Transcriptome Analysis Reveals the Requirement of the TGFβ Pathway in Ascidian Tail Regression
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Collection and Fertilization
2.2. Drug Treatment and IMAGING
2.3. RNA Extraction and Transcriptome Sequencing
2.4. Transcriptome Analysis
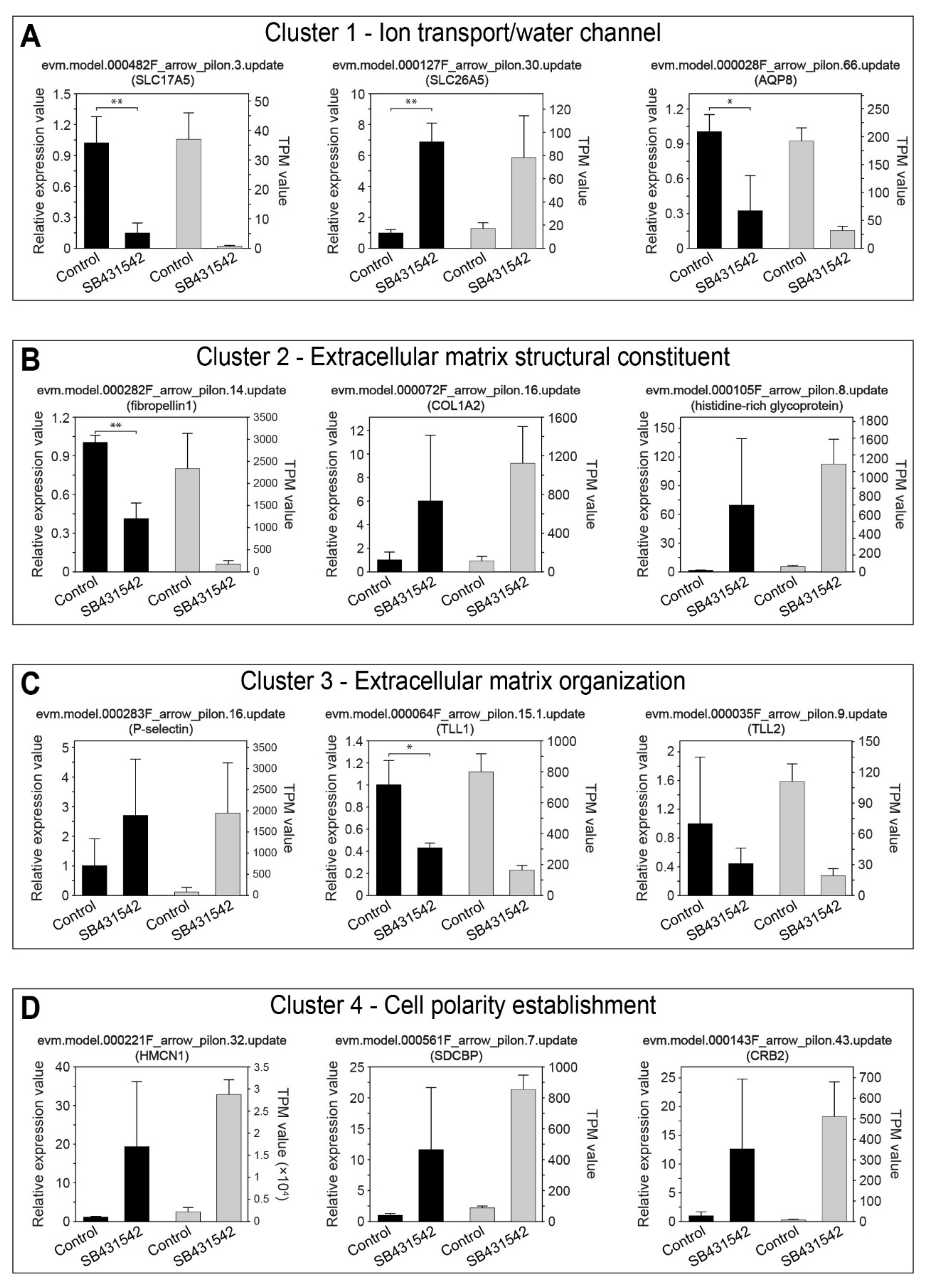
2.5. Quantitative Real-Time PCR (qRT-PCR) and Analysis
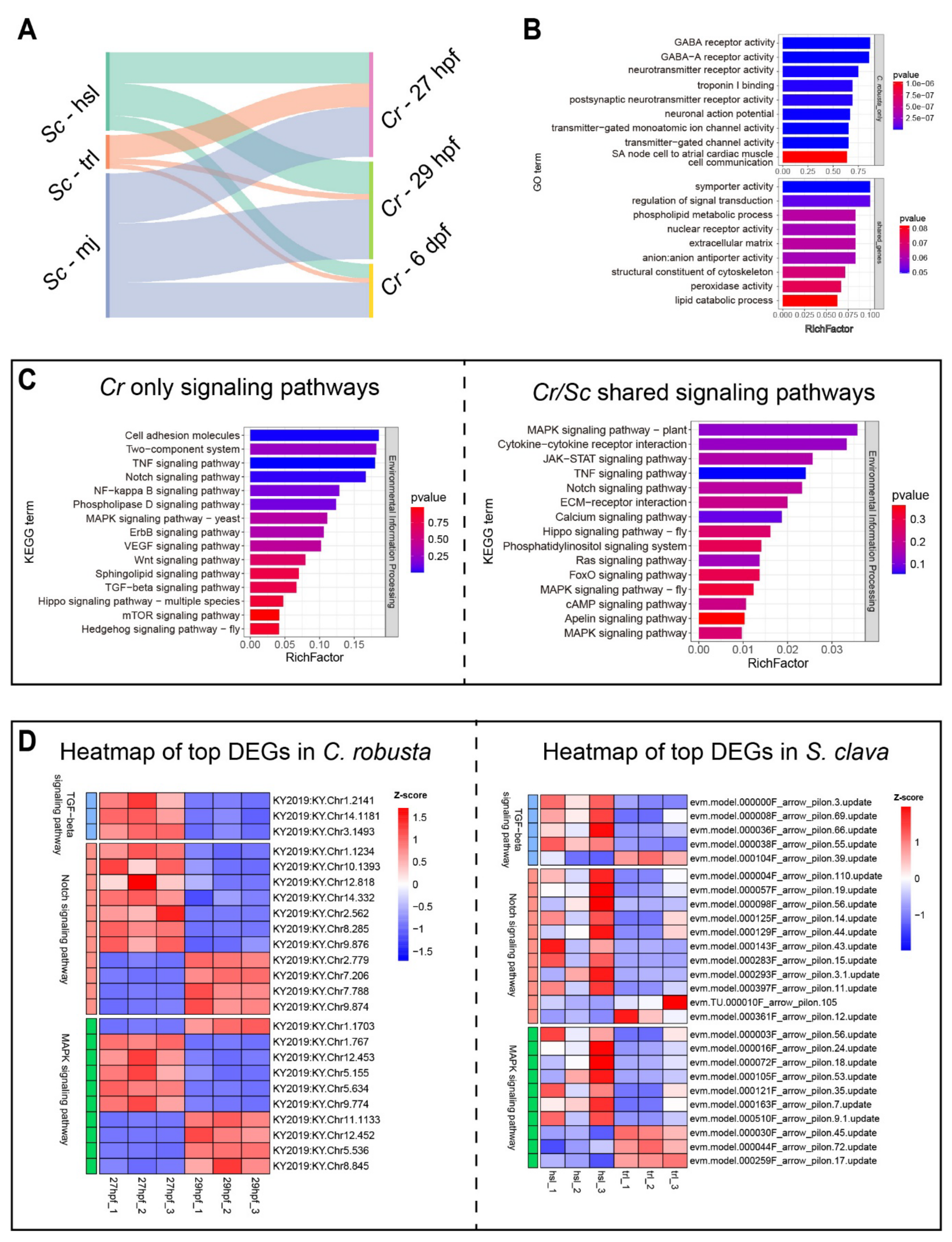
2.6. Cross-Species Analysis Between S. clava and C. robusta
2.7. Statistics
3. Results
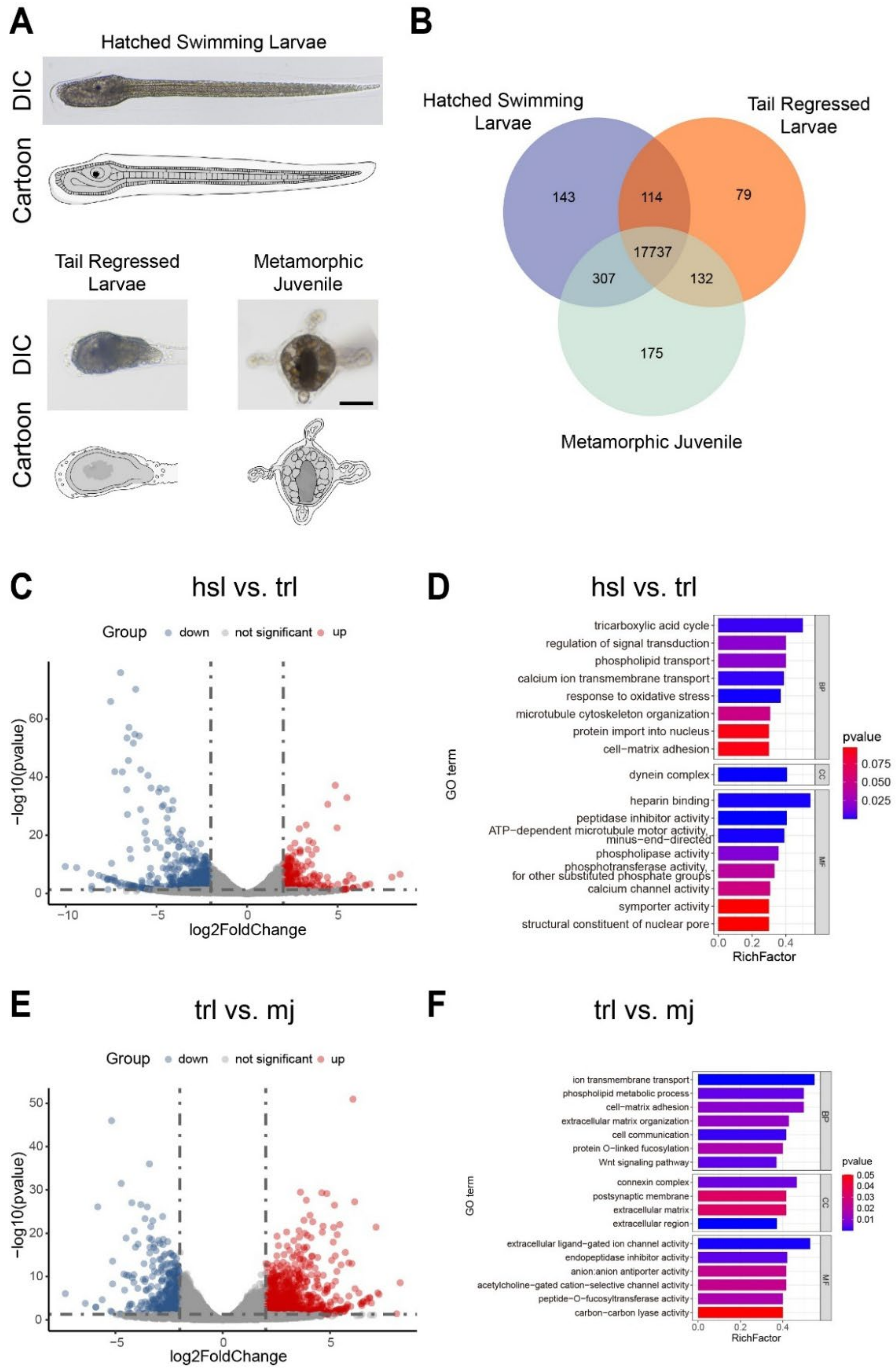
3.1. Morphological Observation and Gene Expression Profile of S. clava Tail Regression
3.2. Screening of Potential Regulatory Signaling Pathways During Tail Regression
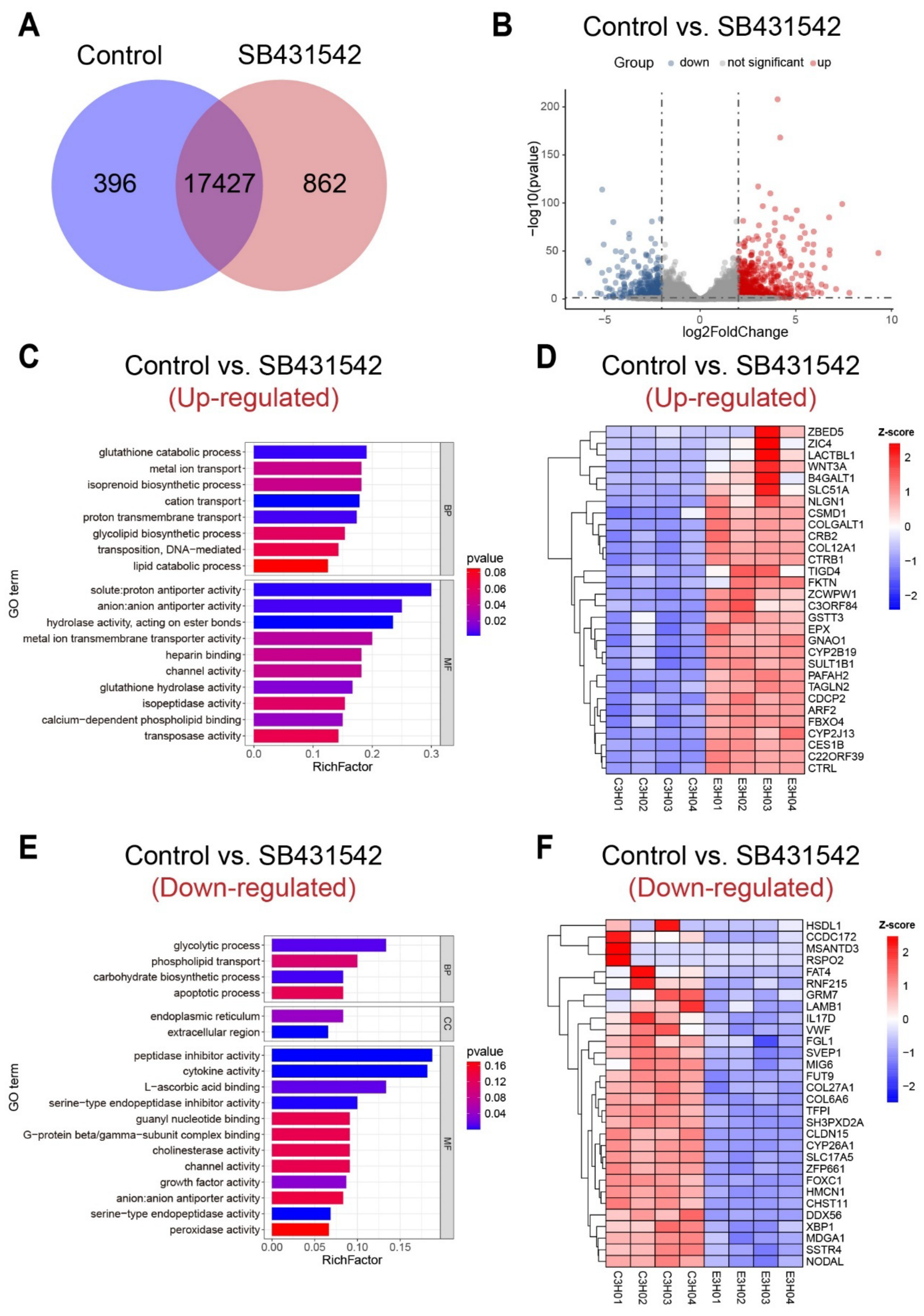
3.3. Inhibition of TGFβ Signaling Pathway Cause Failure of Tail Regression
3.4. Transcriptomic Profiling Reveals the Downstream Regulatory Patterns of TGFβ Pathway
3.5. Multi-Species Comparison Explains the Conservation and Divergence of Tail Regression in Ascidians
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilbert, L.I.; Tata, J.R.; Atkinson, B.G. Metamorphosis: Postembryonic Reprogramming of Gene Expression in Amphibian and Insect Cells; Academic Press: Cambridge, MA, USA, 1996. [Google Scholar]
- Brown, D. Amphibian metamorphosis: From morphology to molecular biology. BioEssays 2000, 22, 775. [Google Scholar] [CrossRef]
- Hall, B.K.; Wake, M.H. The Origin and Evolution of Larval Forms; Gulf Professional Publishing: Oxford, UK, 1999. [Google Scholar]
- Laudet, V. The origins and evolution of vertebrate metamorphosis. Curr. Biol. 2011, 21, R726–R737. [Google Scholar] [CrossRef]
