Highlights
What are the main findings?
- 1.
- In open mitosis, ESCRTs type III are recruited to the anaphase/telophase chromatin core by BAF1 and LEM2; CHMP7 and IST1 bring in VPS4 and Spastin to couple nuclear envelope reformation with spindle microtubule disassembly. Canonical ESCRTs type I and II have not yet been identified as part of this nuclear envelope ESCRT module.
- 2.
- Beyond mitosis, the same axis operates during interphase nuclear envelope rupture and repair. ESCRT factors also contribute to nuclear pore complex quality control.
What are the implication of the main findings?
- 3.
- Correct ESCRTs type III assembly in mitosis is essential for nuclear reformation and genome integrity. ESCRT dysfunction causes DNA damage, spindle clearance defects, and chromosomal instability, mechanistically linking ESCRTs to disease including cancer and neurodegeneration.
- 4.
- ESCRT-dependent nuclear envelope surveillance pathways are emerging as therapeutic entries. Moreover, candidate new factors, such as the ESCRT type I like factor AKTIP, suggest additional druggable nodes.
Abstract
The Endosomal Sorting Complex Required for Transport (ESCRT) is a highly conserved machinery best known for its role in endosomal trafficking and membrane remodeling. Increasing evidence shows that ESCRT components are also key regulators during open mitosis, where precise membrane dynamics are essential for nuclear envelope reformation and spindle disassembly. In this review, we explore how the ESCRT machinery coordinates mitotic processes under physiological conditions and how their dysregulation contributes to genomic instability, altered cell division, and disease. We highlight recent findings on the spatiotemporal control of ESCRT recruitment at mitotic membranes, the interplay with chromatin and nuclear envelope-associated factors, and the consequences of defective ESCRT function in pathological contexts such as cancer and neurodegeneration. By connecting molecular mechanisms with cellular outcomes, we provide an integrated view of how the ESCRT machinery acts as critical guardian of mitotic fidelity and offer some routes for the identification of potential therapeutic targets in human disease.
1. The Organization of the ESCRT Machinery
The ESCRT machinery is multi-complex, working at several sites and involved in different cellular processes. These include the intracellular trafficking and, more specifically, the formation of multivesicular bodies [1], the final phase of cell division [2], that requires the physical separation of the daughter cells, and the repair of nuclear envelopes, which happens post-mitotically and in interphase ruptured nuclei [3,4]. The ESCRT factors composing the machinery are conserved through evolution. The first studies describing ESCRTs started in the year 2000 in eukaryotes, prevalently in yeast, followed by studies in mammals. Recent analyses highlight ESCRT activity in procaryotes too [5,6].
The ESCRT factors are regrouped into four types, from 0 to IV. The type 0 and type I ESCRT factors define, in principle, the site of action of the machinery (Figure 1). ESCRTs type II are a bridge to the recruitment of ESCRTs type III or late ESCRT factors. The latter are needed for the action of the machinery; namely, they are directly involved in membrane remodeling and scission. In human cells, the main components of the ESCRT machinery are as follows: type 0—HRS-HGS, STAM1 and STAM2. ESCRTs; type I—TSG101, VPS28, VPS37, in its four isoforms VPS37 A to D, MVB12 A and B, UBAP1 and Alix; type II—VPS25, VPS36 and VPS22; and ESCRTs type III—CHMP1A and B, CHMP2A and B, CHMP3, CHMP4A to C, CHMP5, CHMP6, CHMP7, and IST1 (or CHMP8). Physical and in silico prediction studies have defined a rod shaped hetero-tetrameric structure for ESCRTs type I, and a Y-shaped structure for ESCRTs type II [7,8,9]. Single ESCRT III factors are small proteins containing five to six alpha helices that form polymers and have a dynamic structural behavior. Indeed, they are detected as soluble factors, or as filaments [10,11] and, on membranes, they can typically spiralize [12].
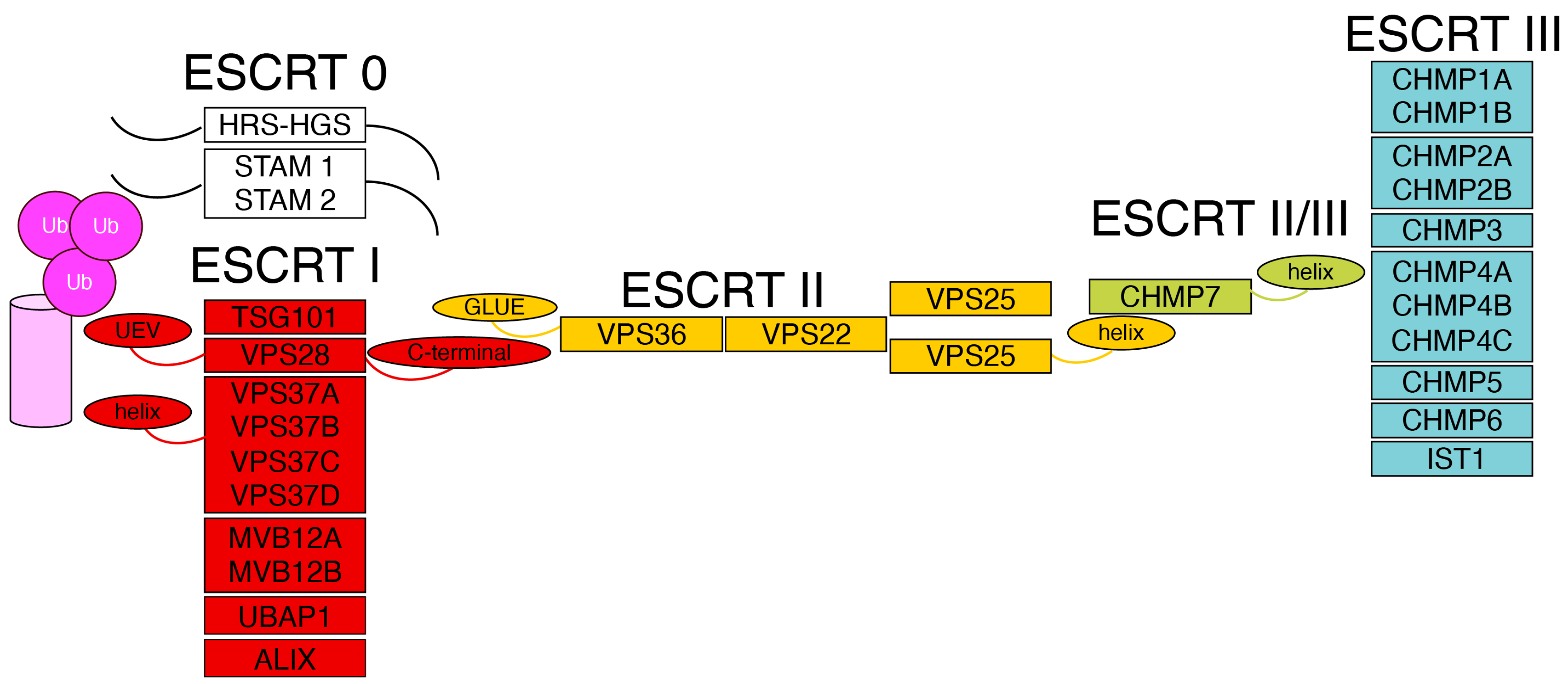
Figure 1.
The organization of the ESCRT machinery Schematic representation of the ESCRT complexes. The N-terminal regions of the ESCRTs type 0, HRS-HGS and STAM, are involved in the recognition of ubiquitinated cargo proteins (magenta) and in the recruitment of the ESCRT I complex (red). The winged-helix domain of the ESCRT I subunit VPS37 and the UEV domain of TSG101 interact with ubiquitinated cargo. ESCRTs type I recruit ESCRTs type II (yellow) through an interaction between the C-terminal region of the ESCRT type I VPS28 and the GLUE domain of the ESCRT type II VPS36. ESCRTs type II assemble into a Y-shaped structure and, via the winged-helix domain of VPS25, recruit the ESCRT subunit CHMP7 (light green). CHMP7, in turn, recruits ESCRTs type III (light blue) through its winged-helix domain.
VPS4 is an ATPase, which is associated with the ESCRT machinery. VPS4 is recruited by ESCRTs type III via an interaction between MIT (Microtubule-Interacting and Trafficking) and MIM (MIT Interacting Motifs) domains [13]. VPS4 has a pivotal role in the control of the machinery: it regulates the disassembly and recycling of the ESCRT type III filaments [14,15]. The factor CC2D1B is also associated with the ESCRT machinery and controls the accumulation of ESCRTs type III during nuclear envelope reformation and repair [16].
2. The Function of the ESCRT Machinery Across Species
To interpret the function of the ESCRT machinery, it is relevant to analyze its components through an evolutionary lens. Indeed, this perspective highlights multiple concepts which help in interpretating the roles played by this machinery. This applies both to the known functions and to those which are currently only being hypothesized. The first element that strikes, when comparatively analyzing the genes encoding the components of the ESCRT machinery in different species, is their conservation. Indeed, ESCRT factors’ encoding genes are retrieved from Archaea to mammals, from Escherichia coli to Drosophila melanogaster [17,18]. The second aspect emerging from the evolutionary analysis is that the ESCRT machinery-associated ATPase VPS4 is present in most of the organisms, as much as at least one representative of the ESCRTs type III. On the other hand, this is not the case for ESCRTs type I and II. Indeed, for example, ESCRTs type I are abundant in humans, but can be absent in less complex organisms [13,19,20]. This evolutionary conservation of the ESCRTs type III core components and of the ATPases indicates a pivotal role for the machinery in the life of a cell, and, on the other end, they suggest common mechanisms of action for ESCRTs type III and VSP4 across the different species (Table 1).
