Integrative Analysis of Metabolome and Proteome in the Cerebrospinal Fluid of Patients with Multiple System Atrophy
Abstract
1. Introduction
2. Characteristic Features of MSA
3. Biochemical Analysis of CSF for the Diagnosis of MSA
4. Metabolomics Approaches in CSF with MSA
5. Analysis of Proteomes in CSF with MSA
6. Integrated Omics to Enhance Diagnostic Accuracy
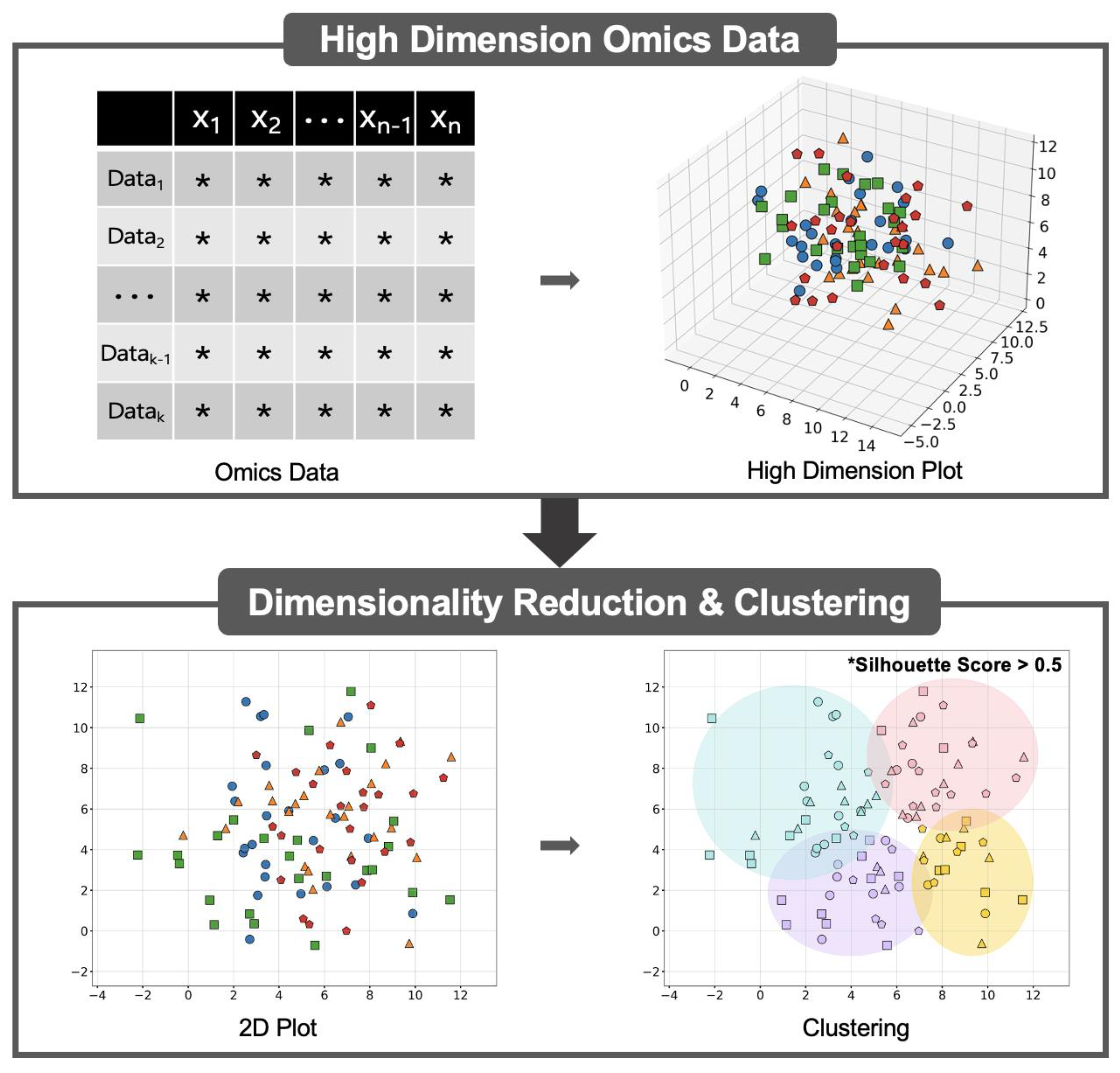
7. Future Directions Using Machine Learning
8. Summary
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Fanciulli, A.; Wenning, G.K. Multiple-system atrophy. N. Engl. J. Med. 2015, 372, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Swan, L.; Dupont, J. Multiple system atrophy. Phys. Ther. 1999, 79, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Kaindlstorfer, C.; Granata, R.; Wenning, G.K. Tremor in Multiple System Atrophy—A review. Tremor Other Hyperkinetic Mov. 2013, 3. [Google Scholar] [CrossRef]
- Jellinger, K.A.; Lantos, P.L. Papp-Lantos inclusions and the pathogenesis of multiple system atrophy: An update. Acta Neuropathol. 2010, 119, 657–667. [Google Scholar] [CrossRef]
- Shahnawaz, M.; Mukherjee, A.; Pritzkow, S.; Mendez, N.; Rabadia, P.; Liu, X.; Hu, B.; Schmeichel, A.; Singer, W.; Wu, G.; et al. Discriminating alpha-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 2020, 578, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xiao, Y.; Hou, Y.; Li, C.; Zhang, L.; Ou, R.; Wei, Q.; Lin, J.; Yang, T.; Che, N.; et al. Comparison of spontaneous brain activity in distinguishing parkinsonian variant of multiple system atrophy from Parkinson’s disease at an early stage. Front. Aging Neurosci. 2024, 16, 1427991. [Google Scholar] [CrossRef] [PubMed]
- Wenning, G.K.; Ben-Shlomo, Y.; Hughes, A.; Daniel, S.E.; Lees, A.; Quinn, N.P. What clinical features are most useful to distinguish definite multiple system atrophy from Parkinson’s disease? J. Neurol. Neurosurg. Psychiatry 2000, 68, 434–440. [Google Scholar] [CrossRef]
- Bargar, C.; De Luca, C.M.G.; Devigili, G.; Elia, A.E.; Cilia, R.; Portaleone, S.M.; Wang, W.; Tramacere, I.; Bistaffa, E.; Cazzaniga, F.A.; et al. Discrimination of MSA-P and MSA-C by RT-QuIC analysis of olfactory mucosa: The first assessment of assay reproducibility between two specialized laboratories. Mol. Neurodegener. 2021, 16, 82. [Google Scholar] [CrossRef]
- Palma, J.A.; Norcliffe-Kaufmann, L.; Kaufmann, H. Diagnosis of multiple system atrophy. Auton. Neurosci. 2018, 211, 15–25. [Google Scholar] [CrossRef]
- Abdo, W.F.; De Jong, D.; Hendriks, J.C.; Horstink, M.W.; Kremer, B.P.; Bloem, B.R.; Verbeek, M.M. Cerebrospinal fluid analysis differentiates multiple system atrophy from Parkinson’s disease. Mov. Disord. 2004, 19, 571–579. [Google Scholar] [CrossRef]
- Lee, P.H.; Lee, G.; Paik, M.J. Polyunsaturated fatty acid levels in the cerebrospinal fluid of patients with Parkinson’s disease and multiple system atrophy. Mov. Disord. 2008, 23, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Paik, M.J.; Ahn, Y.H.; Lee, P.H.; Kang, H.; Park, C.B.; Choi, S.; Lee, G. Polyamine patterns in the cerebrospinal fluid of patients with Parkinson’s disease and multiple system atrophy. Clin. Chim. Acta 2010, 411, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Hwang, J.S.; George, N.P.; Jang, Y.E.; Kwon, M.; Lee, S.S.; Lee, G. Integrative Metabolome and Proteome Analysis of Cerebrospinal Fluid in Parkinson’s Disease. Int. J. Mol. Sci. 2024, 25, 11406. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef]
- Hwang, J.S.; Kim, S.G.; George, N.P.; Kwon, M.; Jang, Y.E.; Lee, S.S.; Lee, G. Biological Function Analysis of MicroRNAs and Proteins in the Cerebrospinal Fluid of Patients with Parkinson’s Disease. Int. J. Mol. Sci. 2024, 25, 13260. [Google Scholar] [CrossRef]
- Feldner-Busztin, D.; Firbas Nisantzis, P.; Edmunds, S.J.; Boza, G.; Racimo, F.; Gopalakrishnan, S.; Limborg, M.T.; Lahti, L.; de Polavieja, G.G. Dealing with dimensionality: The application of machine learning to multi-omics data. Bioinformatics 2023, 39, btad021. [Google Scholar] [CrossRef]
- Shim, W.; Paik, M.J.; Nguyen, D.T.; Lee, J.K.; Lee, Y.; Kim, J.H.; Shin, E.H.; Kang, J.S.; Jung, H.S.; Choi, S.; et al. Analysis of changes in gene expression and metabolic profiles induced by silica-coated magnetic nanoparticles. ACS Nano 2012, 6, 7665–7680. [Google Scholar] [CrossRef]
