A Rapid, Reliable and Reproducible Protocol for DNA Degradation in Genetic Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample and Sample Preparation
2.2. Experimental Set-Up
2.3. Analysis
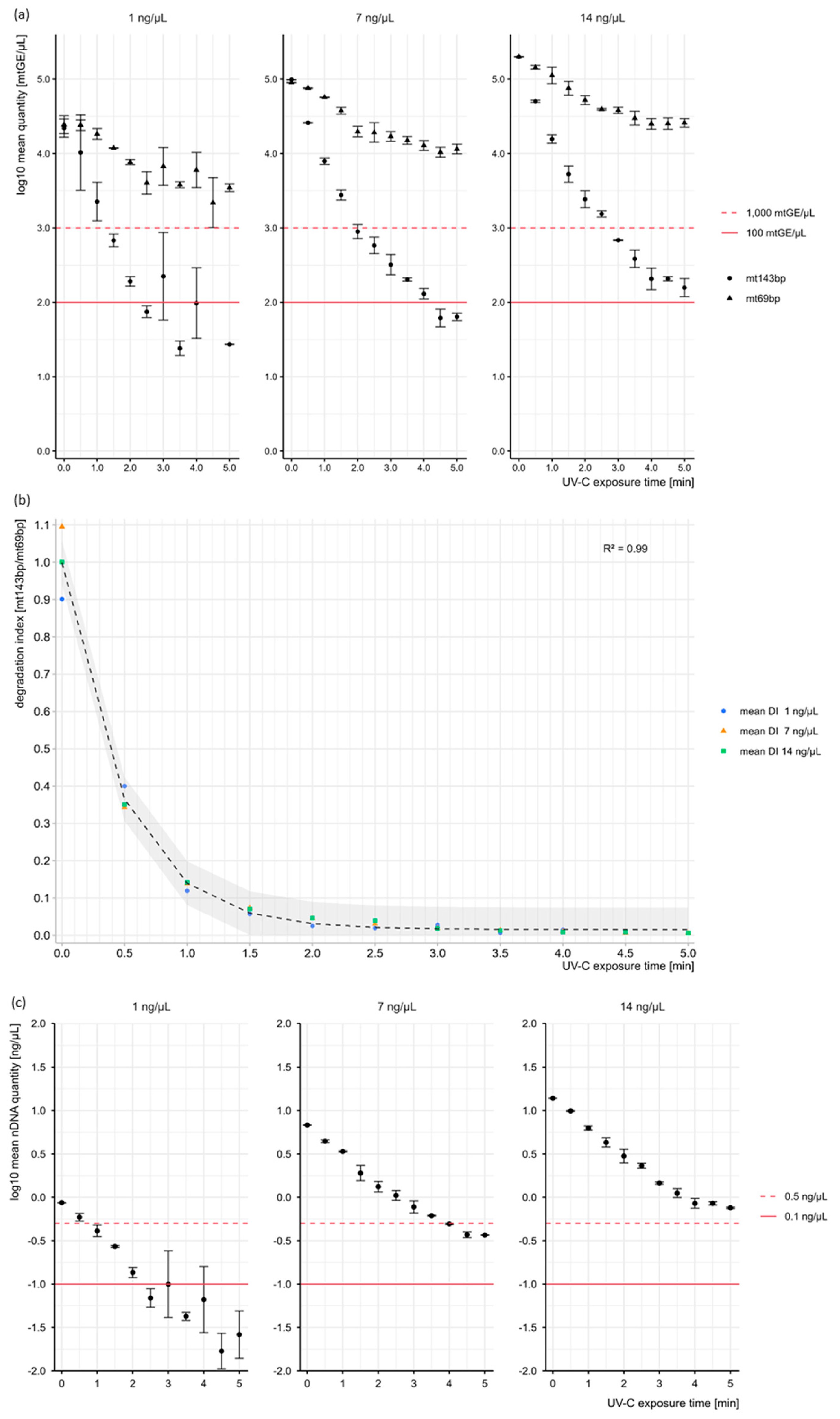
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alaeddini, R.; Walsh, S.J.; Abbas, A. Forensic implications of genetic analyses from degraded DNA—A review. Forensic Sci. Int. Genet. 2010, 4, 148–157. [Google Scholar] [CrossRef]
- Gill, P. Role of Short Tandem Repeat DNA in Forensic Casework in the UK—Past, Present, and Future Perspectives. BioTechniques 2002, 32, 366–385. [Google Scholar] [CrossRef]
- Whitaker, J.P.; Clayton, T.M.; Urquhart, A.J.; Millican, E.S.; Downes, T.J.; Kimpton, C.P.; Gill, P. Short Tandem Repeat Typing of Bodies from a Mass Disaster: High Success Rate and Characteristic Amplification Patterns in Highly Degraded Samples. Biotechniques 1995, 18, 670–677. [Google Scholar] [PubMed]
- Schneider, P.M.; Bender, K.; Mayr, W.; Parson, W.; Hoste, B.; Decorte, R.; Cordonnier, J.; Vanek, D.; Morling, N.; Karjalainen, M.; et al. STR analysis of artificially degraded DNA—Results of a collaborative European exercise. Forensic Sci. Int. 2004, 139, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Hering, S.; Augustin, C.; Edelmann, J.; Heidel, M.; Chamaon, K.; Dressler, J.; Szibor, R. Complex variability of intron 40 of the von Willebrand factor (vWF) gene. Int. J. Leg. Med. 2008, 122, 67–71. [Google Scholar] [CrossRef] [PubMed]
- da Costa Francez, P.; Rodrigues, E.; de Velasco, A.; dos Santos, S. Insertion-deletion polymorphisms--utilization on forensic analysis. Int. J. Leg. Med. 2012, 126, 491–496. [Google Scholar] [CrossRef]
- Coble, M.; Just, R.; O’Callaghan, J.; Letmanyi, I.; Peterson, C.; Irwin, J.; Parsons, T. Single nucleotide polymorphisms over the entire mtDNA genome that increase the power of forensic testing in Caucasians. Int. J. Leg. Med. 2004, 118, 137–146. [Google Scholar] [CrossRef]
- Phillips, C. Using online databases for developing SNP markers of forensic interest. In Forensic DNA Typing Protocols; Carracedo, A., Ed.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2005; Volume 297, pp. 83–106. [Google Scholar]