The current knowledge on ESCRT biology in humans originates from studies in yeast. Indeed, from yeast to humans, the main functions of the ESCRT machinery are conserved. One such function is the control of multivesicular bodies formation, which regulate the recycling of cellular receptors. Indeed, in this process, receptors are ubiquitinated and endocytosed from the plasma membrane; at this stage they interact with early ESCRT factors and are internalized as endosomal intraluminal vesicles [1]. These multivesicular bodies are successively fused with lysosomes. The destiny of receptors can be their degradation or their release as exosome components. If the first route influences the cell in an autocrine manner, exosomes exert a paracrine, cell-to-cell communication effect [21] (Figure 2).
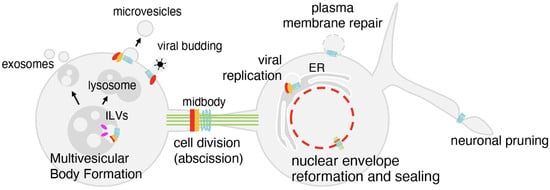
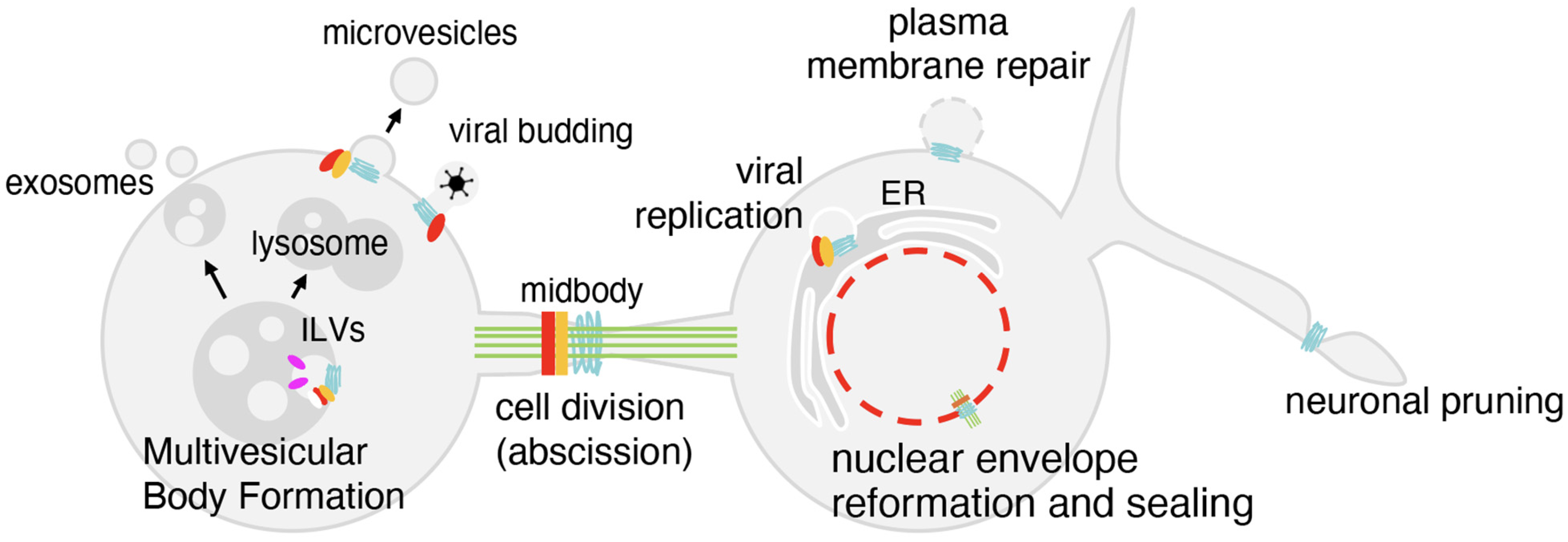
Figure 2.
The functions of the ESCRT machinery. At endosomes, the ESCRT machinery controls intraluminal vesicle (ILV) formation, leading either to lysosomal degradation or exosome release. During cytokinesis, ESCRTs type I (red), II (orange) and III (light blue) are recruited to the midbody to finalize abscission. At the nuclear envelope, ESCRTs type III cooperate to seal membranes after mitosis and repair ruptures. ESCRTs also act at the plasma membrane to close lesions, at the neuronal branches to drive pruning, and are hijacked by viruses to promote budding.

Table 1.
ESCRT proteins in humans and in model organisms. ESCRT proteins show a high degree of conservation. The table reports ESCRT protein names in yeast (S. cerevisiae and S. pombe), fly, and human. Alternative ESCRT protein names, when present, are given in parentheses; - indicates that no ortholog has been identified.
Table 1.
ESCRT proteins in humans and in model organisms. ESCRT proteins show a high degree of conservation. The table reports ESCRT protein names in yeast (S. cerevisiae and S. pombe), fly, and human. Alternative ESCRT protein names, when present, are given in parentheses; - indicates that no ortholog has been identified.
| S. cerevisiae | S. pombe | D. melanogaster | H. sapiens | |
|---|---|---|---|---|
| ESCRT 0 | Vps27 [22] | Vps27 (Sst4) [23] | Hrs [24] | HRS (HGS) [25] |
| Hse1 [22] | Hse1 [23] | Stam [26] | STAM 1 [27] | |
| STAM2 [27] | ||||
| ESCRT I | Vps23 (Stp22) [7] | Sst6 [28] | Erupted (Tsg101) [29] | TSG101 [30] |
| Vps28 [7] | Vps28 [23] | Vps28 [31,32] | VPS28 [33] | |
| Vps37/Srn2 [7] | - | Vps37a [34,35] | VPS37A [36] | |
| Vps37b [34,35] | VPS37B [36] | |||
| VPS37C [37] | ||||
| VPS37D [36] | ||||
| Mvb12 [7] | - | Mvb12 [38] | MVB12A [39,40] | |
| MVB12B [39,40] | ||||
| ESCRT II | Vps36 [9] | Vps36 [28] | Vps36 [41,42] | VPS36 (EAP45) [8,43,44] |
| Snf8 (Vps22) [9] | Dot2 [45] | Lsn/Vps22 [31,46] | VPS22 (EAP30) [43] | |
| Vps25 [9] | Vps25 [47] | Vps25 [31,48] | VPS25 (EAP20) [43] | |
| ESCRT III | Vps2 (Did4) [49] | Vps2 (Did4) [47] | Vps2 [50] | CHMP2A [49] |
| CHMP2B [49] | ||||
| Vps24 (Did3) [49] | Vps24 [28,51] | Vps24 [31] | CHMP3 [49] | |
| Snf7 (Did1, Vps32) [49] | Vps32 [28,51] | Shrb [31,52] | CHMP4A [11] | |
| CHMP4B [11] | ||||
| CHMP4C [11] | ||||
| Vps60 [49] | Vps60 [53] | Vps60 [54,55] | CHMP5 [55] | |
| Vps20 [49] | Vps20 [23] | Vps20 [56] | CHMP6 [57] | |
| Did2 (Vps46) [49] | Did2 [58] | Chmp1 [55] | CHMP1A [59,60] | |
| CHMP1B [59,60] | ||||
| Chm7 [61,62] | Cmp7 [58,63] | CG5498 * | CHMP7 [64,65,66] | |
| Ist1 [67] | Ist1 [58,68] | Ist1 [54,55] | IST1 (CHMP8) [69] | |
| ESCRT related protein | Vps4 [70] | Vps4 [51] | Vps4 [71,72] | VPS4A [15,73] VPS4B [15,73] |
* predicted ortholog (Flybase).
A second key biological process regulated by the ESCRT machinery and conserved through evolution is cell division (Figure 2). Indeed, simple organisms such as Archaea and the Red alga share with humans the implication of the ESCRT machinery in this process [74,75,76]. Here, during the final stages of cytokinesis, in the abscission phase, ESCRTs type I, II, and III are recruited at the midbody, which is the central region of the 1um wide microtubule-rich intercellular bridge linking the daughter cells [2]. In humans, ESCRTs type I and II are recruited to the midbody by CEP55 [77]. In the late abscission stage, ESCRTs type III, together with the microtubule severing enzyme Spastin, finalize the disassembly of the intercellular bridge completing the separation of the daughter cells [2,60]. This process is prototypical of the ESCRT machinery; it contributes to cell division of unicellular organisms and also has a potential significant role in determining the characteristics of the daughter cells in multicellular organisms, including their differentiative properties, their stemness potential, and their senescence [78,79].
Beyond multivesicular body formation and abscission, the ESCRT machinery is involved in many other functions (Figure 2). It contributes, for example, to neuronal pruning, impacting organismal development [71].
The ESCRT machinery is also exploited by invading organisms [80]. The need for the ESCRT machinery has been described, for example, for the budding of HIV from the cell [33], or for the egress of EBV particles from the nucleus during infection [81]. Studies based on ESCRT–virus interaction have highlighted the pivotal role of short peptides as, for example, that of the PTAP motif of the HIV1 protein GAG which interacts with the ESCRT type I TSG101 [82,83]. It has also been suggested that the GAG protein interacts with type III ESCRTs, including CHMP2, CHMP4, and CHMP6 [84,85].
If ESCRTs are hijacked by invading microorganisms, the machinery also plays a role in protecting the cell from invasion. Indeed, the machinery can, for example, repair cell membrane ruptures caused by bacteria [86,87].
Finally, the ESCRT machinery plays a critical role at the nuclear envelope, mitotically and in interphase nuclei [3,4], as detailed in the following sections.
3. The Process of Open Mitosis
Mitosis is a complex phenomenon crucial for eukaryotic cells. Indeed, during cell division, the mother cell not only must ensure the distribution of cytoplasmic material, but, importantly, must also divide the genetic material equally and correctly into the daughter cells. This process is made even more delicate by the necessity to guarantee in the newly formed cells the separation of the genetic material between nucleus and cytoplasm.