- Shin, T.H.; Lee, D.Y.; Manavalan, B.; Basith, S.; Na, Y.C.; Yoon, C.; Lee, H.S.; Paik, M.J.; Lee, G. Silica-coated magnetic nanoparticles activate microglia and induce neurotoxic D-serine secretion. Part. Fibre Toxicol. 2021, 18, 30. [Google Scholar] [CrossRef]
- Dong, X.; Liu, C.; Dozmorov, M. Review of multi-omics data resources and integrative analysis for human brain disorders. Brief. Funct. Genom. 2021, 20, 223–234. [Google Scholar] [CrossRef]
- Shin, T.H.; Nithiyanandam, S.; Lee, D.Y.; Kwon, D.H.; Hwang, J.S.; Kim, S.G.; Jang, Y.E.; Basith, S.; Park, S.; Mo, J.S.; et al. Analysis of Nanotoxicity with Integrated Omics and Mechanobiology. Nanomaterials 2021, 11, 2385. [Google Scholar] [CrossRef]
- Anwar, M.Y.; Highland, H.; Buchanan, V.L.; Graff, M.; Young, K.; Taylor, K.D.; Tracy, R.P.; Durda, P.; Liu, Y.; Johnson, C.W.; et al. Machine learning-based clustering identifies obesity subgroups with differential multi-omics profiles and metabolic patterns. Obesity 2024, 32, 2024–2034. [Google Scholar] [CrossRef] [PubMed]
- DeGroat, W.; Abdelhalim, H.; Peker, E.; Sheth, N.; Narayanan, R.; Zeeshan, S.; Liang, B.T.; Ahmed, Z. Multimodal AI/ML for discovering novel biomarkers and predicting disease using multi-omics profiles of patients with cardiovascular diseases. Sci. Rep. 2024, 14, 26503. [Google Scholar] [CrossRef]
- Shin, T.H.; Lee, G. Reduced lysosomal activity and increased amyloid beta accumulation in silica-coated magnetic nanoparticles-treated microglia. Arch. Toxicol. 2024, 98, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Cui, S.; Huang, P.; Gao, C.; Zhang, P.; Liu, J.; Li, H.; Huang, M.; Shen, X.; Liu, Z.; et al. Predicting the Prognosis of Multiple System Atrophy Using Cluster and Principal Component Analysis. J. Park. Dis. 2023, 13, 937–946. [Google Scholar] [CrossRef] [PubMed]
- Claassen, D.O. Multiple System Atrophy. Continuum 2022, 28, 1350–1363. [Google Scholar] [CrossRef]
- Poewe, W.; Stankovic, I.; Halliday, G.; Meissner, W.G.; Wenning, G.K.; Pellecchia, M.T.; Seppi, K.; Palma, J.A.; Kaufmann, H. Multiple system atrophy. Nat. Rev. Dis. Primers 2022, 8, 56. [Google Scholar] [CrossRef]
- Ndayisaba, A.; Halliday, G.M.; Khurana, V. Multiple System Atrophy: Pathology, Pathogenesis, and Path Forward. Annu. Rev. Pathol. 2024, 20, 245–273. [Google Scholar] [CrossRef]
- Ben-Shlomo, Y.; Wenning, G.K.; Tison, F.; Quinn, N.P. Survival of patients with pathologically proven multiple system atrophy: A meta-analysis. Neurology 1997, 48, 384–393. [Google Scholar] [CrossRef]
- Schrag, A.; Wenning, G.K.; Quinn, N.; Ben-Shlomo, Y. Survival in multiple system atrophy. Mov. Disord. 2008, 23, 294–296. [Google Scholar] [CrossRef]
- Lee, H.J.; Ricarte, D.; Ortiz, D.; Lee, S.J. Models of multiple system atrophy. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Kaji, S.; Maki, T.; Ishimoto, T.; Yamakado, H.; Takahashi, R. Insights into the pathogenesis of multiple system atrophy: Focus on glial cytoplasmic inclusions. Transl. Neurodegener. 2020, 9, 7. [Google Scholar] [CrossRef]
- Watanabe, H.; Shima, S.; Mizutani, Y.; Ueda, A.; Ito, M. Multiple System Atrophy: Advances in Diagnosis and Therapy. J. Mov. Disord. 2023, 16, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Rukmani, M.R.; Yadav, R.; Bhaskarapillai, B.; Pal, P.K.; Sathyaprabha, T.N. Parkinsonian and Cerebellar Phenotypes of Probable MSA: An Insight in to Prognostic Factors Based on Autonomic Functions. Ann. Indian Acad. Neurol. 2020, 23, 289–295. [Google Scholar] [PubMed]
- Abdo, W.F.; van de Warrenburg, B.P.; Kremer, H.P.; Bloem, B.R.; Verbeek, M.M. CSF biomarker profiles do not differentiate between the cerebellar and parkinsonian phenotypes of multiple system atrophy. Park. Relat. Disord. 2007, 13, 480–482. [Google Scholar] [CrossRef] [PubMed]
- Al-Chalabi, A.; Durr, A.; Wood, N.W.; Parkinson, M.H.; Camuzat, A.; Hulot, J.S.; Morrison, K.E.; Renton, A.; Sussmuth, S.D.; Landwehrmeyer, B.G.; et al. Genetic variants of the alpha-synuclein gene SNCA are associated with multiple system atrophy. PLoS ONE 2009, 4, e7114. [Google Scholar] [CrossRef] [PubMed]
- Reis, P.M.; Holec, S.A.M.; Ezeiruaku, C.; Frost, M.P.; Brown, C.K.; Liu, S.L.; Olson, S.H.; Woerman, A.L. Structurally targeted mutagenesis identifies key residues supporting alpha -synuclein misfolding in multiple system atrophy. bioRxiv 2024. bioRxiv:2024.07.04.602104. [Google Scholar]
