.; Stolz, L.; Khera, A.; Kathiresan, S.; Bellinger, A.; Fiedorek, F. Design of Heart-2: A Phase 1b Clinical Trial of VERVE-102, an in Vivo Base Editing Medicine Delivered by a GalNAc-LNP and Targeting PCSK9 to Durably Lower LDL Cholesterol. In Circulation; Lippincott Williams & WilkinsHagerstown: Hagerstown, MD, USA, 2024. [Google Scholar]
- Frangoul, H.; Altshuler, D.; Cappellini, M.D.; Chen, Y.-S.; Domm, J.; Eustace, B.K.; Foell, J.; de la Fuente, J.; Grupp, S.; Handgretinger, R.; et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 2021, 384, 252–260. [Google Scholar] [CrossRef]
- Dibas, A.; Batabyal, S.; Kim, S.; Carlson, M.; Mohanty, S.; Sharif, N.A. Efficacy of Intravitreal Multi-Characteristic Opsin (MCO-010) Optogenetic Gene Therapy in a Mouse Model of Leber Congenital Amaurosis. J. Ocul. Pharmacol. Ther. 2024, 40, 702–708. [Google Scholar] [CrossRef] [PubMed]
- GenSight Biologics Announces 1 Year Safety Data and Efficacy Signals from PIONEER Phase I/II Clinical Trial of GS030, an Optogenetic Treatment Candidate for Retinitis Pigmentosa—GenSight Biologics. Available online: https://www.gensight-biologics.com/2023/02/13/gensight-biologics-announces-1-year-safety-data-and-efficacy-signals-from-pioneer-phase-i-ii-clinical-trial-of-gs030-an-optogenetic-treatment-candidate-for-retinitis-pigmentosa/ (accessed on 14 November 2025).
- Dulla, K.; Aguila, M.; Lane, A.; Jovanovic, K.; Parfitt, D.A.; Schulkens, I.; Chan, H.L.; Schmidt, I.; Beumer, W.; Vorthoren, L.; et al. Splice-Modulating Oligonucleotide QR-110 Restores CEP290 mRNA and Function in Human c.2991+1655A>G LCA10 Models. Mol. Ther. Nucleic Acids 2018, 12, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Maurer, M.S.; Kale, P.; Fontana, M.; Berk, J.L.; Grogan, M.; Gustafsson, F.; Hung, R.R.; Gottlieb, R.L.; Damy, T.; González-Duarte, A.; et al. Patisiran Treatment in Patients with Transthyretin Cardiac Amyloidosis. N. Engl. J. Med. 2023, 389, 1553–1565. [Google Scholar] [CrossRef] [PubMed]
- Fontana, M.; Berk, J.L.; Gillmore, J.D.; Witteles, R.M.; Grogan, M.; Drachman, B.; Damy, T.; Garcia-Pavia, P.; Taubel, J.; Solomon, S.D.; et al. Vutrisiran in Patients with Transthyretin Amyloidosis with Cardiomyopathy. N. Engl. J. Med. 2025, 392, 33–44. [Google Scholar] [CrossRef]
- Benson, M.D.; Waddington-Cruz, M.; Berk, J.L.; Polydefkis, M.; Dyck, P.J.; Wang, A.K.; Planté-Bordeneuve, V.; Barroso, F.A.; Merlini, G.; Obici, L.; et al. Inotersen Treatment for Patients with Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 22–31. [Google Scholar] [CrossRef]