- Dong, B. Cellular processes and gene regulatory network of notochord development in a marine model animal: Ciona intestinalis. Sci. Bull. 2015, 60, 1167–1179. [Google Scholar] [CrossRef]
- Cloney, R.A. Ascidian larvae and the events of metamorphosis. Am. Zool. 1982, 22, 817–826. [Google Scholar]
- Chambon, J.P.; Nakayama, A.; Takamura, K.; McDougall, A.; Satoh, N. ERK- and JNK-signalling regulate gene networks that stimulate metamorphosis and apoptosis in tail tissues of ascidian tadpoles. Development 2007, 134, 1203–1219. [Google Scholar] [CrossRef] [PubMed]
- Comes, S.; Locascio, A.; Silvestre, F.; d’Ischia, M.; Russo, G.L.; Tosti, E.; Branno, M.; Palumbo, A. Regulatory roles of nitric oxide during larval development and metamorphosis in Ciona intestinalis. Dev. Biol. 2007, 306, 772–784. [Google Scholar] [CrossRef]
- Hozumi, A.; Matsunobu, S.; Mita, K.; Treen, N.; Sugihara, T.; Horie, T.; Sakuma, T.; Yamamoto, T.; Shiraishi, A.; Hamada, M.; et al. GABA-Induced GnRH Release Triggers Chordate Metamorphosis. Curr. Biol. 2020, 30, 1555–1561.e4. [Google Scholar] [CrossRef]
- Lash, J.W.; Cloney, R.A.; Minor, R.R. The Effect of Cytochalasin B upon Tail Resorption and Metamorphosis in Ten Species of Ascidians. Biol. Bull. 1973, 145, 360–372. [Google Scholar] [CrossRef]
- Yamaji, S.; Hozumi, A.; Matsunobu, S.; Sasakura, Y. Orchestration of the distinct morphogenetic movements in different tissues drives tail regression during ascidian metamorphosis. Dev. Biol. 2020, 465, 66–78. [Google Scholar] [CrossRef]