- Kon, T.; Ichimata, S.; Di Luca, D.G.; Martinez-Valbuena, I.; Kim, A.; Yoshida, K.; Alruwaita, A.A.; Kleiner, G.; Strafella, A.P.; Forrest, S.L.; et al. Multiple system atrophy with amyloid-beta-predominant Alzheimer’s disease neuropathologic change. Brain Commun. 2024, 6, fcae141. [Google Scholar] [CrossRef]
- Stefanova, N.; Reindl, M.; Neumann, M.; Haass, C.; Poewe, W.; Kahle, P.J.; Wenning, G.K. Oxidative stress in transgenic mice with oligodendroglial alpha-synuclein overexpression replicates the characteristic neuropathology of multiple system atrophy. Am. J. Pathol. 2005, 166, 869–876. [Google Scholar] [CrossRef]
- Ahmed, Z.; Asi, Y.T.; Lees, A.J.; Revesz, T.; Holton, J.L. Identification and quantification of oligodendrocyte precursor cells in multiple system atrophy, progressive supranuclear palsy and Parkinson’s disease. Brain Pathol. 2013, 23, 263–273. [Google Scholar] [CrossRef]
- Maeda, N.; Honda, H.; Suzuki, S.O.; Fujii, N.; Kira, J.I.; Iwaki, T. Mitochondrial dysfunction and altered ribostasis in hippocampal neurons with cytoplasmic inclusions of multiple system atrophy. Neuropathology 2018, 38, 361–371. [Google Scholar] [CrossRef]
- Foti, S.C.; Hargreaves, I.; Carrington, S.; Kiely, A.P.; Houlden, H.; Holton, J.L. Cerebral mitochondrial electron transport chain dysfunction in multiple system atrophy and Parkinson’s disease. Sci. Rep. 2019, 9, 6559. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Chen, T.H.; Koga, S.; Bredenberg, J.M.; Faroqi, A.H.; Delenclos, M.; Bu, G.; Wszolek, Z.K.; Carr, J.A.; Ross, O.A.; et al. Alpha-synuclein-associated changes in PINK1-PRKN-mediated mitophagy are disease context dependent. Brain Pathol. 2023, 33, e13175. [Google Scholar] [CrossRef]
- Papp, M.I.; Lantos, P.L. The distribution of oligodendroglial inclusions in multiple system atrophy and its relevance to clinical symptomatology. Brain 1994, 117 Pt 2, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, J.T.; Tanglay, O.; Li, A.A.; Strobbe, A.Y.G.; Kim, W.S.; Halliday, G.M.; Fu, Y. Role of Oligodendrocyte Lineage Cells in Multiple System Atrophy. Cells 2023, 12, 739. [Google Scholar] [CrossRef]
- Tokutake, T.; Kasuga, K.; Tsukie, T.; Ishiguro, T.; Shimohata, T.; Onodera, O.; Ikeuchi, T. Clinical correlations of cerebrospinal fluid biomarkers including neuron-glia 2 and neurofilament light chain in patients with multiple system atrophy. Park. Relat. Disord. 2022, 102, 30–35. [Google Scholar] [CrossRef]
- Compta, Y.; Dias, S.P.; Giraldo, D.M.; Perez-Soriano, A.; Munoz, E.; Saura, J.; Fernandez, M.; Bravo, P.; Camara, A.; Pulido-Salgado, M.; et al. Cerebrospinal fluid cytokines in multiple system atrophy: A cross-sectional Catalan MSA registry study. Park. Relat. Disord. 2019, 65, 3–12. [Google Scholar] [CrossRef]
- Laurens, B.; Constantinescu, R.; Freeman, R.; Gerhard, A.; Jellinger, K.; Jeromin, A.; Krismer, F.; Mollenhauer, B.; Schlossmacher, M.G.; Shaw, L.M.; et al. Fluid biomarkers in multiple system atrophy: A review of the MSA Biomarker Initiative. Neurobiol. Dis. 2015, 80, 29–41. [Google Scholar] [CrossRef]
- Marques, T.M.; Kuiperij, H.B.; Bruinsma, I.B.; van Rumund, A.; Aerts, M.B.; Esselink, R.A.J.; Bloem, B.R.; Verbeek, M.M. MicroRNAs in Cerebrospinal Fluid as Potential Biomarkers for Parkinson’s Disease and Multiple System Atrophy. Mol. Neurobiol. 2017, 54, 7736–7745. [Google Scholar] [CrossRef]
- Lenska-Mieciek, M.; Madetko-Alster, N.; Alster, P.; Krolicki, L.; Fiszer, U.; Koziorowski, D. Inflammation in multiple system atrophy. Front. Immunol. 2023, 14, 1214677. [Google Scholar] [CrossRef]
- Kwon, D.H.; Hwang, J.S.; Kim, S.G.; Jang, Y.E.; Shin, T.H.; Lee, G. Cerebrospinal Fluid Metabolome in Parkinson’s Disease and Multiple System Atrophy. Int. J. Mol. Sci. 2022, 23, 1879. [Google Scholar] [CrossRef]
- Goldstein, D.S.; Holmes, C.; Sharabi, Y. Cerebrospinal fluid biomarkers of central catecholamine deficiency in Parkinson’s disease and other synucleinopathies. Brain 2012, 135, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Polinsky, R.J.; Holmes, K.V.; Brown, R.T.; Weise, V. CSF acetylcholinesterase levels are reduced in multiple system atrophy with autonomic failure. Neurology 1989, 39, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Reiber, H. Dynamics of brain-derived proteins in cerebrospinal fluid. Clin. Chim. Acta 2001, 310, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.H.; Tennagels, S.; Gold, R.; Gerwert, K.; Beyer, L.; Tonges, L. Update on CSF Biomarkers in Parkinson’s Disease. Biomolecules 2022, 12, 329. [Google Scholar] [CrossRef]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Kuiper, M.A.; Teerlink, T.; Visser, J.J.; Bergmans, P.L.; Scheltens, P.; Wolters, E.C. L-glutamate, L-arginine and L-citrulline levels in cerebrospinal fluid of Parkinson’s disease, multiple system atrophy, and Alzheimer’s disease patients. J. Neural Transm. 2000, 107, 183–189. [Google Scholar] [CrossRef]