- Dixon, L.; Murray, C.; Archer, E.; Dobbins, A.; Koumi, P.; Gill, P. Validation of a 21-locus autosomal SNP multiplex for forensic identification purposes. Forensic Sci. Int. 2005, 154, 62–77. [Google Scholar] [CrossRef]
- Robin, E.; Wong, R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell. Physiol. 1988, 136, 507–513. [Google Scholar] [CrossRef]
- Lutz, S.; Weisser, H.J.; Heizmann, J.; Pollak, S. Location and frequency of polymorphic positions in the mtDNA control region of individuals from Germany. Int. J. Leg. Med. 1998, 111, 67–77. [Google Scholar] [CrossRef]
- Holland, M.M.; Parsons, T.J. Mitochondrial DNA Sequence Analysis—Validation and Use for Forensic Casework. Forensic Sci. Rev. 1999, 11, 21–50. [Google Scholar] [PubMed]
- Eichmann, C.; Parson, W. ‘Mitominis’: Multiplex PCR analysis of reduced size amplicons for compound sequence analysis of the entire mtDNA control region in highly degraded samples. Int. J. Leg. Med. 2008, 122, 385–388. [Google Scholar] [CrossRef] [PubMed]
- Naue, J.; Xavier, C.; Hörer, S.; Parson, W.; Lutz-Bonengel, S. Assessment of mitochondrial DNA copy number variation relative to nuclear DNA quantity between different tissues. Mitochondrion 2024, 74, 101823. [Google Scholar] [CrossRef] [PubMed]
- Templeton, J.E.; Brotherton, P.M.; Llamas, B.; Soubrier, J.; Haak, W.; Cooper, A.; Austin, J.J. DNA capture and next-generation sequencing can recover whole mitochondrial genomes from highly degraded samples for human identification. Investig. Genet. 2013, 4, 26. [Google Scholar] [CrossRef]
- Eduardoff, M.; Xavier, C.; Strobl, C.; Casas-Vargas, A.; Parson, W. Optimized mtDNA Control Region Primer Extension Capture Analysis for Forensically Relevant Samples and Highly Compromised mtDNA of Different Age and Origin. Genes 2017, 8, 237. [Google Scholar] [CrossRef]
- Gorden, E.M.; Greytak, E.M.; Sturk-Andreaggi, K.; Cady, J.; McMahon, T.P.; Armentrout, S.; Marshall, C. Extended kinship analysis of historical remains using SNP capture. Forensic Sci. Int. Genet. 2022, 57, 102636. [Google Scholar] [CrossRef]
- Xavier, C.; Eduardoff, M.; Strobl, C.; Parson, W. SD quants-Sensitive detection tetraplex-system for nuclear and mitochondrial DNA quantification and degradation inference. Forensic Sci. Int. Genet. 2019, 42, 39–44. [Google Scholar] [CrossRef]