- Lin, B.; Shi, W.; Lu, Q.; Shito, T.T.; Yu, H.; Dong, B. Establishment of a developmental atlas and transgenetic tools in the ascidian Styela clava. Mar. Life Sci. Technol. 2023, 5, 435–454. [Google Scholar] [CrossRef]
- Dupont, L.; Viard, F.; Dowell, M.J.; Wood, C.; Bishop, J.D. Fine- and regional-scale genetic structure of the exotic ascidian Styela clava (Tunicata) in southwest England, 50 years after its introduction. Mol. Ecol. 2009, 18, 442–453. [Google Scholar] [CrossRef] [PubMed]
- Goldstien, S.J.; Schiel, D.R.; Gemmell, N.J. Regional connectivity and coastal expansion: Differentiating pre-border and post-border vectors for the invasive tunicate Styela clava. Mol. Ecol. 2010, 19, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Cinar, M.E. The alien ascidian Styela clava now invading the Sea of Marmara (Tunicata: Ascidiacea). ZooKeys 2016, 563, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Goldstien, S.J.; Dupont, L.; Viard, F.; Hallas, P.J.; Nishikawa, T.; Schiel, D.R.; Gemmell, N.J.; Bishop, J.D. Global phylogeography of the widely introduced North West Pacific ascidian Styela clava. PLoS ONE 2011, 6, e16755. [Google Scholar] [CrossRef]
- Mastrototaro, F.; Gasparini, F.; Montesanto, F. The clubbed tunicate Styela clava has arrived in the Lagoon of Venice. Eur. Zool. J. 2022, 89, 502–509. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, J.; Lu, Q.; Ren, P.; Guo, X.; Wang, J.; Li, X.; Chang, Y.; Duan, S.; Wang, S.; et al. Genomic basis of environmental adaptation in the leathery sea squirt (Styela clava). Mol. Ecol. Resour. 2020, 20, 1414–1431. [Google Scholar] [CrossRef]
- Zhang, J.; Wei, J.; Yu, H.; Dong, B. Genome-Wide Identification, Comparison, and Expression Analysis of Transcription Factors in Ascidian Styela clava. Int. J. Mol. Sci. 2021, 22, 4317. [Google Scholar] [CrossRef]