Mitosis has been cytologically known since the 19th century, and its complexity has made it an object of intense study, which has involved different disciplines in an increasingly interconnected manner, from physics to biochemistry, from high-resolution microscopy to structural biology. Importantly, if the bases of mitosis were described already by W. Flemming long ago, there are still protein complexes and biological processes yet to be defined, or redefined [88].
Mitosis is typically divided into the prophase, metaphase, anaphase, and telophase. In eukaryotic cells, two principal modes of mitosis have evolved: open and closed [89]. In open mitosis, typical of multicellular organisms, the nuclear envelope breaks down before chromosome segregation, allowing the mitotic spindle to assemble in the cytoplasm and capture chromosomes directly. The nuclear envelope then re-forms around the two sets of separated chromatids at the end of division [90]. In contrast, closed mitosis, which is characteristic of many lower eukaryotes such as fungi, occurs entirely within an intact nucleus. Here, spindle fibers form inside the nuclear compartment, and chromosome segregation takes place without nuclear envelope disassembly [91]. From an evolutionary perspective, closed mitosis is considered the more ancient mechanism, while open mitosis appears to have been independently modified several times during evolution [89].
Of course, there are numerous reviews dedicated to mitosis, and this is not the scope of this paper, but we want to highlight some elements of open mitosis which are instrumental to the analysis of the function exerted by the ESCRT machinery in this process.
In open mitosis (from now on mitosis), prophase and metaphase are cytologically recognizable by the observation of microtubule spindle assembly and by the compaction of chromatin/DNA into chromosomes/chromatids. Anaphase and telophase are recognized by the observation of the separation of chromatids and by the reassembly of the nuclear envelope as a continuous membrane around the progressively reorganizing chromatin [90,92].
The starting of mitosis, the prophase, coincides with the organization of chromatin into compact chromosomes. Human chromosomes vary significantly in length. The largest chromosome, Chromosome 1, indeed has a length of approximately 85 mm when unfolded (249 million base pairs), while the smallest, Chromosome 21, is only 16 mm (48 million base pairs). In this case, it would be better to specify the range of chromosome lengths rather than the average value (48 mm). To reach the condensed format, the extended DNA fiber is wrapped around nucleosomes and extensively folded. The topological organization of mitotic chromosome loops has been experimentally demonstrated and simulated with polymers. It has been shown that chromosome compaction requires condensin, a ring-shaped protein complex, which has a key role in promoting the formation of chromosome bodies as also suggested by the fact that it is able to induce chromosome condensation even in the absence of nucleosomes [93]. Beyond condensin, the protein Ki-67, contributes to the structural organization of chromosomes. This aspect has been related to the surfactant properties of Ki-67, which indicate that this factor can act as a biological moiety for the dispersion of mitotic chromosomes. This biological function is supported also by cytological data which have highlighted Ki-67 distribution in brush-like structures around chromosomes [94].
If chromosome compaction and individualization is a determinant of mitotic prophase and a critical element for their correct distribution in the daughter cells, the dynamic organization of the spindle microtubules represents a second central aspect in the starting of the mitotic process. The spindle was first described in plant cells by Eduard Strasburger, who also coined the term spindle apparatus, and in animal cells by Oscar Hertwig and Eduard van Beneden in 1875 and 1876 [95,96]. Flemming, in 1882, introduced the term mitosis and provided the first detailed description of the spindle [97]. Since then, many studies have highlighted its role, composition, and dynamic nature. Microtubules are dimers of alpha- and beta-tubulin assembled into a polar protofilament with minus and plus ends. Nucleation of spindle microtubules, in all human cells but in meiotic cells, originates from centrosomes [98]. During metaphase, chromosomes attach to the spindle fibers to become organized on the metaphase plate. The interaction happens between the kinetochores, multi-protein structures present on each chromatid, attached to specialized chromatin, and the microtubules. Kinetochores contain the motor enzymes which represent essential elements for microtubule and chromosome dynamics. These include dyneins and kinesins [99]. By a search-and-capture process along with dynamic rearrangements, chromosomes position at microtubule plus ends. When all kinetochores are properly attached, a cell cycle checkpoint—the spindle checkpoint—is released and the mitotic process proceeds further into anaphase [100]. While chromosomes are distributed at the spindle midplane during metaphase, sister chromatids migrate in opposite directions to create the conditions required for the formation of the daughter nuclei and daughter cells during anaphase and successive telophase. Beyond that of chromosomes, the spindle also guides the segregation of centrosomes, of the Golgi apparatus, and, in certain cells, of the mitochondria [101].
The accuracy of chromosome segregation is striking. In budding yeast, only one error occurs in about 100,000 divisions [102]. In mammalian cells, in particular in cultured cell lines, the accuracy is lower [103]. In any case, the achievement is all the more remarkable when considering the scale: chromosomes are thousands of times larger than individual cytoskeletal motors, and mitotic movements span distances far exceeding the size of the molecular machines involved. The spindle must therefore orchestrate minute nanocomponents to move and partition massive structures with strict fidelity [99].
During telophase, chromatids tend to decompactify and be reorganized to ensure correct chromatin distribution, which includes the peripheral organization of heterochromatin and the redistribution of euchromatin into territories which will determine the transcriptional and epigenetic profiles of single cells [104,105]. Interestingly, in human cells, in anaphase and telophase, chromosome telomeres are found close to lamina-associated proteins [106,107,108]. This is believed to guide—at least partly—interphase chromatin organization [109,110].
In the final stages of mitosis, the spindle microtubules must be disassembled to permit the full reformation of the nuclear envelope. On the other hand, at the start of open mitosis, the nuclear envelope must be disassembled [90].
Disassembly and reassembly of the nuclear envelope in mitosis
In eukaryotic cells, the nuclear envelope separating the nuclear DNA from the cytoplasm is composed of an inner and an outer nuclear envelope. Beneath the nuclear envelope inner membrane resides the nuclear lamina, which plays an important role in the mechano-stability of the nucleus. The nuclear lamina is composed of nuclear lamins. The principal lamins are A-type and B-type and they are structurally organized in microfilaments of 3–5 nm diameter [111,112,113]. The nuclear envelope and lamin-associated proteins include LAP1 [114], LAP2/emerin/MAN1 (LEM)-domain proteins including LAP2β [115], emerin [116], MAN1 [117], LEM2 [118] and ANKLE2 [119], lamin B receptor (LBR) [120] and Sad1p/UNC-84 (SUN) domain proteins SUN1/2 [121,122].
The disassembly of the nuclear envelope in open mitosis begins with the release of nucleoporins, the constituents of nuclear pore complexes [123]. Successively, the tearing of the nuclear envelope follows, which is helped by the microtubules residing out of the nucleus and the disassembly is completed by the depolymerization of the nuclear lamina underlying the inner nuclear envelope of the nuclear envelope. During prophase, A-type lamins are released in the nucleoplasm, B-type lamins are reorganized, and nuclear envelope proteins are redistributed in the endoplasmic reticulum [124,125].
Lamin release and nuclear envelope disassembly are controlled by phosphorylation. Indeed, lamins, nuclear envelope components, nuclear pore complex components, and nuclear envelope/chromatin linking factors, such as BAF1, are phosphorylated by multiple kinases including CDK1, PKC, VRK1, aurora kinases, PLK1, and NIMA-related kinases [123]. CDK1 and PKC, by phosphorylating serine residues, cause the disassembly of the lamin filaments [126,127].
In the final stages of mitosis, the reverse process happens, i.e., nuclear envelope reassembly. During anaphase and telophase, while the chromatids slide along microtubules with the help of kinetochore motors to opposite poles, the nuclear envelope reassembles in the daughter nuclei. During this process, the multiple factors associated and composing the nuclear envelope wrap the decondensing chromatids. These include lamin A/C and lamin-associated proteins such as LAP2α, LAP2 β, LEM2 and emerin, and the chromatin-binding factor BAF1, lamin B, lamin B receptor and the components of the nuclear pore complexes [107,128,129,130,131,132,133,134]. How the recruitment of these factors and the completion of nuclear envelope assembly are intermingled with the ESCRT machinery will be discussed in the successive section.
As for nuclear envelope disassembly, the level of phosphorylation of nuclear envelope components controls nuclear envelope reassembly. Dephosphorylation is operated by the cell cycle-controlled PP1 and PP2A [135]. The process also involves the factor Repo-man which brings PP1 onto anaphase chromosomes [136].
4. The Roles of the ESCRT Machinery in Mitosis
In open mitosis, the steps involving the ESCRT machinery have been studied in depth. According to the current literature, canonical ESCRTs type I and II have not yet been identified at the nuclear envelope, while ESCRTs type III are typically recruited.
During prophase, the ESCRT type III, CHMP4C, has been shown to localize at kinetochores; its presence is then reduced at kinetochores aligned on the metaphase plate [137,138]. CHMP4C plays a role in the mitotic spindle checkpoint, independently of its membrane-related functions during cytokinesis [138]. CHMP4C is indeed required for the stable attachment of microtubules to kinetochores through the recruitment of the NDC80 complex proteins Hec1 and Nuf2, thereby ensuring proper chromosome alignment and segregation [137,138]. Despite its mitotic function, CHMP4C does not participate in nuclear envelope or membrane reassembly during the temporal window from anaphase to late telophase [139].
From anaphase to telophase, a specific region at chromatin is involved in ESCRT aggregation and nuclear envelope reformation. This region, named core, is placed on the sides of anaphase chromatin and close to spindle microtubules. The core region is subdivided into each anaphase chromatin disk in inner and outer core regions—the inner core faces the mid-spindle and the outer core faces the spindle poles (Figure 3) [3,4,128,140].