- Xavier, C.; de la Puente, M.; Mosquera-Miguel, A.; Freire-Aradas, A.; Kalamara, V.; Vidaki, A.; Gross, T.E.; Revoir, A.; Pośpiech, E.; Kartasińska, E.; et al. Development and validation of the VISAGE AmpliSeq basic tool to predict appearance and ancestry from DNA. Forensic Sci. Int. Genet. 2020, 48, 102336. [Google Scholar] [CrossRef]
- Grubwieser, P.; Mühlmann, R.; Berger, B.; Niederstätter, H.; Pavlic, M.; Parson, W. A new “miniSTR-multiplex” displaying reduced amplicon lengths for the analysis of degraded DNA. Int. J. Leg. Med. 2006, 120, 115–120. [Google Scholar] [CrossRef]
- Kiefer, J. Effects of Ultraviolet Radiation on DNA. In Chromosomal Alterations Methods, Results and Importance in Human Health, 1st ed.; Vijayalaxmi, G.O.a., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 39–53. [Google Scholar]
- Douki, T.; Court, M.; Sauvaigo, S.; Odin, F.; Cadet, J. Formation of the main UV-induced thymine dimeric lesions within isolated and cellular DNA as measured by high performance liquid chromatography-tandem mass spectrometry. J. Biol. Chem. 2000, 275, 11678–11685. [Google Scholar] [CrossRef]
- Rosenstein, B.S.; Mitchell, D.L. Action spectra for the induction of pyrimidine(6-4)pyrimidone photoproducts and cyclobutane pyrimidine dimers in normal human skin fibroblasts. Photochem. Photobiol. 1987, 45. [Google Scholar] [CrossRef] [PubMed]
- Rosenstein, B.S.; Ducore, J.M. Induction of DNA strand breaks in normal human fibroblasts exposed to monochromatic ultraviolet and visible wavelengths in the 240–546 nm range. Photochem. Photobiol. 1983, 38, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Douki, T. Formation of UV-induced DNA damage contributing to skin cancer development. Photochem. Photobiol. Sci. 2018, 17, 1816–1841. [Google Scholar] [CrossRef] [PubMed]
- Kielbassa, C.; Roza, L.; Epe, B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis 1997, 18, 811–816. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Precision ID mtDNA Panels with the HID Ion S5™/HID Ion GeneStudio™ S5 System APPLICATION GUIDE. Available online: https://documents.thermofisher.com/TFS-Assets/LSG/manuals/MAN0017770_PrecisionID_mtDNA_Panels_S5_UG.pdf (accessed on 22 October 2025).