- Martignoni, E.; Blandini, F.; Petraglia, F.; Pacchetti, C.; Bono, G.; Nappi, G. Cerebrospinal fluid norepinephrine, 3-methoxy-4-hydroxyphenylglycol and neuropeptide Y levels in Parkinson’s disease, multiple system atrophy and dementia of the Alzheimer type. J. Neural Transm. Park. Dis. Dement. Sect. 1992, 4, 191–205. [Google Scholar] [CrossRef]
- Compta, Y.; Giraldo, D.M.; Muñoz, E.; Antonelli, F.; Fernández, M.; Bravo, P.; Soto, M.; Cámara, A.; Torres, F.; Martí, M.J. Cerebrospinal fluid levels of coenzyme Q10 are reduced in multiple system atrophy. Park. Relat. Disord. 2018, 46, 16–23. [Google Scholar] [CrossRef]
- Nagao, R.; Mizutani, Y.; Shima, S.; Ueda, A.; Ito, M.; Yoshimoto, J.; Watanabe, H. Correlations between serotonin impairments and clinical indices in multiple system atrophy. Eur. J. Neurol. 2024, 31, e16158. [Google Scholar] [CrossRef]
- Kuiper, M.A.; Visser, J.J.; Bergmans, P.L.; Scheltens, P.; Wolters, E.C. Decreased cerebrospinal fluid nitrate levels in Parkinson’s disease, Alzheimer’s disease and multiple system atrophy patients. J. Neurol. Sci. 1994, 121, 46–49. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Zhang, B.; Lin, X.; Fu, X.; An, Y.; Zou, Y.; Wang, J.X.; Wang, Z.; Yu, T. Lactate metabolism in human health and disease. Signal Transduct. Target. Ther. 2022, 7, 305. [Google Scholar] [CrossRef]
- Jia, L.; Liao, M.; Mou, A.; Zheng, Q.; Yang, W.; Yu, Z.; Cui, Y.; Xia, X.; Qin, Y.; Chen, M.; et al. Rheb-regulated mitochondrial pyruvate metabolism of Schwann cells linked to axon stability. Dev. Cell 2021, 56, 2980–2994.e2986. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.; Robay, D.; Hindupur, S.K.; Pohlmann, J.; Colombi, M.; El-Shemerly, M.Y.; Maira, S.-M.; Moroni, C.; Lane, H.A.; Hall, M.N. Dual Inhibition of the Lactate Transporters MCT1 and MCT4 Is Synthetic Lethal with Metformin due to NAD+ Depletion in Cancer Cells. Cell Rep. 2018, 25, 3047–3058.e3044. [Google Scholar] [CrossRef] [PubMed]
- Manzar, H.; Abdulhussein, D.; Yap, T.E.; Cordeiro, M.F. Cellular Consequences of Coenzyme Q10 Deficiency in Neurodegeneration of the Retina and Brain. Int. J. Mol. Sci. 2020, 21, 9299. [Google Scholar] [CrossRef]
- Mitsui, J.; Matsukawa, T.; Yasuda, T.; Ishiura, H.; Tsuji, S. Plasma Coenzyme Q10 Levels in Patients With Multiple System Atrophy. JAMA Neurol. 2016, 73, 977–980. [Google Scholar] [CrossRef]
- Schottlaender, L.V.; Bettencourt, C.; Kiely, A.P.; Chalasani, A.; Neergheen, V.; Holton, J.L.; Hargreaves, I.; Houlden, H. Coenzyme Q10 Levels Are Decreased in the Cerebellum of Multiple-System Atrophy Patients. PLoS ONE 2016, 11, e0149557. [Google Scholar] [CrossRef]
- Bartošová, T.; Klempíř, J.; Hansíková, H. Coenzyme Q10: A Biomarker in the Differential Diagnosis of Parkinsonian Syndromes. Antioxidants 2023, 12, 2104. [Google Scholar] [CrossRef]
- Rangel-Barajas, C.; Coronel, I.; Florán, B. Dopamine Receptors and Neurodegeneration. Aging Dis. 2015, 6, 349–368. [Google Scholar]
- Monzio Compagnoni, G.; Di Fonzo, A. Understanding the pathogenesis of multiple system atrophy: State of the art and future perspectives. Acta Neuropathol. Commun. 2019, 7, 113. [Google Scholar] [CrossRef]
- Anderson, N.L.; Anderson, N.G. Proteome and proteomics: New technologies, new concepts, and new words. Electrophoresis 1998, 19, 1853–1861. [Google Scholar] [CrossRef]
- Blackstock, W.P.; Weir, M.P. Proteomics: Quantitative and physical mapping of cellular proteins. Trends Biotechnol. 1999, 17, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, H.G. Two-dimensional polyacrylamide gel electrophoresis for separation of ribosomal proteins. Methods Enzymol. 1974, 30, 497–505. [Google Scholar] [PubMed]
- Li, K.W.; Ganz, A.B.; Smit, A.B. Proteomics of neurodegenerative diseases: Analysis of human post-mortem brain. J. Neurochem. 2019, 151, 435–445. [Google Scholar] [CrossRef]
- Demartini, D.R.; Schilling, L.P.; da Costa, J.C.; Carlini, C.R. Alzheimer’s and Parkinson’s diseases: An environmental proteomic point of view. J. Proteomics. 2014, 104, 24–36. [Google Scholar] [CrossRef]