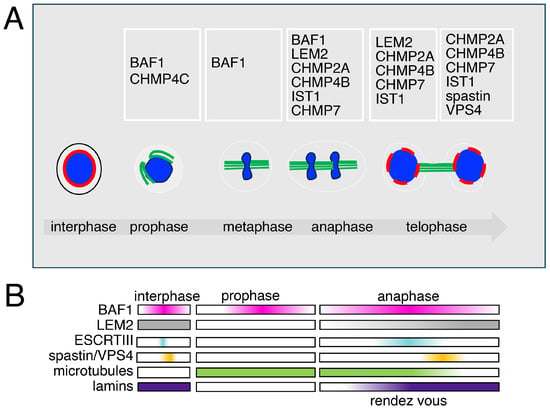
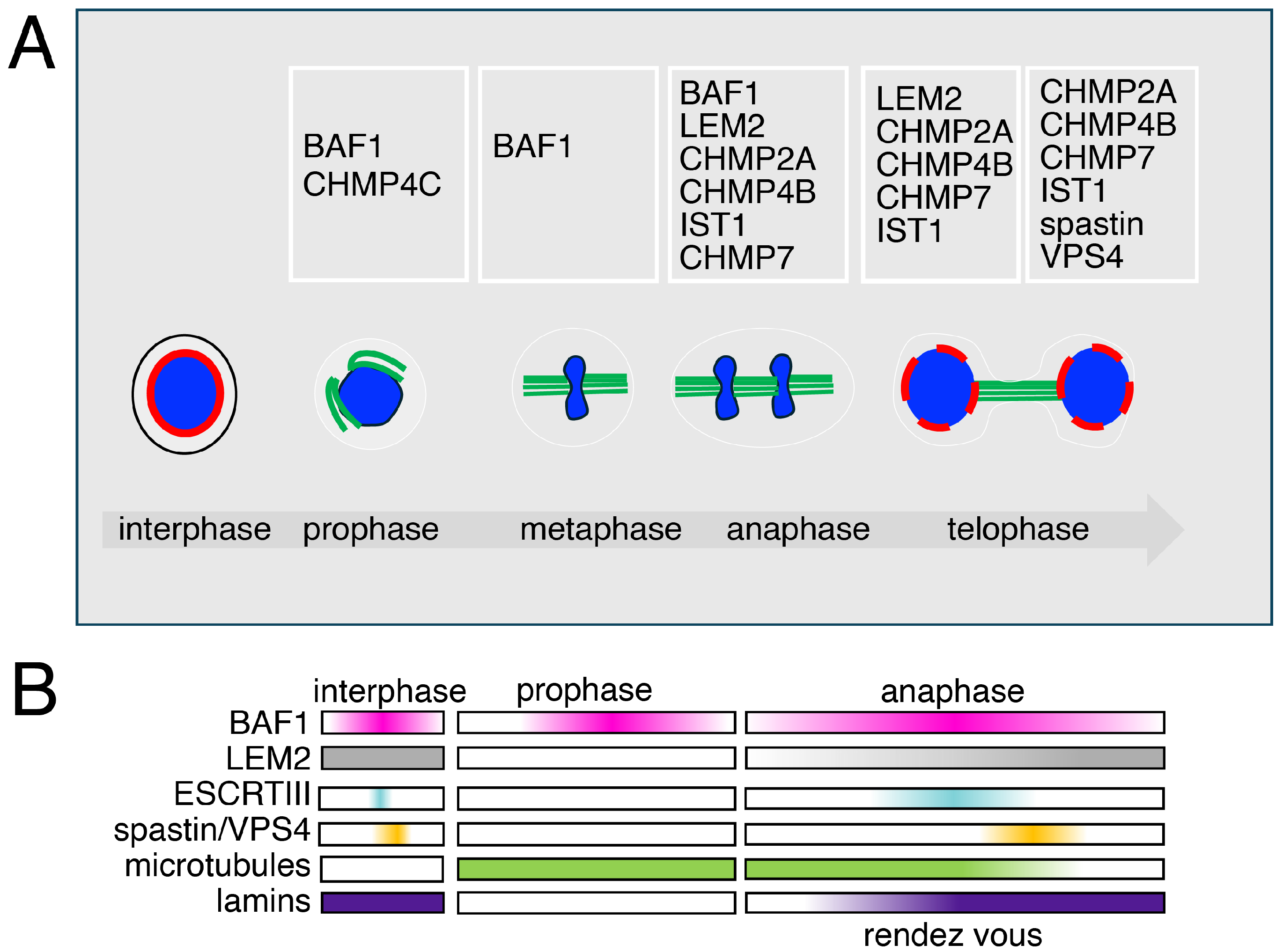
Figure 3.
The different roles and dynamics of the ESCRT machinery in mitosis (A) Open mitosis is characterized in eukaryotes by the disassembly of the nuclear envelope components. BAF1 is found at spindle poles when cells enter mitosis, preceding the recruitment of the type III ESCRTs. CHMP4C, an ESCRT type III protein, is found at the kinetochores in prophase. During ana-telophase, ESCRTs type III are recruited at the chromatin core by BAF1 and LEM2 to promote nuclear envelope sealing. CHMP7, CHMP4B, CHMP2A, and IST1 coordinate the activity of VPS4 and Spastin, ensuring both ESCRT disassembly and spindle microtubule clearance. This dual action allows proper nuclear reformation and the reassembly of lamins and chromatin reorganization, a mechanism conserved from yeast to mammalian cells. (B) The dynamics of the recruitment of the chromatin core-associated factors.
The early literature, mainly based on cytological observations, including time lapse, FRAP and FRET analyses, defined the presence of an immobile aggregate at the core of chromatin in ana-telophase [140]. These studies also integrated with electron microscopy showed the presence in this aggregate of BAF1 and of nuclear envelope-associated proteins. The aggregate was also defined to be in close association with the microtubule spindle fibers [140]. These studies have now been integrated with those that define a BAF1-LEM2-ESCRT connection and give a phase separation mechanistic interpretation for the formation of the aggregate. Indeed, the recruitment, or rendez vous, of factors at the core has been shown to be coordinated by the non-ESCRT factors BAF1 and LEM2. To mechanistically explain the rendezvous, the main hypothesis based on the phase separation process, is enchained by the presence of BAF1 and LEM2, which would promote the recruitment of ESCRTs type III including CHMP2A, CHMP4B, CHMP7, and IST1 (CHMP8) [65,129,141].
As at other ESCRT-controlled sites, the ESCRTs type III working at the nuclear envelope recruit downstream ATPases. At the nuclear envelope, these are VPS4, regulating the disassembly of ESCRT polymers, and Spastin, an ATPase operating for the disassembly of microtubules. According to biochemical and genetic studies, VPS4 would be directly recruited by the ESCRT CHMP7, while IST1 (CHMP8) would be responsible for the recruitment of Spastin. In the absence of Spastin, spindle microtubules are not disassembled, impairing the correct finalization of mitosis, including nuclear envelope resealing and chromatin reorganization. In such conditions, DNA-damaged foci and an overall defective phenotype characterize the cell [4].
In sum, the generally accepted mechanistic scheme foresees that the ESCRT machinery at the nuclear envelope has, mitotically, a double function—that of repairing the nuclear envelope to avoid its along with that of contributing to the elimination of microtubule spindle fibers which hinder nuclear envelope reassembly.
While the structural organization and dynamics of the ESCRT III biochemical complexes in anaphase and telophase have been thoroughly characterized and it has been defined that most factors but CHMP7 function as a partial replica of the same complex operating at other cell sites, the earlier moments of the assembly of the machinery during nuclear envelope reformation are different and less well-defined. In fact, the absence of ESCRTs type I and II in this process appears as an exception for the ESCRT organization in higher organisms. Moreover, if we consider an extension of the time window for the analysis of the ESCRT dynamics at the nuclear envelope, some poorly explored elements emerge. Namely, a more defined understanding of the phase preceding the rendezvous of ESCRT factors at anaphase core chromatin.
Following this line of reasoning, it is interesting to take into consideration a recent ESCRT study in which the authors have extended the analysis of the role of the ESCRT machinery taking a view that bypasses that of the anaphase and telophase stages. The work focuses on analyses performed in the fission yeast Schizosaccharomyces japonicus. The study re-links together the ana-telophase with interphase and takes into consideration the reassembly of the nuclear envelope and also the reorganization of heterochromatin. The authors propose a model in which the chromatin-binding factor Nur1 and the lamin-interacting protein Lem2 form a complex with heterochromatin [142]. Partly in analogy with what happens with LEM and BAF1 in human cells, the Lem2-Nur1 complex favors the local concentration of the yeast ortholog of CHMP7, Cmp7. Again, as in mammalian cells, Cmp7 recruits the ATPase Vps4 (ortholog of mammalian VPS4) which regulates the release of Cmp7 from Lem2 and presumably, that of Lem2 from heterochromatin. In other words, Cmp7 and VPS4 regulate the attachment of heterochromatin to the nuclear envelope via Lem2, an interaction which must be restored systematically at the end of mitosis to ensure the correct reorganization of (hetero)chromatin during interphase. Consistently, in Cmp7 depleted cells, the Lem2-Nur1 machinery remains associated with heterochromatin, compromising the correct reestablishment of post-mitotic nuclear organization and compartmentalization [3,4,142].
Intriguingly, a factor, named AKTIP, associated with ESCRTs type I, has been identified for its enrichment at the nuclear envelope of interphase nuclei [143,144]. Although it has not yet been described in nuclear envelope reassembly, this protein shares similarity with the ESCRT type I TSG101 and its reduction impacts on the recruitment of ESCRTs type III at the midbody during abscission [145]. The properties of AKTIP induce to hypothesize the possibility that new factors are still to be identified which could contribute to the nuclear reformation, and that redundancy of mechanisms can be foreseen for this activity. Interestingly, AKTIP possesses a further characteristic which connects it not only with the ESCRT machinery but also with heterochromatin. Indeed, AKTIP depletion generates a significant alteration of telomeres, which represent a particularly sensitive heterochromatic portion of the human genome [146].