- Coelho, T.; Marques, W., Jr.; Dasgupta, N.R.; Chao, C.-C.; Parman, Y.; França, M.C., Jr.; Guo, Y.-C.; Wixner, J.; Ro, L.-S.; Calandra, C.R.; et al. Eplontersen for Hereditary Transthyretin Amyloidosis with Polyneuropathy. JAMA 2023, 330, 1448–1458. [Google Scholar] [CrossRef]
- Riedl, M.A.; Tachdjian, R.; Lumry, W.R.; Craig, T.; Karakaya, G.; Gelincik, A.; Stobiecki, M.; Jacobs, J.S.; Gokmen, N.M.; Reshef, A.; et al. Efficacy and Safety of Donidalorsen for Hereditary Angioedema. N. Engl. J. Med. 2024, 391, 21–31. [Google Scholar] [CrossRef]
- Craig, T.J.; Reshef, A.; Li, H.H.; Jacobs, J.S.; Bernstein, J.A.; Farkas, H.; Yang, W.H.; Stroes, E.S.G.; Ohsawa, I.; Tachdjian, R.; et al. Efficacy and Safety of Garadacimab, a Factor XIIa Inhibitor for Hereditary Angioedema Prevention (VANGUARD): A Global, Multicentre, Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet 2023, 401, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- BioMarin Pharmaceutical. A Phase 1/2 Open-Label, Dose-Escalation Study to Determine the Safety Tolerability & Efficacy of BMN 331 an AAV Vector-Mediated Gene Transfer of Human SERPING1 Gene in Subjects with HAE Due to Human C1-INH Deficiency; BioMarin Pharmaceutical: San Rafael, CA, USA, 2024. Available online: https://clinicaltrials.gov/study/NCT05121376 (accessed on 27 December 2025).
- Verve Therapeutics, Inc. A Phase 1b Single Ascending Dose Study to Evaluate the Safety of VERVE-201 in Patients with Refractory Hyperlipidemia; Verve Therapeutics, Inc.: Boston, MA, USA, 2025. Available online: https://clinicaltrials.gov/study/NCT06451770 (accessed on 12 November 2025).
- CRISPR Therapeutics Reports Positive Additional Phase 1 Data for CTX310TM Targeting ANGPTL3 and Provides Update on In Vivo Cardiovascular Pipeline. Available online: https://crisprtx.com/about-us/press-releases-and-presentations/crispr-therapeutics-reports-positive-additional-phase-1-data-for-ctx310-targeting-angptl3-and-provides-update-on-in-vivo-cardiovascular-pipeline (accessed on 14 November 2025).
- Ray, K.K.; Wright, R.S.; Kallend, D.; Koenig, W.; Leiter, L.A.; Raal, F.J.; Bisch, J.A.; Richardson, T.; Jaros, M.; Wijngaard, P.L.J.; et al. Two Phase 3 Trials of Inclisiran in Patients with Elevated LDL Cholesterol. N. Engl. J. Med. 2020, 382, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Ikaria Bioscience Pty Ltd. A Phase 1, Randomized, Single-Blinded, Placebo-Controlled, Single-Ascending-Dose Study to Evaluate the Safety, Tolerability, Pharmacokinetics, and Pharmacodynamic Effects of RN0191 in Adult Subjects with Elevated Low-Density Lipoprotein-Cholesterol; Ikaria Bioscience Pty Ltd.: Hampton, NJ, USA, 2024. Available online: https://clinicaltrials.gov/study/NCT05905068 (accessed on 27 December 2025).