- Seifar, F.; Fox, E.J.; Shantaraman, A.; Liu, Y.; Dammer, E.B.; Modeste, E.; Duong, D.M.; Yin, L.; Trautwig, A.N.; Guo, Q.; et al. Large-scale deep proteomic analysis in Alzheimer’s disease brain regions across race and ethnicity. Alzheimers Dement. 2024, 20, 8878–8897. [Google Scholar] [CrossRef]
- Vilkaite, G.; Vogel, J.; Mattsson-Carlgren, N. Integrating amyloid and tau imaging with proteomics and genomics in Alzheimer’s disease. Cell Rep. Med. 2024, 5, 101735. [Google Scholar] [CrossRef]
- Reumer, A.; Maes, E.; Mertens, I.; Cho, W.C.; Landuyt, B.; Valkenborg, D.; Schoofs, L.; Baggerman, G. Colorectal cancer biomarker discovery and validation using LC-MS/MS-based proteomics in blood: Truth or dare? Expert Rev. Proteomics 2014, 11, 449–463. [Google Scholar] [CrossRef]
- Kane, M.A.; Chen, N.; Sparks, S.; Napoli, J.L. Quantification of endogenous retinoic acid in limited biological samples by LC/MS/MS. Biochem. J. 2005, 388, 363–369. [Google Scholar] [CrossRef]
- Zhu, S.; Backstrom, D.; Forsgren, L.; Trupp, M. Alterations in Self-Aggregating Neuropeptides in Cerebrospinal Fluid of Patients with Parkinsonian Disorders. J. Park. Dis. 2022, 12, 1169–1189. [Google Scholar] [CrossRef]
- Katzdobler, S.; Nübling, G.; Klietz, M.; Fietzek, U.M.; Palleis, C.; Bernhardt, A.M.; Wegner, F.; Huber, M.; Rogozinski, S.; Schneider, L.S.; et al. GFAP and NfL as fluid biomarkers for clinical disease severity and disease progression in multiple system atrophy (MSA). J. Neurol. 2024, 271, 6991–6999. [Google Scholar] [CrossRef]
- Magdalinou, N.K.; Paterson, R.W.; Schott, J.M.; Fox, N.C.; Mummery, C.; Blennow, K.; Bhatia, K.; Morris, H.R.; Giunti, P.; Warner, T.T.; et al. A panel of nine cerebrospinal fluid biomarkers may identify patients with atypical parkinsonian syndromes. J. Neurol. Neurosurg. Psychiatry 2015, 86, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Santaella, A.; Kuiperij, H.B.; van Rumund, A.; Esselink, R.A.J.; Bloem, B.R.; Verbeek, M.M. Cerebrospinal fluid myelin basic protein is elevated in multiple system atrophy. Park. Relat. Disord. 2020, 76, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Hall, S.; Janelidze, S.; Surova, Y.; Widner, H.; Zetterberg, H.; Hansson, O. Cerebrospinal fluid concentrations of inflammatory markers in Parkinson’s disease and atypical parkinsonian disorders. Sci. Rep. 2018, 8, 13276. [Google Scholar] [CrossRef] [PubMed]
- Mondello, S.; Constantinescu, R.; Zetterberg, H.; Andreasson, U.; Holmberg, B.; Jeromin, A. CSF α-synuclein and UCH-L1 levels in Parkinson’s disease and atypical parkinsonian disorders. Park. Relat. Disord. 2014, 20, 382–387. [Google Scholar] [CrossRef]
- Herbert, M.K.; Eeftens, J.M.; Aerts, M.B.; Esselink, R.A.; Bloem, B.R.; Kuiperij, H.B.; Verbeek, M.M. CSF levels of DJ-1 and tau distinguish MSA patients from PD patients and controls. Park. Relat. Disord. 2014, 20, 112–115. [Google Scholar] [CrossRef]
- Santaella, A.; Kuiperij, H.B.; van Rumund, A.; Esselink, R.A.J.; van Gool, A.J.; Bloem, B.R.; Verbeek, M.M. Inflammation biomarker discovery in Parkinson’s disease and atypical parkinsonisms. BMC Neurol. 2020, 20, 26. [Google Scholar] [CrossRef]
- Wang, Y.; Hancock, A.M.; Bradner, J.; Chung, K.A.; Quinn, J.F.; Peskind, E.R.; Galasko, D.; Jankovic, J.; Zabetian, C.P.; Kim, H.M.; et al. Complement 3 and factor h in human cerebrospinal fluid in Parkinson’s disease, Alzheimer’s disease, and multiple-system atrophy. Am. J. Pathol. 2011, 178, 1509–1516. [Google Scholar] [CrossRef]
- Shi, M.; Bradner, J.; Hancock, A.M.; Chung, K.A.; Quinn, J.F.; Peskind, E.R.; Galasko, D.; Jankovic, J.; Zabetian, C.P.; Kim, H.M.; et al. Cerebrospinal fluid biomarkers for Parkinson disease diagnosis and progression. Ann. Neurol. 2011, 69, 570–580. [Google Scholar] [CrossRef]
- Hall, S.; Janelidze, S.; Zetterberg, H.; Brix, B.; Mattsson, N.; Surova, Y.; Blennow, K.; Hansson, O. Cerebrospinal fluid levels of neurogranin in Parkinsonian disorders. Mov. Disord. 2020, 35, 513–518. [Google Scholar] [CrossRef]
- Kaiserova, M.; Chudackova, M.; Mensikova, K.; Vastik, M.; Kurcova, S.; Prikrylova Vranova, H.; Stejskal, D.; Kanovsky, P. Cerebrospinal Fluid Levels of Chromogranin A in Parkinson’s Disease and Multiple System Atrophy. Brain Sci. 2021, 11, 141. [Google Scholar] [CrossRef]