Another aspect, that is not the object of this review, yet is important, is the control by the ESCRT machinery operating at the nuclear envelope on defective nuclear pore complexes [147]. In yeast, the assembly and surveillance of nuclear pore complexes is operated by an LEM factor, Heh2, which recruits ESCRTs type III [148]. The latter in turn recruit the Vps4 ATPase which contributes to the regulation and clearance of defective nuclear pore complexes [148,149,150].
Another pivotal activity of the ESCRT machinery is linked with the mechanism of repair of lysosome membranes [151,152]. ESCRTs type I, II, and III are involved in the repair of the damaged lysosomes: the process is started by the recruitment of TSG101 and ALIX, which then cooperate in the recruitment of the ESCRTs type III and of VPS4A [151]. Recently, experimental evidence has also shown that lysosomes play important regulatory roles in cell metabolism and during cell division [153,154]. Indeed, in mitosis, although cells can block or reduce the activity of processes not involved in cell division (i.e., transcription, translation, and autophagy), those involving lysosomes remain active [155]. Intriguingly, blocking lysosomal leakage at mitotic entry causes telomeric segregation defects, whereas telomere deprotection can trigger lysosome leakage [156]. Moreover, ESCRTs type III have been linked to vesicle-mediated nucleocytoplasmic transport. Finally, beyond their roles discussed so far, ESCRTs type III can act at the nuclear membrane during the nuclear egress of the Herpes Simplex Virus 1 (HSV-1) [157].
5. The ESCRT Activity at Nuclear Envelope Ruptures
A role for the ESCRT subunits at the nuclear envelope has been defined beyond their implication in mitosis. This role is exerted at nuclear envelope ruptures that can be generated by mechanical stress induced on the interphase cell and to its nucleus by internal or external forces [158,159]. The consequences of nuclear envelope ruptures are detrimental to cell viability because of two main reasons: the exposure of nuclear DNA to cytoplasmic factors, and epigenetic alterations due to the disorganization of the nuclear structure [160].
When, upon rupture, genomic DNA is exposed to the cytoplasm, DNA damage can occur as a consequence of the activity of cytoplasmic exonucleases, for example, TREX1 [161,162]. Damaged DNA enchains a response in the cell, in which ATM and ATR proteins are recruited at the site of damage and the sensor cGAS binds to cytoplasm-exposed DNA [163]. cGAS in turn activates the immune signaling cascade controlled by STING [164,165]. The activation of such a cellular response on the one side creates an overall alteration of the biology of the cell and, on the other hand, shows the importance for the cell to be able to repair ruptures and limit genomic DNA vulnerability [166].
For the second aspect, i.e., the epigenetic alterations, these have been analyzed in depth in lamin mutants which display fragile nuclei. The lamin D50 progeroid dominant mutation, for example, has been linked with alterations of epigenetic marks generated by nuclear envelope alterations [166,167,168,169].
In the process of nuclear rupture/alteration repair, the ESCRT subunits act, in part, as in post-mitotic nuclear envelope reassembly. The main route identified for this process is based on the recruitment of BAF1 on double-stranded DNA which is exposed to the cytoplasm [170,171]. Upon BAF1 accumulation at the site of rupture, cGAS is moved away to make room for DNA repair enzymes such as PARP1 [172,173]. For nuclear envelope repair, lamins and lamin-associated factors are recruited at the site of damage to create a plaque. This structure is a partial replica of the macromolecular aggregate condensating at the core of anaphase chromatin. Indeed, lamins, LAP2α and β, emerin, MAN1, LEM2 and ANKLE2 compose the plaque [171,174]. It is at this plaque that the ESCRT type III and the ATPase VPS4 are recruited [158]. Here, again, according to a scheme partly replicating the mitotic one, the ESCRT III subunits control the process, including CHMP7, CHMP4B, CHMP2A, and CHMP3 [4,158,159]. As post-mitotically, the BAF1-LEM2-CHMP7 interaction plays a critical role in interphase nuclear envelope repair [171,174]. Differently from what happens during mitotic nuclear envelope reassembly, the nuclear pore complex components are not recruited ruptured nuclear envelope sites of interphase nuclei [174,175].
6. The Implication of ESCRTs in Disease
Being involved in so many cellular processes, it is not surprising that the ESCRT factors have been associated with multiple pathological conditions, from cancer to neurological disease, from viral infection to cataract (Table 2).

Table 2.
ESCRT components and diseases. ESCRT factors have been implicated in multiple pathological conditions, the most relevant of which are reported in the table. For each disease, the ESCRT factors involved, the specific alterations associated with the pathological condition, and the corresponding dysregulated pathway are indicated. ↑ increased; ↓ decreased; - wild type protein involved.
The correlation with cancer can be assigned to the role of the ESCRT machinery in cell division. Indeed, a defect in this process generates genome instability, including aneuploidy and single-chromosome aberrations [210]. Several studies have highlighted the implication of the ESCRT type I TSG101 in cancer [177]. Namely, TSG101 has been linked with hepatocellular carcinoma, with ovarian cancer [176] and, in Drosophila, a connection has been suggested between TSG101 and neoplasia [29]. Interestingly, the TSG101-like factor AKTIP acts as a co-driver of cancer aggressiveness when tested in p53 knockout, cancer-modeling mice [211]. The ESCRTs type III CHMP3 and CHMP1A have also been associated with cancer. A CHMP4C-associated polymorphism (rs35094336, CHMP4CT232) was identified in humans and associated with defective abscission, DNA damage, genome instability, and loss of p53 [139].
Beyond cancer, a conspicuous body of literature links defects in ESCRT factors with alterations of neuronal development and with neurodegenerative diseases [212]. Drosophila model systems of Huntington disease, Amyotrophic lateral sclerosis, and of Frontotemporal lobar degeneration have highlighted the putative role of ESCRTs along with that of the ESCRT-associated ATPase VPS4 in neurodegeneration [72,194].
Association of ESCRT factors with neurodegeneration has also been established in mammalian—human, mouse, rat and monkey—cells. Among the ESCRT-controlled processes, autophagy and endosomal trafficking were frequently indicated as the mechanisms associating ESCRT defects with neurodegeneration [213]. ESCRT type I TSG101 and ESCRTs type III CHMP3 and CHMP2B were, for example, associated with defective removal of Huntingtin-positive aggregates [193].
The role of the ESCRT machinery at the nuclear envelope has been discovered later in time as compared to its role in trafficking and in abscission. As a consequence, the pathological implications of the ESCRT machinery, particularly in the context of nuclear envelope reformation, have been less extensively explored. However, a link has been established between the Hutchinson–Gilford progeria syndrome, a nuclear envelope-associated disease, and the ESCRT machinery [157]. It can be foreseen that a larger body of evidence will link in the future nuclear envelopathies with ESCRT factors. Importantly a connection has been established between nuclear envelope defects, ESCRTs and cancer invasion. Indeed, it has been suggested that the fragility of the nuclear envelope could contribute to the mechanical properties of cells when these are under the pressure of narrow spaces as it happens in tumor metastasis [158,159].
One final aspect relating the nuclear envelope role of the ESCRT machinery pertains the regulation of the nuclear pore complexes [214]. Abundant evidence links neurodegenerative disease to defects of nuclear pore complexes caused by mutation in their single components, the nucleoporins [214]. Amyotrophic lateral sclerosis has been, for example, associated with mutations of NUP50 [215], while ataxia and microcephaly have been linked with mutations of TPR [216]. Interestingly, it has been demonstrated that the ESCRT CHMP7 contributes to the accumulation and compartmentalization of defective nuclear pore complexes at the nuclear envelope in a dedicated structure named SINC [62]. Taken together, these data indicate that since the ESCRT machinery is needed to prevent the detrimental effects of altered nuclear pore complexes, ESCRTs subunits are expected be, on the one hand, also directly responsible for damage and, on the other hand, able to represent a route for new therapeutic strategies in related pathological conditions. In general terms, the latter concept could be extended to other ESCRT-based mechanisms and to the relative disease conditions. One relevant example is the usage of a peptide targeting the α-synuclein-ESCRT interaction to treat Parkinson neurodegeneration [217].
In sum, ESCRTs are conserved through evolution, play multiple and critical roles in the cell and in the organism, and represent a new druggable route for human diseases. These aspects taken together make this field of study of particularly large and deep significance.
7. Simple Summary
The ESCRT machinery is a conserved complex involved in endosomal trafficking and membrane remodeling. Recent evidence shows that it also regulates key steps of open mitosis, including nuclear envelope reformation and spindle disassembly. Its proper ac-tivity ensures mitotic fidelity, while its dysfunction causes genomic instability and con-tributes to diseases such as cancer and neurodegeneration. This review summarizes recent insights into how ESCRT components coordinate mitotic membrane dynamics under physiological and pathological conditions.
Author Contributions
Conceptualization, I.S. and M.L.T.; data curation, R.B.; writing—original draft preparation, I.S. and M.L.T.; visualization, F.C. and R.B.; funding acquisition, I.S. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge the support of NBFC to IS, funded by the National Recovery and Resilience Plan (NRRP), Mission 4 Component 2 Investment 1.4-Call for tender No. 3138 of 16 December 2021, rectified by Decree n. 3175 of 18 December 2021 of Italian Ministry of University and Research funded by the European Union-NextGenerationEU; Award Number: Pro, Project code CN_00000033, Concession Decree No. 1034 of 17 June 2022 adopted by the Italian Ministry of University and Research, Project title “National Biodiversity Future Center-NBFC” Sapienza CN5-Spoke 7. AIRC IG-24614 to IS. Istituto Pasteur Fondazione Cenci Bolognetti Anna Tramontano call 2020 to IS. MUR-Sapienza, (RP1201729E377B2D, RP11916B7F20A9E3, RP12218167BDB0A9) to IS. Sapienza RM12117A5D970AB9 and GA122181AFEB4283 to IS as co-PI. Sapienza AR22117A575BCFA5 to MLT. Italian ministry of health 2022 Programma di ricerca e formazione to IS. Singapore ministry of health HLCA22Feb-0029 to IS as co-PI.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the writing of the manuscript.