- Rosenson, R.S.; Gaudet, D.; Hegele, R.A.; Ballantyne, C.M.; Nicholls, S.J.; Lucas, K.J.; Martin, J.S.; Zhou, R.; Muhsin, M.; Chang, T.; et al. Zodasiran, an RNAi Therapeutic Targeting ANGPTL3, for Mixed Hyperlipidemia. N. Engl. J. Med. 2024, 391, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, D.; Karwatowska-Prokopczuk, E.; Baum, S.J.; Hurh, E.; Kingsbury, J.; Bartlett, V.J.; Figueroa, A.L.; Piscitelli, P.; Singleton, W.; Witztum, J.L.; et al. Vupanorsen, an N-Acetyl Galactosamine-Conjugated Antisense Drug to ANGPTL3 mRNA, Lowers Triglycerides and Atherogenic Lipoproteins in Patients with Diabetes, Hepatic Steatosis, and Hypertriglyceridaemia. Eur. Heart J. 2020, 41, 3936–3945. [Google Scholar] [CrossRef]
| Feature | Transient Delivery | Sustained Delivery |
|---|---|---|
| Active Duration | Hours to Days | Months to Permanent, |
| Primary Vehicles | mRNA, RNPs, AdVs, | Integrating LVs, episomal AAV, |
| Off-Target Risk | Lower (limited active time) | Higher (prolonged activity), |
| Immunogenicity | Generally lower/transient | Potentially persistent/toxic |
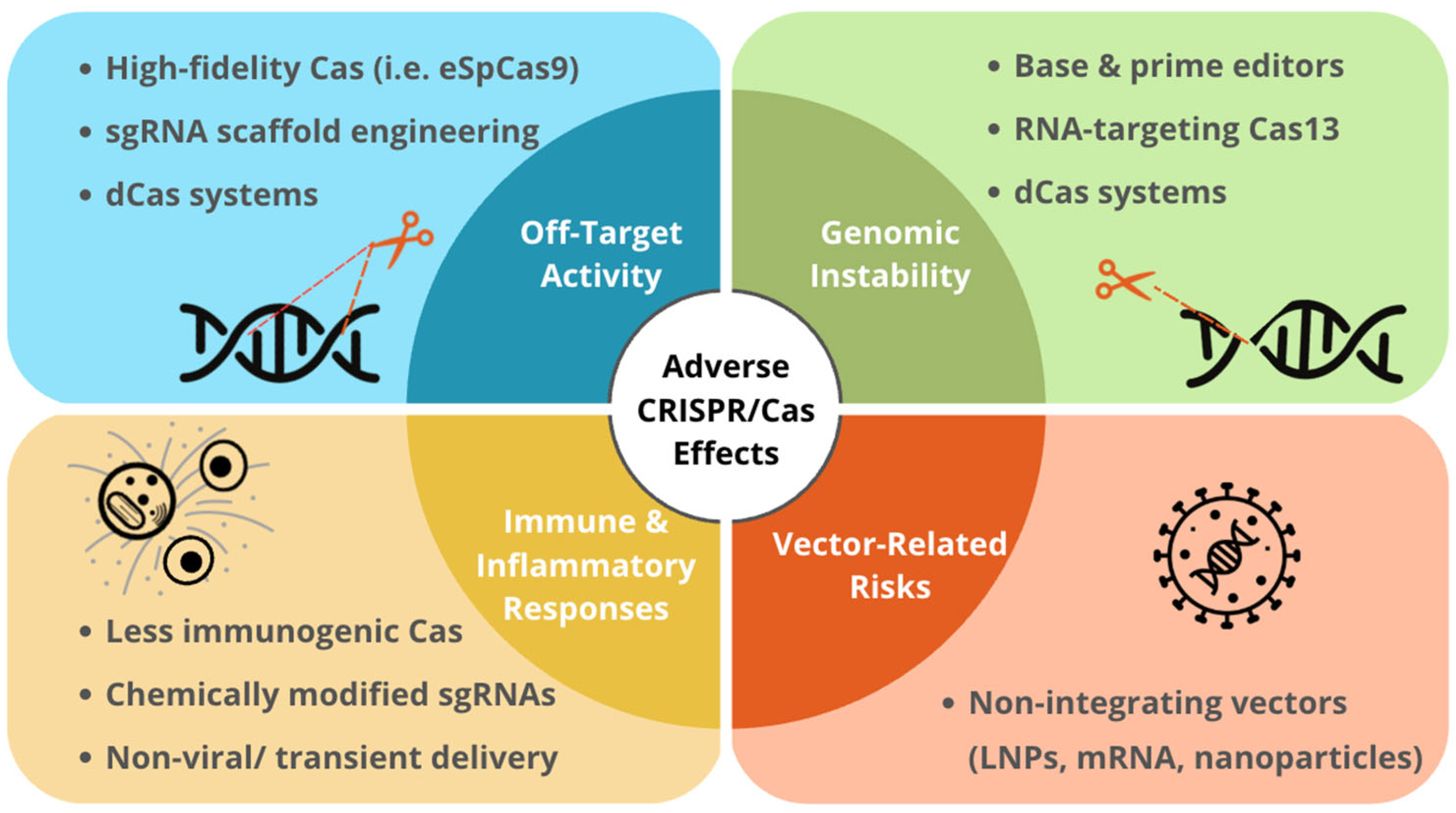
| Adverse Effect | Mechanism | Consequences | Modifications to Address Adverse Effects |
|---|---|---|---|
| Double Stranded Breaks (DSB) | CRISPR/Cas9 endonuclease activity forms DSBs | Indels Chromosomal translocations | Base editors and prime editors that do not create DSBs Cas13 RNA editor instead editing DNA |
| Off-Target Genomic Editing | Nonspecific sgRNA binding | Editing at other loci | High fidelity Cas enzymes sgRNA design |
| Off-Target Transcriptomic Effects | Off-target base editing Off-target transcription repressor-fused dCas activity | Unintended changes in gene expression | High fidelity Cas enzymes sgRNA design |
| Genomic Vector Integration | Integration of AAVs into host genome | Introduction of new mutations Low oncogenic risks | Alternative delivery vectors Non-DSB editing approaches |