- Hotta, K.; Mitsuhara, K.; Takahashi, H.; Inaba, K.; Oka, K.; Gojobori, T.; Ikeo, K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 2007, 236, 1790–1805. [Google Scholar] [CrossRef]
- Hotta, K.; Dauga, D.; Manni, L. The ontology of the anatomy and development of the solitary ascidian Ciona: The swimming larva and its metamorphosis. Sci. Rep. 2020, 10, 17916. [Google Scholar] [CrossRef]
- Kobayashi, K.; Satou, Y. Microinjection of Exogenous Nucleic Acids into Eggs: Ciona Species. In Transgenic Ascidians; Sasakura, Y., Ed.; Springer: Singapore, 2018; pp. 5–13. [Google Scholar]
- Ikushima, H.; Todo, T.; Ino, Y.; Takahashi, M.; Miyazawa, K.; Miyazono, K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell 2009, 5, 504–514. [Google Scholar] [CrossRef]
- Wei, J.; Liu, P.; Liu, F.; Jiang, A.; Qiao, J.; Pu, Z.; Wang, B.; Zhang, J.; Jia, D.; Li, Y.; et al. EDomics: A comprehensive and comparative multi-omics database for animal evo-devo. Nucleic Acids Res. 2023, 51, D913–D923. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wilkinson, L. ggplot2: Elegant Graphics for Data Analysis by Wickham, H. Biometrics 2011, 67, 678–679. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Zambelli, F.; Mastropasqua, F.; Picardi, E.; D’Erchia, A.M.; Pesole, G.; Pavesi, G. RNentropy: An entropy-based tool for the detection of significant variation of gene expression across multiple RNA-Seq experiments. Nucleic Acids Res. 2018, 46, e46. [Google Scholar] [CrossRef]