- Hall, A.; Ballantyne, J. Characterization of UVC-induced DNA damage in bloodstains: Forensic implications. Anal. Bioanal. Chem. 2004, 380, 72–83. [Google Scholar] [CrossRef]
- Rahi, G.S.; Adams, J.L.; Yuan, J.; Devone, D.J.; Lodhi, K.M. Whole human blood DNA degradation associated with artificial ultraviolet and solar radiations as a function of exposure time. Forensic Sci. Int. 2021, 319, 110674. [Google Scholar] [CrossRef]
- Douki, T. Low ionic strength reduces cytosine photoreactivity in UVC-irradiated isolated DNA. Photochem. Photobiol. Sci. 2006, 5, 1045–1051. [Google Scholar] [CrossRef]
| UV-C Exposure [Min] | mt143bp [mtGE/µL] | mt69bp [mtGE/µL] | nuRNU [ng/µL] | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Sd | Mean | Sd | Mean | Sd | ||||
| 0 | 98,556 | - a | 89,995 | - a | 6.79 | - a | |||
| 0.5 | 25,875 | ± | 210 | 75,524 | ± | 1016 | 4.44 | ± | 0.17 |
| 1 | 7880 | ± | 802 | 56,881 | ± | 371 | 3.38 | ± | 0.06 |
| 1.5 | 2763 | ± | 432 | 37,587 | ± | 4067 | 1.92 | ± | 0.39 |
| 2 | 892 | ± | 191 | 19,735 | ± | 3155 | 1.33 | ± | 0.18 |
| 2.5 | 581 | ± | 148 | 19,167 | ± | 5692 | 1.05 | ± | 0.14 |
| 3 | 320 | ± | 99 | 16,978 | ± | 2543 | 0.78 | ± | 0.13 |
| 3.5 | 203 | ± | 10 | 15,032 | ± | 1840 | 0.61 | ± | 0.01 |
| 4 | 130 | ± | 21 | 12,786 | ± | 1914 | 0.49 | ± | 0.01 |
| 4.5 | 62 | ± | 17 | 10,413 | ± | 1614 | 0.37 | ± | 0.03 |
| 5 | 64 | ± | 7 | 11,521 | ± | 1730 | 0.37 | ± | 0 |
| UV-C Exposure [min] | Quantity Loss [%] | ||
|---|---|---|---|
| mt143bp | mt69bp | nuRNU | |
| 0.5 | 73.75 | 16.08 | 34.64 |
| 1 | 92.00 | 36.80 | 50.22 |
| 1.5 | 97.20 | 58.23 | 71.70 |
| 2 | 99.10 | 78.07 | 80.39 |
| 2.5 | 99.41 | 78.70 | 84.47 |
| 3 | 99.68 | 81.13 | 88.54 |
| 3.5 | 99.79 | 83.30 | 90.97 |
| 4 | 99.87 | 85.79 | 92.73 |
| 4.5 | 99.94 | 88.43 | 94.53 |
| 5.0 | 99.94 | 87.20 | 94.59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ewers, L.; Parson, W. A Rapid, Reliable and Reproducible Protocol for DNA Degradation in Genetic Applications. Cells 2025, 14, 1683. https://doi.org/10.3390/cells14211683
Ewers L, Parson W. A Rapid, Reliable and Reproducible Protocol for DNA Degradation in Genetic Applications. Cells. 2025; 14(21):1683. https://doi.org/10.3390/cells14211683
Chicago/Turabian StyleEwers, Lena, and Walther Parson. 2025. "A Rapid, Reliable and Reproducible Protocol for DNA Degradation in Genetic Applications" Cells 14, no. 21: 1683. https://doi.org/10.3390/cells14211683
APA StyleEwers, L., & Parson, W. (2025). A Rapid, Reliable and Reproducible Protocol for DNA Degradation in Genetic Applications. Cells, 14(21), 1683. https://doi.org/10.3390/cells14211683