- Jabbari, E.; Woodside, J.; Guo, T.; Magdalinou, N.K.; Chelban, V.; Athauda, D.; Lees, A.J.; Foltynie, T.; Houlden, H.; Church, A.; et al. Proximity extension assay testing reveals novel diagnostic biomarkers of atypical parkinsonian syndromes. J. Neurol. Neurosurg. Psychiatry 2019, 90, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Schulz, I.; Kruse, N.; Gera, R.G.; Kremer, T.; Cedarbaum, J.; Barbour, R.; Zago, W.; Schade, S.; Otte, B.; Bartl, M.; et al. Systematic Assessment of 10 Biomarker Candidates Focusing on α-Synuclein-Related Disorders. Mov. Disord. 2021, 36, 2874–2887. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, J.; Constantinescu, J.; Nellgård, B.; Jakobsson, P.; Brum, W.S.; Gobom, J.; Forsgren, L.; Dalla, K.; Constantinescu, R.; Zetterberg, H.; et al. Cerebrospinal Fluid Biomarkers of Synaptic Dysfunction are Altered in Parkinson’s Disease and Related Disorders. Mov. Disord. 2023, 38, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Furiya, Y.; Hirano, M.; Kurumatani, N.; Nakamuro, T.; Matsumura, R.; Futamura, N.; Ueno, S. Alpha-1-antichymotrypsin gene polymorphism and susceptibility to multiple system atrophy (MSA). Brain Res. Mol. Brain Res. 2005, 138, 178–181. [Google Scholar] [CrossRef]
- Liu, Y.; Given, K.S.; Dickson, E.L.; Owens, G.P.; Macklin, W.B.; Bennett, J.L. Concentration-dependent effects of CSF1R inhibitors on oligodendrocyte progenitor cells ex vivo and in vivo. Exp. Neurol. 2019, 318, 32–41. [Google Scholar] [CrossRef]
- Hoffmann, A.; Miron, V.E. CNS macrophage contributions to myelin health. Immunol. Rev. 2024, 327, 53–70. [Google Scholar] [CrossRef]
- Smith, C.J.; Allard, D.E.; Wang, Y.; Howard, J.F., Jr.; Montgomery, S.A.; Su, M.A. IL-10 Paradoxically Promotes Autoimmune Neuropathy through S1PR1-Dependent CD4(+) T Cell Migration. J. Immunol. 2018, 200, 1580–1592. [Google Scholar] [CrossRef]
- Dace, D.S.; Khan, A.A.; Stark, J.L.; Kelly, J.; Cross, A.H.; Apte, R.S. Interleukin-10 Overexpression Promotes Fas-Ligand-Dependent Chronic Macrophage-Mediated Demyelinating Polyneuropathy. PLoS ONE 2009, 4, e7121. [Google Scholar] [CrossRef]
- Dai, X.; Bu, X.; Gao, Y.; Guo, J.; Hu, J.; Jiang, C.; Zhang, Z.; Xu, K.; Duan, J.; He, S.; et al. Energy status dictates PD-L1 protein abundance and anti-tumor immunity to enable checkpoint blockade. Mol. Cell 2021, 81, 2317–2331.e2316. [Google Scholar] [CrossRef]
- Tsai, S.-J. Role of interleukin 8 in depression and other psychiatric disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2021, 106, 110173. [Google Scholar] [CrossRef]
- Siebers, K.; Fink, B.; Zakrzewicz, A.; Agné, A.; Richter, K.; Konzok, S.; Hecker, A.; Zukunft, S.; Küllmar, M.; Klein, J.; et al. Alpha-1 Antitrypsin Inhibits ATP-Mediated Release of Interleukin-1β via CD36 and Nicotinic Acetylcholine Receptors. Front. Immunol. 2018, 9, 877. [Google Scholar] [CrossRef]
- Sanabria, E.R.; Su, H.; Yaari, Y. Initiation of network bursts by Ca2+-dependent intrinsic bursting in the rat pilocarpine model of temporal lobe epilepsy. J. Physiol. 2001, 532, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Fremont, R.; Calderon, D.P.; Maleki, S.; Khodakhah, K. Abnormal high-frequency burst firing of cerebellar neurons in rapid-onset dystonia-parkinsonism. J. Neurosci. 2014, 34, 11723–11732. [Google Scholar] [CrossRef] [PubMed]
- Cain, S.M.; Tyson, J.R.; Choi, H.B.; Ko, R.; Lin, P.J.; LeDue, J.M.; Powell, K.L.; Bernier, L.P.; Rungta, R.L.; Yang, Y. CaV3.2 drives sustained burst-firing, which is critical for absence seizure propagation in reticular thalamic neurons. Epilepsia 2018, 59, 778–791. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; George, N.P.; Hwang, J.S.; Park, S.; Kim, M.O.; Lee, S.H.; Lee, G. Human Bone Marrow-Derived Mesenchymal Stem Cell Applications in Neurodegenerative Disease Treatment and Integrated Omics Analysis for Successful Stem Cell Therapy. Bioengineering 2023, 10, 621. [Google Scholar] [CrossRef]
- Lee, D.Y.; Lee, S.E.; Kwon, D.H.; Nithiyanandam, S.; Lee, M.H.; Hwang, J.S.; Basith, S.; Ahn, J.H.; Shin, T.H.; Lee, G. Strategies to Improve the Quality and Freshness of Human Bone Marrow-Derived Mesenchymal Stem Cells for Neurological Diseases. Stem Cells Int. 2021, 2021, 8444599. [Google Scholar] [CrossRef]
- Shin, T.H.; Lee, D.Y.; Basith, S.; Manavalan, B.; Paik, M.J.; Rybinnik, I.; Mouradian, M.M.; Ahn, J.H.; Lee, G. Metabolome Changes in Cerebral Ischemia. Cells 2020, 9, 1630. [Google Scholar] [CrossRef]