References
- Katzmann, D.J.; Babst, M.; Emr, S.D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 2001, 106, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Morita, E.; Sandrin, V.; Chung, H.Y.; Morham, S.G.; Gygi, S.P.; Rodesch, C.K.; Sundquist, W.I. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007, 26, 4215–4227. [Google Scholar] [CrossRef] [PubMed]
- Olmos, Y.; Hodgson, L.; Mantell, J.; Verkade, P.; Carlton, J.G. ESCRT-III controls nuclear envelope reformation. Nature 2015, 522, 236–239. [Google Scholar] [CrossRef] [PubMed]
- Vietri, M.; Schink, K.O.; Campsteijn, C.; Wegner, C.S.; Schultz, S.W.; Christ, L.; Thoresen, S.B.; Brech, A.; Raiborg, C.; Stenmark, H. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature 2015, 522, 231–235. [Google Scholar] [CrossRef]
- Lindås, A.C.; Karlsson, E.A.; Lindgren, M.T.; Ettema, T.J.; Bernander, R. A unique cell division machinery in the Archaea. Proc. Natl. Acad. Sci. USA 2008, 105, 18942–18946. [Google Scholar] [CrossRef]
- Williams, T.A.; Low, H.H. The evolution and mechanism of bacterial and archaeal ESCRT-III-like systems. Curr. Opin. Struct. Biol. 2025, 93, 103111. [Google Scholar] [CrossRef]
- Kostelansky, M.S.; Schluter, C.; Tam, Y.Y.; Lee, S.; Ghirlando, R.; Beach, B.; Conibear, E.; Hurley, J.H. Molecular architecture and functional model of the complete yeast ESCRT-I heterotetramer. Cell 2007, 129, 485–498. [Google Scholar] [CrossRef]
- Teo, H.; Gill, D.J.; Sun, J.; Perisic, O.; Veprintsev, D.B.; Vallis, Y.; Emr, S.D.; Williams, R.L. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 2006, 125, 99–111. [Google Scholar] [CrossRef]
- Hierro, A.; Sun, J.; Rusnak, A.S.; Kim, J.; Prag, G.; Emr, S.D.; Hurley, J.H. Structure of the ESCRT-II endosomal trafficking complex. Nature 2004, 431, 221–225. [Google Scholar] [CrossRef]
- Stoten, C.L.; Carlton, J.G. ESCRT-dependent control of membrane remodelling during cell division. Semin. Cell Dev. Biol. 2018, 74, 50–65. [Google Scholar] [CrossRef]
- Shim, S.; Kimpler, L.A.; Hanson, P.I. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 2007, 8, 1068–1079. [Google Scholar] [CrossRef]
- Goliand, I.; Adar-Levor, S.; Segal, I.; Nachmias, D.; Dadosh, T.; Kozlov, M.M.; Elia, N. Resolving ESCRT-III Spirals at the Intercellular Bridge of Dividing Cells Using 3D STORM. Cell Rep. 2018, 24, 1756–1764. [Google Scholar] [CrossRef]
- Saksena, S.; Wahlman, J.; Teis, D.; Johnson, A.E.; Emr, S.D. Functional reconstitution of ESCRT-III assembly and disassembly. Cell 2009, 136, 97–109. [Google Scholar] [CrossRef]
- Mierzwa, B.E.; Chiaruttini, N.; Redondo-Morata, L.; von Filseck, J.M.; König, J.; Larios, J.; Poser, I.; Müller-Reichert, T.; Scheuring, S.; Roux, A.; et al. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 2017, 19, 787–798. [Google Scholar] [CrossRef]
- Adell, M.A.; Vogel, G.F.; Pakdel, M.; Müller, M.; Lindner, H.; Hess, M.W.; Teis, D. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J. Cell Biol. 2014, 205, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Ventimiglia, L.N.; Cuesta-Geijo, M.A.; Martinelli, N.; Caballe, A.; Macheboeuf, P.; Miguet, N.; Parnham, I.M.; Olmos, Y.; Carlton, J.G.; Weissenhorn, W.; et al. CC2D1B Coordinates ESCRT-III Activity during the Mitotic Reformation of the Nuclear Envelope. Dev. Cell 2018, 47, 547–563.e6. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Tobiasson, V.; Wolf, Y.I.; Lu, Z.; Liu, Y.; Zhang, S.; Krupovic, M.; Li, M.; Koonin, E.V. Diversity, origin, and evolution of the ESCRT systems. mBio 2024, 15, e0033524. [Google Scholar] [CrossRef] [PubMed]
- Nachmias, D.; Frohn, B.P.; Sachse, C.; Mizrahi, I.; Elia, N. ESCRTs - a multi-purpose membrane remodeling device encoded in all life forms. Trends Microbiol. 2025, 33, 665–687. [Google Scholar] [CrossRef]
- Chiaruttini, N.; Redondo-Morata, L.; Colom, A.; Humbert, F.; Lenz, M.; Scheuring, S.; Roux, A. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 2015, 163, 866–879. [Google Scholar] [CrossRef]
- Schöneberg, J.; Pavlin, M.R.; Yan, S.; Righini, M.; Lee, I.H.; Carlson, L.A.; Bahrami, A.H.; Goldman, D.H.; Ren, X.; Hummer, G.; et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. Science 2018, 362, 1423–1428. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Bilodeau, P.S.; Winistorfer, S.C.; Kearney, W.R.; Robertson, A.D.; Piper, R.C. Vps27-Hse1 and ESCRT-I complexes cooperate to increase efficiency of sorting ubiquitinated proteins at the endosome. J. Cell Biol. 2003, 163, 237–243. [Google Scholar] [CrossRef]
- Onishi, M.; Iida, M.; Koga, T.; Yamada, S.; Hirata, A.; Iwaki, T.; Takegawa, K.; Fukui, Y.; Tachikawa, H. Schizosaccharomyces pombe Sst4p, a conserved Vps27/Hrs homolog, functions downstream of phosphatidylinositol 3-kinase Pik3p to mediate proper spore formation. Eukaryot. Cell 2007, 6, 2343–2353. [Google Scholar] [CrossRef]
- Huang, H.R.; Chen, Z.J.; Kunes, S.; Chang, G.D.; Maniatis, T. Endocytic pathway is required for Drosophila Toll innate immune signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 8322–8327. [Google Scholar] [CrossRef] [PubMed]
- Raiborg, C.; Bache, K.G.; Gillooly, D.J.; Madshus, I.H.; Stang, E.; Stenmark, H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 2002, 4, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Chanut-Delalande, H.; Jung, A.C.; Baer, M.M.; Lin, L.; Payre, F.; Affolter, M. The Hrs/Stam complex acts as a positive and negative regulator of RTK signaling during Drosophila development. PLoS ONE 2010, 5, e10245. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, E.; Kawahata, K.; Kato, M.; Kitamura, N.; Komada, M. STAM proteins bind ubiquitinated proteins on the early endosome via the VHS domain and ubiquitin-interacting motif. Mol. Biol. Cell 2003, 14, 3675–3689. [Google Scholar] [CrossRef]
- Iwaki, T.; Onishi, M.; Ikeuchi, M.; Kita, A.; Sugiura, R.; Giga-Hama, Y.; Fukui, Y.; Takegawa, K. Essential roles of class E Vps proteins for sorting into multivesicular bodies in Schizosaccharomyces pombe. Microbiology 2007, 153, 2753–2764. [Google Scholar] [CrossRef]
- Moberg, K.H.; Schelble, S.; Burdick, S.K.; Hariharan, I.K. Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev. Cell 2005, 9, 699–710. [Google Scholar] [CrossRef]
- Bishop, N.; Woodman, P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 2001, 276, 11735–11742. [Google Scholar] [CrossRef]
- Vaccari, T.; Rusten, T.E.; Menut, L.; Nezis, I.P.; Brech, A.; Stenmark, H.; Bilder, D. Comparative analysis of ESCRT-I, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J. Cell Sci. 2009, 122, 2413–2423. [Google Scholar] [CrossRef]
- Sevrioukov, E.A.; Moghrabi, N.; Kuhn, M.; Kramer, H. A mutation in dVps28 reveals a link between a subunit of the endosomal sorting complex required for transport-I complex and the actin cytoskeleton in Drosophila. Mol. Biol. Cell 2005, 16, 2301–2312. [Google Scholar] [CrossRef] [PubMed]
- Stuchell, M.D.; Garrus, J.E.; Müller, B.; Stray, K.M.; Ghaffarian, S.; McKinnon, R.; Kräusslich, H.G.; Morham, S.G.; Sundquist, W.I. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 2004, 279, 36059–36071. [Google Scholar] [CrossRef]
- Fink, J.K. Hereditary spastic paraplegia: Clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol. 2013, 126, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Hodgson, J.J.; Buchon, N.; Blissard, G.W. Identification of Cellular Genes Involved in Baculovirus GP64 Trafficking to the Plasma Membrane. J. Virol. 2022, 96, e0021522. [Google Scholar] [CrossRef] [PubMed]
- Bache, K.G.; Slagsvold, T.; Cabezas, A.; Rosendal, K.R.; Raiborg, C.; Stenmark, H. The growth-regulatory protein HCRP1/hVps37A is a subunit of mammalian ESCRT-I and mediates receptor down-regulation. Mol. Biol. Cell 2004, 15, 4337–4346. [Google Scholar] [CrossRef]
- Eastman, S.W.; Martin-Serrano, J.; Chung, W.; Zang, T.; Bieniasz, P.D. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J. Biol. Chem. 2005, 280, 628–636. [Google Scholar] [CrossRef]
- Li, Z.; Blissard, G. The vacuolar protein sorting genes in insects: A comparative genome view. Insect Biochem. Mol. Biol. 2015, 62, 211–225. [Google Scholar] [CrossRef]