- Jiang, A.; Han, K.; Wei, J.; Su, X.; Wang, R.; Zhang, W.; Liu, X.; Qiao, J.; Liu, P.; Liu, Q.; et al. Spatially resolved single-cell atlas of ascidian endostyle provides insight into the origin of vertebrate pharyngeal organs. Sci. Adv. 2024, 10, eadi9035. [Google Scholar] [CrossRef]
- Yamada, S.; Hotta, K.; Yamamoto, T.S.; Ueno, N.; Satoh, N.; Takahashi, H. Interaction of notochord-derived fibrinogen-like protein with Notch regulates the patterning of the central nervous system of Ciona intestinalis embryos. Dev. Biol. 2009, 328, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Hamada, M.; Goricki, S.; Byerly, M.S.; Satoh, N.; Jeffery, W.R. Evolution of the chordate regeneration blastema: Differential gene expression and conserved role of notch signaling during siphon regeneration in the ascidian Ciona. Dev. Biol. 2015, 405, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Denker, E.; Sehring, I.M.; Dong, B.; Audisso, J.; Mathiesen, B.; Jiang, D. Regulation by a TGFbeta-ROCK-actomyosin axis secures a non-linear lumen expansion that is essential for tubulogenesis. Development 2015, 142, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Vizzini, A.; Bonura, A.; La Paglia, L.; Fiannaca, A.; La Rosa, M.; Urso, A.; Arizza, V. ceRNA Network Regulation of TGF-beta, WNT, FOXO, Hedgehog Pathways in the Pharynx of Ciona robusta. Int. J. Mol. Sci. 2021, 22, 3497. [Google Scholar] [CrossRef]
- Kaplan, N.A.; Wang, W.; Christiaen, L. Initial characterization of Wnt-Tcf functions during Ciona heart development. Dev. Biol. 2019, 448, 199–209. [Google Scholar] [CrossRef]
- Matis, M.; Axelrod, J.D. Regulation of PCP by the Fat signaling pathway. Genes. Dev. 2013, 27, 2207–2220. [Google Scholar] [CrossRef]