- Shin, T.H.; Lee, S.; Choi, K.R.; Lee, D.Y.; Kim, Y.; Paik, M.J.; Seo, C.; Kang, S.; Jin, M.S.; Yoo, T.H.; et al. Quality and freshness of human bone marrow-derived mesenchymal stem cells decrease over time after trypsinization and storage in phosphate-buffered saline. Sci. Rep. 2017, 7, 1106. [Google Scholar] [CrossRef]
- Ding, C.; He, X.; Zha, H.; Simon, H.D. Adaptive dimension reduction for clustering high dimensional data. In Proceedings of the 2002 IEEE International Conference on Data Mining, Maebashi City, Japan, 9–12 December 2002; pp. 147–154. [Google Scholar] [CrossRef]
- Rappoport, N.; Shamir, R. Multi-omic and multi-view clustering algorithms: Review and cancer benchmark. Nucleic Acids Res. 2018, 46, 10546–10562. [Google Scholar] [CrossRef]
- Saxena, A.; Prasad, M.; Gupta, A.; Bharill, N.; Patel, O.P.; Tiwari, A.; Er, M.J.; Ding, W.; Lin, C.-T. A review of clustering tech-niques and developments. Neurocomputing 2017, 267, 664–681. [Google Scholar] [CrossRef]
- Zubair, M.; Iqbal, M.D.A.; Shil, A.; Chowdhury, M.J.M.; Moni, M.A.; Sarker, I.H. An Improved K-means Clustering Algorithm Towards an Efficient Data-Driven Modeling. Ann. Data Sci. 2024, 11, 1525–1544. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Campello, R.; Moulavi, D.; Sander, J. Density-based clustering based on hierarchical density estimates. In Proceedings of the Advances in Knowledge Discovery and Data Mining, Gold Coast, Australia, 14–17 April 2013; Volume 7819, pp. 160–172. [Google Scholar]
- Ankerst, M.; Breunig, M.M.; Kriegel, H.-P.; Sander, J. OPTICS: Ordering points to identify the clustering structure. ACM Sigmod Rec. 1999, 28, 49–60. [Google Scholar] [CrossRef]
- Shahapure, K.R.; Nicholas, C. Cluster quality analysis using silhouette score. In Proceedings of the 7th International Conference on Data Science and Advanced Analytics (DSAA), Sydney, Australia, 6–9 October 2020; pp. 747–748. [Google Scholar]

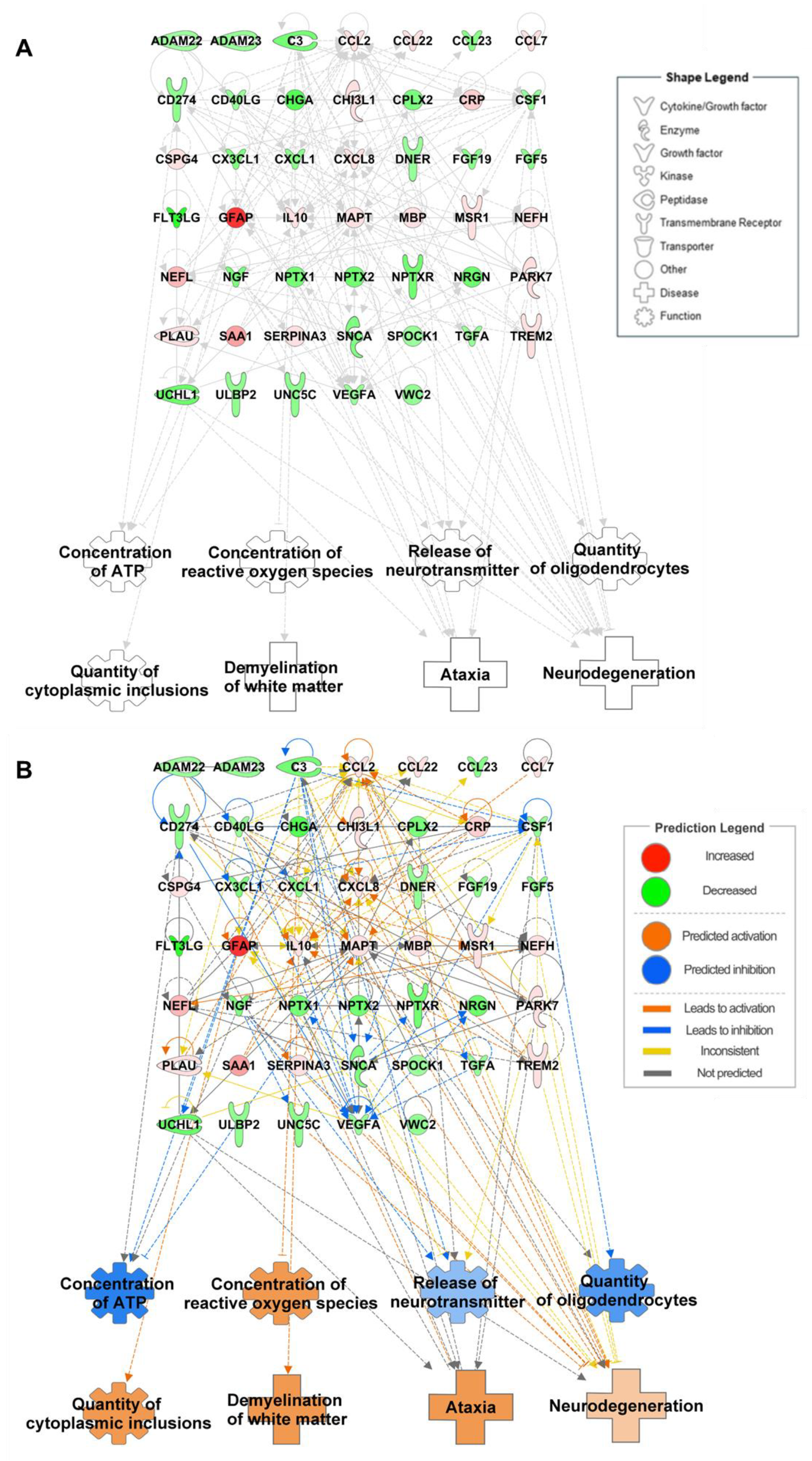

| Metabolite | IPA Name | Expression Level | Fold Change | Analysis Method a | p Value | References |
|---|---|---|---|---|---|---|
| Primary Metabolites | ||||||
| 3,4-dihydroxyphenylalanine | Levodopa | Decrease | 0.77 | LC | <0.0001 | [51] |
| Dopamine | Dopamine | Decrease | 0.77 | LC | <0.05 | [51] |
| L-citrulline | Citrulline | Increase | 1.31 | HPLC | <0.05 | [56] |
| Lactic Acid | Lactic acid | Increase | 1.05 | Amino acid analyser | 0.04 | [34] |
| Norepinephrine | Norepinephrine | Decrease | 0.56 | HPLC | <0.005 | [57] |
| Eicosapentaenoic acid | Icosapent | Increase | 1.08 | GC-MS | 0.006 | [11] |
| Coenzyme Q10 | Coenzyme Q10 | Decrease | 0.73 | ELISA | 0.036 | [58] |
| Secondary Metabolites | ||||||
| N1-acetylcadaverine | N-acetylcadaverine | Increase | 3.36 | GC-MS | <0.001 | [12] |
| N1-acetylspermidine | N1-acetylspermidine | Decrease | 0.68 | GC-MS | <0.001 | [12] |
| N8-acetylsperimidine | N(8)-acetylspermidine | Increase | 2.08 | GC-MS | <0.005 | [12] |
| N1-acetylputrescine | N-acetylputrescine | Decrease | 0.66 | GC-MS | <0.001 | [12] |
| Cadaverine | Cadaverine | Increase | 1.70 | GC-MS | <0.007 | [12] |
| Homovanillic acid | Homovanillic acid | Decrease | 0.61 | HPLC | 0.003 | [34] |
| Neuropeptide Y | Neuropeptide Y | Decrease | 0.51 | NPY-ir Assay | <0.01 | [57] |
| 3,4-dihydroxyphenylglycol | Dihydroxyphenylethylene glycol | Decrease | 0.70 | LC | <0.0001 | [51] |
| 3-methoxy-4-hydroxyphenylglycol | Methoxyhydroxyphenylglycol | Decrease | 0.64 | HPLC | <0.05 | [57] |
| 5-hydroxyindoleacetic acid | 5-hydroxyindole-3- acetic acid | Decrease | 0.54 | HPLC | <0.0001 | [59] |
| Nitrate | Nitrate | Decrease | 0.66 | ELISA | 0.01 | [60] |
| 3,4-dihydroxyphenylacetic acid | 3,4-dihydroxyphenylacetic acid | Decrease | 0.46 | LC | <0.0001 | [51] |
| Protein a | IPA Name | Accession Number b | Expression Level | Fold Change | Analysis Method c | p Value | References |
|---|---|---|---|---|---|---|---|
| NFL | NEFL | P07196 | Increase | 6.51 | SIMOA | <0.0001 | [80] |
| GFAP | GFAP | P14136 | Increase | 1.88 | SIMOA | <0.01 | [80] |
| SYUA | SNCA | P37840 | Decrease | 0.75 | Immunoassay | <0.05 | [81] |
| TAU | MAPT | P10636 | Increase | 1.48 | Innotest hTau assay | <0.0001 | [10] |
| MBP | MBP | P02686 | Increase | 1.6 | ELISA | <0.001 | [82] |
| CH3L1 | CHI3L1 | P36222 | Increase | 1.54 | Immunoassay | <0.05 | [81] |
| CCL2 | CCL2 | P13500 | Increase | 1.27 | Immunoassay | <0.05 | [81] |