- Morita, E.; Sandrin, V.; Alam, S.L.; Eckert, D.M.; Gygi, S.P.; Sundquist, W.I. Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe 2007, 2, 41–53. [Google Scholar] [CrossRef]
- Oestreich, A.J.; Davies, B.A.; Payne, J.A.; Katzmann, D.J. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol. Biol. Cell 2007, 18, 646–657. [Google Scholar] [CrossRef]
- Yang, X.; Mao, F.; Lv, X.; Zhang, Z.; Fu, L.; Lu, Y.; Wu, W.; Zhou, Z.; Zhang, L.; Zhao, Y. Drosophila Vps36 regulates Smo trafficking in Hedgehog signaling. J. Cell Sci. 2013, 126, 4230–4238. [Google Scholar] [CrossRef][Green Version]
- Herz, H.M.; Woodfield, S.E.; Chen, Z.; Bolduc, C.; Bergmann, A. Common and distinct genetic properties of ESCRT-II components in Drosophila. PLoS ONE 2009, 4, e4165. [Google Scholar] [CrossRef]
- Babst, M.; Katzmann, D.J.; Snyder, W.B.; Wendland, B.; Emr, S.D. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 2002, 3, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Slagsvold, T.; Aasland, R.; Hirano, S.; Bache, K.G.; Raiborg, C.; Trambaiolo, D.; Wakatsuki, S.; Stenmark, H. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J. Biol. Chem. 2005, 280, 19600–19606. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Mancuso, J.J.; Uzawa, S.; Cronembold, D.; Cande, W.Z. The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev. Cell 2005, 9, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Irion, U.; St Johnston, D. bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 2007, 445, 554–558. [Google Scholar] [CrossRef]
- Bhutta, M.S.; Roy, B.; Gould, G.W.; McInerny, C.J. A complex network of interactions between mitotic kinases, phosphatases and ESCRT proteins regulates septation and membrane trafficking in S. pombe. PLoS ONE 2014, 9, e111789. [Google Scholar] [CrossRef]
- Thompson, B.J.; Mathieu, J.; Sung, H.H.; Loeser, E.; Rorth, P.; Cohen, S.M. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev. Cell 2005, 9, 711–720. [Google Scholar] [CrossRef]
- Babst, M.; Katzmann, D.J.; Estepa-Sabal, E.J.; Meerloo, T.; Emr, S.D. Escrt-III: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 2002, 3, 271–282. [Google Scholar] [CrossRef]
- Aoyama, N.; Yamakawa, T.; Sasamura, T.; Yoshida, Y.; Ohori, M.; Okubo, H.; Iida, E.; Sasaki, N.; Ueda, R.; Matsuno, K. Loss- and gain-of-function analyses of vacuolar protein sorting 2 in Notch signaling of Drosophila melanogaster. Genes Genet. Syst. 2013, 88, 45–57. [Google Scholar] [CrossRef]
- Frost, A.; Elgort, M.G.; Brandman, O.; Ives, C.; Collins, S.R.; Miller-Vedam, L.; Weibezahn, J.; Hein, M.Y.; Poser, I.; Mann, M.; et al. Functional repurposing revealed by comparing S. pombe and S. cerevisiae genetic interactions. Cell 2012, 149, 1339–1352. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, N.T.; Brenman, J.E.; Jan, Y.N.; Gao, F.B. The coiled-coil protein shrub controls neuronal morphogenesis in Drosophila. Curr. Biol. 2006, 16, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Roguev, A.; Krogan, N.J.; Russell, P. Genetic Interaction Landscape Reveals Critical Requirements for Schizosaccharomyces pombe Brc1 in DNA Damage Response Mutants. G3 Genes Genomes Genet. 2015, 5, 953–962. [Google Scholar] [CrossRef] [PubMed]
- Baumers, M.; Klose, S.; Bruser, C.; Haag, C.; Hansch, S.; Pannen, H.; Weidtkamp-Peters, S.; Feldbrugge, M.; Klein, T. The auxiliary ESCRT complexes provide robustness to cold in poikilothermic organisms. Biol. Open 2019, 8, bio043422. [Google Scholar] [CrossRef]
- Marie, P.P.; Fan, S.J.; Mason, J.; Wells, A.; Mendes, C.C.; Wainwright, S.M.; Scott, S.; Fischer, R.; Harris, A.L.; Wilson, C.; et al. Accessory ESCRT-III proteins are conserved and selective regulators of Rab11a-exosome formation. J. Extracell. Vesicles 2023, 12, e12311, Erratum in J. Extracell. Vesicles 2023, 12, e12321. https://doi.org/10.1002/jev2.12311. [Google Scholar] [CrossRef]
- Troost, T.; Jaeckel, S.; Ohlenhard, N.; Klein, T. The tumour suppressor Lethal (2) giant discs is required for the function of the ESCRT-III component Shrub/CHMP4. Cell Sci. 2012, 125, 763–776. [Google Scholar] [CrossRef]
- Yorikawa, C.; Shibata, H.; Waguri, S.; Hatta, K.; Horii, M.; Katoh, K.; Kobayashi, T.; Uchiyama, Y.; Maki, M. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem. J. 2005, 387, 17–26. [Google Scholar] [CrossRef]
- Ader, N.R.; Chen, L.; Surovtsev, I.V.; Chadwick, W.L.; Rodriguez, E.C.; King, M.C.; Lusk, C.P. An ESCRT grommet cooperates with a diffusion barrier to maintain nuclear integrity. Nat. Cell Biol. 2023, 25, 1465–1477. [Google Scholar] [CrossRef]
- Reid, E.; Connell, J.; Edwards, T.L.; Duley, S.; Brown, S.E.; Sanderson, C.M. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 2005, 14, 19–38. [Google Scholar] [CrossRef]
- Yang, D.; Rismanchi, N.; Renvoisé, B.; Lippincott-Schwartz, J.; Blackstone, C.; Hurley, J.H. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 2008, 15, 1278–1286. [Google Scholar] [CrossRef]
- Bauer, I.; Brune, T.; Preiss, R.; Kolling, R. Evidence for a Nonendosomal Function of the Saccharomyces cerevisiae ESCRT-III-Like Protein Chm7. Genetics 2015, 201, 1439–1452. [Google Scholar] [CrossRef]
- Webster, B.M.; Thaller, D.J.; Jäger, J.; Ochmann, S.E.; Borah, S.; Lusk, C.P. Chm7 and Heh1 collaborate to link nuclear pore complex quality control with nuclear envelope sealing. EMBO J. 2016, 35, 2447–2467. [Google Scholar] [CrossRef]
- Rhind, N.; Chen, Z.; Yassour, M.; Thompson, D.A.; Haas, B.J.; Habib, N.; Wapinski, I.; Roy, S.; Lin, M.F.; Heiman, D.I.; et al. Comparative functional genomics of the fission yeasts. Science 2011, 332, 930–936. [Google Scholar] [CrossRef]
- Horii, M.; Shibata, H.; Kobayashi, R.; Katoh, K.; Yorikawa, C.; Yasuda, J.; Maki, M. CHMP7, a novel ESCRT-III-related protein, associates with CHMP4b and functions in the endosomal sorting pathway. Biochem. J. 2006, 400, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; LaJoie, D.; Chen, O.S.; von Appen, A.; Ladinsky, M.S.; Redd, M.J.; Nikolova, L.; Bjorkman, P.J.; Sundquist, W.I.; Ullman, K.S.; et al. LEM2 recruits CHMP7 for ESCRT-mediated nuclear envelope closure in fission yeast and human cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2166–E2175. [Google Scholar] [CrossRef] [PubMed]
- Olmos, Y.; Perdrix-Rosell, A.; Carlton, J.G. Membrane Binding by CHMP7 Coordinates ESCRT-III-Dependent Nuclear Envelope Reformation. Curr. Biol. 2016, 26, 2635–2641. [Google Scholar] [CrossRef] [PubMed]
- Dimaano, C.; Jones, C.B.; Hanono, A.; Curtiss, M.; Babst, M. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell 2008, 19, 465–474. [Google Scholar] [CrossRef]
- Koch, A.; Krug, K.; Pengelley, S.; Macek, B.; Hauf, S. Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci. Signal. 2011, 4, rs6. [Google Scholar] [CrossRef]
- Bajorek, M.; Morita, E.; Skalicky, J.J.; Morham, S.G.; Babst, M.; Sundquist, W.I. Biochemical analyses of human IST1 and its function in cytokinesis. Mol. Biol. Cell 2009, 20, 1360–1373. [Google Scholar] [CrossRef]
- Babst, M.; Wendland, B.; Estepa, E.J.; Emr, S.D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998, 17, 2982–2993. [Google Scholar] [CrossRef]
- Loncle, N.; Agromayor, M.; Martin-Serrano, J.; Williams, D.W. An ESCRT module is required for neuron pruning. Sci. Rep. 2015, 5, 8461. [Google Scholar] [CrossRef] [PubMed]
- Rusten, T.E.; Vaccari, T.; Lindmo, K.; Rodahl, L.M.; Nezis, I.P.; Sem-Jacobsen, C.; Wendler, F.; Vincent, J.P.; Brech, A.; Bilder, D.; et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr. Biol. 2007, 17, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Morita, E.; Colf, L.A.; Karren, M.A.; Sandrin, V.; Rodesch, C.K.; Sundquist, W.I. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc. Natl. Acad. Sci. USA 2010, 107, 12889–12894. [Google Scholar] [CrossRef] [PubMed]
- Ettema, T.J.; Bernander, R. Cell division and the ESCRT complex: A surprise from the archaea. Commun. Integr. Biol. 2009, 2, 86–88. [Google Scholar] [CrossRef]