- Deng, W.; Nies, F.; Feuer, A.; Bocina, I.; Oliver, D.; Jiang, D. Anion translocation through an Slc26 transporter mediates lumen expansion during tubulogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 14972–14977. [Google Scholar] [CrossRef]
- Verkman, A.S. More than just water channels: Unexpected cellular roles of aquaporins. J. Cell Sci. 2005, 118, 3225–3232. [Google Scholar] [CrossRef]
- Peng, H.; Qiao, J.; Wang, G.; Shi, W.; Xia, F.; Qiao, R.; Dong, B. A collagen-rich arch in the urochordate notochord coordinates cell shaping and multi-tissue elongation. Curr. Biol. 2023, 33, 5390–5403.e3. [Google Scholar] [CrossRef]
- Wei, J.; Wang, G.; Li, X.; Ren, P.; Yu, H.; Dong, B. Architectural delineation and molecular identification of extracellular matrix in ascidian embryos and larvae. Biol. Open 2017, 6, 1383–1390. [Google Scholar] [CrossRef]
- Xu, X.; Vogel, B.E. A secreted protein promotes cleavage furrow maturation during cytokinesis. Curr. Biol. 2011, 21, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Toffoli, B.; Zennaro, C.; Winkler, C.; Giordano Attianese, G.M.P.; Bernardi, S.; Carraro, M.; Gilardi, F.; Desvergne, B. Hemicentin 1 influences podocyte dynamic changes in glomerular diseases. Am. J. Physiol. Renal. Physiol. 2018, 314, F1154–F1165. [Google Scholar] [CrossRef] [PubMed]
- Yaoita, Y. Tail Resorption During Metamorphosis in Xenopus Tadpoles. Front. Endocrinol. 2019, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Matsuda, H.; Fujimoto, K.; Sun, G.; Matsuura, K.; Shi, Y.B. Molecular and genetic studies suggest that thyroid hormone receptor is both necessary and sufficient to mediate the developmental effects of thyroid hormone. Gen. Comp. Endocrinol. 2010, 168, 174–180. [Google Scholar] [CrossRef][Green Version]