| CRP | CRP | P02741 | Increase | 4.53 | Biomarkers Kit | <0.05 | [83] |
| SAA1 | SAA1 | P0DJI8 | Increase | 8.66 | Biomarkers Kit | <0.001 | [83] |
| IL8 | CXCL8 | P10145 | Increase | 1.21 | Biomarkers kit | <0.05 | [83] |
| UCHL1 | UCHL1 | P09936 | Decrease | 0.63 | Sandwich ELISA | <0.05 | [84] |
| PARK7 | PARK7 | Q99497 | Increase | 1.69 | Sandwich ELISA | <0.001 | [85] |
| X3CL1 | CX3CL1 | P78423 | Decrease | 0.75 | Proximity Extension Assay | <0.05 | [86] |
| CCL7 | CCL7 | P80098 | Increase | 1.22 | Multiplex Assay | 0.00001 | [46] |
| IL10 | IL10 | P22301 | Increase | 1.32 | Multiplex Assay | 0.00001 | [46] |
| CCL22 | CCL22 | O00626 | Increase | 1.41 | Multiplex Assay | 0.00001 | [46] |
| CO3 | C3 | P01024 | Decrease | 0.69 | Bead-based Luminex Assay | <0.05 | [87] |
| FLT3L | FLT3LG | P49771 | Decrease | 0.45 | Luminex Assay | <0.001 | [88] |
| FGF19 | FGF19 | O95750 | Decrease | 0.82 | Proximity Extension Assay | <0.05 | [86] |
| CD40L | CD40LG | P29965 | Decrease | 0.92 | Proximity Extension Assay | <0.05 | [86] |
| PD1L1 | CD274 | Q9NZQ7 | Decrease | 0.85 | Proximity Extension Assay | <0.05 | [86] |
| TGFA | TGFA | P01135 | Decrease | 0.91 | Proximity Extension Assay | <0.05 | [86] |
| CSF1 | CSF1 | P09603 | Decrease | 0.91 | Proximity Extension Assay | <0.05 | [86] |
| UROK | PLAU | P00749 | Increase | 1.13 | Proximity Extension Assay | <0.05 | [86] |
| VEGFA | VEGFA | P15692 | Decrease | 0.92 | Proximity Extension Assay | <0.05 | [86] |
| CCL23 | CCL23 | P55773 | Decrease | 0.80 | Proximity Extension Assay | <0.05 | [86] |
| GROA | CXCL1 | P09341 | Decrease | 0.81 | Proximity Extension Assay | <0.05 | [86] |
| DNER | DNER | Q8NFT8 | Decrease | 0.98 | Proximity Extension Assay | <0.05 | [86] |
| NGF | NGF | P01138 | Decrease | 0.72 | Proximity Extension Assay | <0.05 | [86] |
| NEUG | NRGN | Q92686 | Decrease | 0.65 | ELISA | <0.001 | [89] |
| NFH | NEFH | P12036 | Increase | 2.50 | ELISA | <0.001 | [89] |
| CMGA | CHGA | P10645 | Decrease | 0.60 | Sandwich ELISA | 0.014 | [90] |
| FG5 | FGF5 | P12034 | Decrease | 0.83 | Proximity Extension Assay | <0.05 | [91] |
| MSRE | MSR1 | P21757 | Increase | 1.27 | Proximity Extension Assay | <0.05 | [91] |
| VWC2 | VWC2 | Q2TAL6 | Decrease | 0.89 | Proximity Extension Assay | <0.05 | [91] |
| ADA22 | ADAM22 | Q9P0K1 | Decrease | 0.97 | Proximity Extension Assay | <0.05 | [91] |
| UNC5C | UNC5C | O95185 | Decrease | 0.84 | Proximity Extension Assay | <0.05 | [91] |
| ADA23 | ADAM23 | O75077 | Decrease | 0.91 | Proximity Extension Assay | <0.05 | [91] |
| T1CN1 | SPOCK1 | Q08629 | Decrease | 0.95 | Proximity Extension Assay | <0.05 | [91] |
| ULBP2 | ULBP2 | Q9BZM5 | Decrease | 0.86 | Proximity Extension Assay | <0.05 | [91] |
| TREM2 | TREM2 | Q9NZC2 | Increase | 1.75 | Multiplex Assay | <0.001 | [92] |
| CSPG4 | CSPG4 | Q6UVK1 | Increase | 1.22 | Biomarker Assay | 0.0234 | [45] |
| NPTX1 | NPTX1 | Q15818 | Decrease | 0.71 | LC-MS | <0.01 | [93] |
| NPTX2 | NPTX2 | P47972 | Decrease | 0.68 | LC-MS | <0.01 | [93] |
| NPTXR | NPTXR | O95502 | Decrease | 0.68 | LC-MS | <0.01 | [93] |
| CPLX2 | CPLX2 | Q6PUV4 | Decrease | 0.73 | LC-MS | <0.01 | [93] |
| AACT | SERPINA3 | P01011 | Increase | 1.29 | Immunoturbidimetry | <0.05 | [94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
George, N.P.; Kwon, M.; Jang, Y.E.; Kim, S.G.; Hwang, J.S.; Lee, S.S.; Lee, G. Integrative Analysis of Metabolome and Proteome in the Cerebrospinal Fluid of Patients with Multiple System Atrophy. Cells 2025, 14, 265. https://doi.org/10.3390/cells14040265
George NP, Kwon M, Jang YE, Kim SG, Hwang JS, Lee SS, Lee G. Integrative Analysis of Metabolome and Proteome in the Cerebrospinal Fluid of Patients with Multiple System Atrophy. Cells. 2025; 14(4):265. https://doi.org/10.3390/cells14040265
Chicago/Turabian StyleGeorge, Nimisha Pradeep, Minjun Kwon, Yong Eun Jang, Seok Gi Kim, Ji Su Hwang, Sang Seop Lee, and Gwang Lee. 2025. "Integrative Analysis of Metabolome and Proteome in the Cerebrospinal Fluid of Patients with Multiple System Atrophy" Cells 14, no. 4: 265. https://doi.org/10.3390/cells14040265
APA StyleGeorge, N. P., Kwon, M., Jang, Y. E., Kim, S. G., Hwang, J. S., Lee, S. S., & Lee, G. (2025). Integrative Analysis of Metabolome and Proteome in the Cerebrospinal Fluid of Patients with Multiple System Atrophy. Cells, 14(4), 265. https://doi.org/10.3390/cells14040265