| Immunogenicity | Cas protein sgRNAs Delivery vector | Elevated inflammatory markers Hyperactivation of the innate immune system, hemolytic disorders at high doses of AAVs | Alternative delivery vectors Engineering less immunogenic sgRNAs |
| Tumorigenicity | DSBs Off-target editing | Activation of oncogenic genes or suppression of tumor repressor genes | Modifications reduce DSBs and off-target editing |
| Drug Name | Condition Treated | Alternative Therapies |
|---|---|---|
| EDIT-101 [136] | CEP290-associated inherited retinal degeneration (Leber’s Congenital Amaurosis) | MCO-010 * [142], GS030 * [143], QR-110 [144] |
| NTLA-2001 [55] | ATTR amyloidosis | Patisiran [145], Vutrisiran [146], Inotersen [147], Eplontersen [148] |
| NTLA-2002 [141] | Hereditary Angioedema (KLKB1 target) | Donidalorsen [149], Garadacimab * [150], BMN-331 [151] |
| VERVE 102 [145] | LDL-cholesterol excess (PCSK9 target) | VERVE-201 [152], CTX310 [153], Inclisiran [154], RN0191 [155], ARO-ANG3 [156], Vupanorsen [157] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Bao, C.; Channell, C.I.; Tseng, Y.H.; Bailey, J.; Sbaiti, N.; Demirkol, A.; Tsang, S.H. Chronic In Vivo CRISPR-Cas Genome Editing: Challenges, Long-Term Safety, and Outlook. Cells 2026, 15, 156. https://doi.org/10.3390/cells15020156
Bao C, Channell CI, Tseng YH, Bailey J, Sbaiti N, Demirkol A, Tsang SH. Chronic In Vivo CRISPR-Cas Genome Editing: Challenges, Long-Term Safety, and Outlook. Cells. 2026; 15(2):156. https://doi.org/10.3390/cells15020156
Chicago/Turabian StyleBao, Caroline, Catherine I. Channell, Yi Hsuan Tseng, Johnathan Bailey, Naeem Sbaiti, Aykut Demirkol, and Stephen H. Tsang. 2026. "Chronic In Vivo CRISPR-Cas Genome Editing: Challenges, Long-Term Safety, and Outlook" Cells 15, no. 2: 156. https://doi.org/10.3390/cells15020156
APA StyleBao, C., Channell, C. I., Tseng, Y. H., Bailey, J., Sbaiti, N., Demirkol, A., & Tsang, S. H. (2026). Chronic In Vivo CRISPR-Cas Genome Editing: Challenges, Long-Term Safety, and Outlook. Cells, 15(2), 156. https://doi.org/10.3390/cells15020156