- Buchholz, D.R. Xenopus metamorphosis as a model to study thyroid hormone receptor function during vertebrate developmental transitions. Mol. Cell Endocrinol. 2017, 459, 64–70. [Google Scholar] [CrossRef]
- McNabb, F.M. The hypothalamic-pituitary-thyroid (HPT) axis in birds and its role in bird development and reproduction. Crit. Rev. Toxicol. 2007, 37, 163–193. [Google Scholar] [CrossRef]
- Buchholz, D.R. More similar than you think: Frog metamorphosis as a model of human perinatal endocrinology. Dev. Biol. 2015, 408, 188–195. [Google Scholar] [CrossRef]
- Pearce, C.M.; Scheibling, R.E. Induction of Metamorphosis of Larvae of the Green Sea Urchin, Strongylocentrotus droebachiensis, by Coralline Red Algae. Biol. Bull. 1990, 179, 304–311. [Google Scholar] [CrossRef]
- García-Lavandeira, M.; Silva, A.; Abad, M.; Pazos, A.J.; Sánchez, J.L.; Luz Pérez-Parallé, M. Effects of GABA and epinephrine on the settlement and metamorphosis of the larvae of four species of bivalve molluscs. J. Exp. Mar. Biol. Ecol. 2005, 316, 149–156. [Google Scholar] [CrossRef]
- Izutsu, Y. The immune system is involved in Xenopus metamorphosis. Front. Biosci. 2009, 14, 141–149. [Google Scholar] [CrossRef]
- Sasakura, Y.; Hozumi, A. Formation of adult organs through metamorphosis in ascidians. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e304. [Google Scholar] [CrossRef]

| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| SLC26A5 | AAAGCAACGCCAACAGAGG | TCATGTCCAAGACGAAATGAGTAA |
| SLC17A5 | ACTGGCGGATTGCCTCATC | TGGCTGCTGGTACACTTGGTC |
| AQP8 | TTTCGGTCCAGCGGTTGT | ATCTAATGGTCCTTCTCCATCGT |
| fibropellin1 | TATTGTCAGTGCGACAGAGGTG | GACATTTTGCGTGGGGATT |
| COL1A2 | TGTAAACGGAACCAATGGAATG | GCTGACTGTTGTAATCGGCACT |
| CRB2 | CCCGAATACGGAAATCGAGA | CCGAGGGCAAATGTCAGAAC |
| HMCN1 | GACTCGCACCCGTAAGTGTTT | ACGCTGCATTCGCTCCAT |
| SDCBP | AGATAGTAGCGGCCATGTTGG | CGCATTGTCCGTTCACTTCAC |
| TLL1 | CGCGGAAACGCTGTTAGG | CACGGTGGTGATCTTTGTGG |
| TLL2 | AAGCAGTACGAGGGGAAGATTACAT | CTGTTGATGCAGCCGGTGTAA |
| glycoprotein | ATCACCACCATCACCATGGACC | GTGTGGATATCCTCCGTGACCTG |
| P-selectin | GAAGCACTGAGATCCCAAGGAGTTCT | GTCGCAAGGATCGCCACAAATATTT |
| β-actin | AATCGTGACCAACTGGGATG | GCTGGAGTATTGAAGGTTTCGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, W.; Liu, P.; Yang, D.; Zhuang, Y.; Lin, B.; Dong, B. Transcriptome Analysis Reveals the Requirement of the TGFβ Pathway in Ascidian Tail Regression. Cells 2025, 14, 546. https://doi.org/10.3390/cells14070546
Shi W, Liu P, Yang D, Zhuang Y, Lin B, Dong B. Transcriptome Analysis Reveals the Requirement of the TGFβ Pathway in Ascidian Tail Regression. Cells. 2025; 14(7):546. https://doi.org/10.3390/cells14070546
Chicago/Turabian StyleShi, Wenjie, Penghui Liu, Dongyu Yang, Yuan Zhuang, Boyan Lin, and Bo Dong. 2025. "Transcriptome Analysis Reveals the Requirement of the TGFβ Pathway in Ascidian Tail Regression" Cells 14, no. 7: 546. https://doi.org/10.3390/cells14070546
APA StyleShi, W., Liu, P., Yang, D., Zhuang, Y., Lin, B., & Dong, B. (2025). Transcriptome Analysis Reveals the Requirement of the TGFβ Pathway in Ascidian Tail Regression. Cells, 14(7), 546. https://doi.org/10.3390/cells14070546