- Yagisawa, F.; Fujiwara, T.; Takemura, T.; Kobayashi, Y.; Sumiya, N.; Miyagishima, S.Y.; Nakamura, S.; Imoto, Y.; Misumi, O.; Tanaka, K.; et al. ESCRT Machinery Mediates Cytokinetic Abscission in the Unicellular Red Alga. Front. Cell Dev. Biol. 2020, 8, 169. [Google Scholar] [CrossRef]
- Carlton, J.G.; Baum, B. Roles of ESCRT-III polymers in cell division across the tree of life. Curr. Opin. Cell Biol. 2023, 85, 102274. [Google Scholar] [CrossRef]
- Lee, H.H.; Elia, N.; Ghirlando, R.; Lippincott-Schwartz, J.; Hurley, J.H. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science 2008, 322, 576–580. [Google Scholar] [CrossRef]
- Chaigne, A.; Labouesse, C.; White, I.J.; Agnew, M.; Hannezo, E.; Chalut, K.J.; Paluch, E.K. Abscission Couples Cell Division to Embryonic Stem Cell Fate. Dev. Cell 2020, 55, 195–208.e195. [Google Scholar] [CrossRef]
- Gulluni, F.; Prever, L.; Li, H.; Krafcikova, P.; Corrado, I.; Lo, W.T.; Margaria, J.P.; Chen, A.; De Santis, M.C.; Cnudde, S.J.; et al. PI(3,4)P2-mediated cytokinetic abscission prevents early senescence and cataract formation. Science 2021, 374, eabk0410. [Google Scholar] [CrossRef]
- Votteler, J.; Sundquist, W.I. Virus budding and the ESCRT pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef]
- Lee, C.P.; Liu, P.T.; Kung, H.N.; Su, M.T.; Chua, H.H.; Chang, Y.H.; Chang, C.W.; Tsai, C.H.; Liu, F.T.; Chen, M.R. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus. PLoS Pathog. 2012, 8, e1002904. [Google Scholar] [CrossRef] [PubMed]
- Garrus, J.E.; von Schwedler, U.K.; Pornillos, O.W.; Morham, S.G.; Zavitz, K.H.; Wang, H.E.; Wettstein, D.A.; Stray, K.M.; Côté, M.; Rich, R.L.; et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 2001, 107, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Higginson, D.S.; Stray, K.M.; Fisher, R.D.; Garrus, J.E.; Payne, M.; He, G.P.; Wang, H.E.; Morham, S.G.; Sundquist, W.I. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 2003, 162, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Baumgärtel, V.; Ivanchenko, S.; Dupont, A.; Sergeev, M.; Wiseman, P.W.; Kräusslich, H.G.; Bräuchle, C.; Müller, B.; Lamb, D.C. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 2011, 13, 469–474. [Google Scholar] [CrossRef]
- Cashikar, A.G.; Shim, S.; Roth, R.; Maldazys, M.R.; Heuser, J.E.; Hanson, P.I. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife 2014, 3, e02184. [Google Scholar] [CrossRef]
- Wang, C.; Chen, Y.; Hu, S.; Liu, X. Insights into the function of ESCRT and its role in enveloped virus infection. Front. Microbiol. 2023, 14, 1261651. [Google Scholar] [CrossRef]
- Rivera-Cuevas, Y.; Carruthers, V.B. The multifaceted interactions between pathogens and host ESCRT machinery. PLoS Pathog. 2023, 19, e1011344. [Google Scholar] [CrossRef]
- Paweletz, N. Walther Flemming: Pioneer of mitosis research. Nat. Rev. Mol. Cell Biol. 2001, 2, 72–75. [Google Scholar] [CrossRef]
- Boettcher, B.; Barral, Y. The cell biology of open and closed mitosis. Nucleus 2013, 4, 160–165. [Google Scholar] [CrossRef]
- Güttinger, S.; Laurell, E.; Kutay, U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 2009, 10, 178–191. [Google Scholar] [CrossRef]
- Castagnetti, S.; Oliferenko, S.; Nurse, P. Fission yeast cells undergo nuclear division in the absence of spindle microtubules. PLoS Biol. 2010, 8, e1000512. [Google Scholar] [CrossRef]
- Prosser, S.L.; Pelletier, L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef]
- Batty, P.; Gerlich, D.W. Mitotic Chromosome Mechanics: How Cells Segregate Their Genome. Trends Cell Biol. 2019, 29, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Cuylen, S.; Blaukopf, C.; Politi, A.Z.; Müller-Reichert, T.; Neumann, B.; Poser, I.; Ellenberg, J.; Hyman, A.A.; Gerlich, D.W. Ki-67 acts as a biological surfactant to disperse mitotic chromosomes. Nature 2016, 535, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Strasburger, E. Ueber Zellbildung und Zelltheilung; Hermann Dabis: Jena, Germany, 1875. [Google Scholar]
- Strasburger, E. Ueber Zellbildung und Zelltheilung, 2nd ed.; Hermann Dabis: Jena, Germany, 1876. [Google Scholar]
- Flemming, W. Zellsubstanz, Kern und zelltheilung; Vogel: Leipzig, Germany, 1882. [Google Scholar]
- Meunier, S.; Vernos, I. Microtubule assembly during mitosis—From distinct origins to distinct functions? J. Cell Sci. 2012, 125, 2805–2814. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, J.R. Mitosis. Cold Spring Harb. Perspect. Biol. 2016, 8, a023218. [Google Scholar] [CrossRef]
- Lara-Gonzalez, P.; Westhorpe, F.G.; Taylor, S.S. The spindle assembly checkpoint. Curr. Biol. 2012, 22, R966–R980. [Google Scholar] [CrossRef]
- Mascanzoni, F.; Ayala, I.; Colanzi, A. Organelle Inheritance Control of Mitotic Entry and Progression: Implications for Tissue Homeostasis and Disease. Front. Cell Dev. Biol. 2019, 7, 133. [Google Scholar] [CrossRef]
- Hartwell, L.H.; Smith, D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics 1985, 110, 381–395. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Silkworth, W.T.; Nardi, I.K.; Nicholson, J.M.; Compton, D.A.; Cimini, D. The mitotic origin of chromosomal instability. Curr. Biol. 2014, 24, R148–R149. [Google Scholar] [CrossRef]
- Lucini, F.; Petrini, C.; Salviato, E.; Pal, K.; Rosti, V.; Gorini, F.; Santarelli, P.; Quadri, R.; Lembo, G.; Graziano, G.; et al. Biochemical properties of chromatin domains define genome compartmentalization. Nucleic Acids Res. 2024, 52, e54. [Google Scholar] [CrossRef]
- Shoaib, M.; Nair, N.; Sørensen, C.S. Chromatin Landscaping at Mitotic Exit Orchestrates Genome Function. Front. Genet. 2020, 11, 103. [Google Scholar] [CrossRef]
- Crabbe, L.; Cesare, A.J.; Kasuboski, J.M.; Fitzpatrick, J.A.; Karlseder, J. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep. 2012, 2, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Dechat, T.; Gajewski, A.; Korbei, B.; Gerlich, D.; Daigle, N.; Haraguchi, T.; Furukawa, K.; Ellenberg, J.; Foisner, R. LAP2alpha and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 2004, 117, 6117–6128. [Google Scholar] [CrossRef] [PubMed]
- Chojnowski, A.; Ong, P.F.; Wong, E.S.; Lim, J.S.; Mutalif, R.A.; Navasankari, R.; Dutta, B.; Yang, H.; Liow, Y.Y.; Sze, S.K.; et al. Progerin reduces LAP2α-telomere association in Hutchinson-Gilford progeria. Elife 2015, 4, e07759. [Google Scholar] [CrossRef] [PubMed]
- Burla, R.; La Torre, M.; Maccaroni, K.; Verni, F.; Giunta, S.; Saggio, I. Interplay of the nuclear envelope with chromatin in physiology and pathology. Nucleus 2020, 11, 205–218. [Google Scholar] [CrossRef]
- La Torre, M.; Burla, R.; Saggio, I. Preserving Genome Integrity: Unveiling the Roles of ESCRT Machinery. Cells 2024, 13, 1307. [Google Scholar] [CrossRef]
- Aebi, U.; Cohn, J.; Buhle, L.; Gerace, L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 1986, 323, 560–564. [Google Scholar] [CrossRef]
- Shimi, T.; Kittisopikul, M.; Tran, J.; Goldman, A.E.; Adam, S.A.; Zheng, Y.; Jaqaman, K.; Goldman, R.D. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 2015, 26, 4075–4086. [Google Scholar] [CrossRef]
- Turgay, Y.; Eibauer, M.; Goldman, A.E.; Shimi, T.; Khayat, M.; Ben-Harush, K.; Dubrovsky-Gaupp, A.; Sapra, K.T.; Goldman, R.D.; Medalia, O. The molecular architecture of lamins in somatic cells. Nature 2017, 543, 261–264. [Google Scholar] [CrossRef]
- Foisner, R.; Gerace, L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell 1993, 73, 1267–1279. [Google Scholar] [CrossRef]
- Dechat, T.; Vlcek, S.; Foisner, R. Review: Lamina-associated polypeptide 2 isoforms and related proteins in cell cycle-dependent nuclear structure dynamics. J. Struct. Biol. 2000, 129, 335–345. [Google Scholar] [CrossRef]
- Bione, S.; Maestrini, E.; Rivella, S.; Mancini, M.; Regis, S.; Romeo, G.; Toniolo, D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 1994, 8, 323–327. [Google Scholar] [CrossRef]
- Lin, F.; Blake, D.L.; Callebaut, I.; Skerjanc, I.S.; Holmer, L.; McBurney, M.W.; Paulin-Levasseur, M.; Worman, H.J. MAN1, an inner nuclear membrane protein that shares the LEM domain with lamina-associated polypeptide 2 and emerin. J. Biol. Chem. 2000, 275, 4840–4847. [Google Scholar] [CrossRef] [PubMed]
- Brachner, A.; Reipert, S.; Foisner, R.; Gotzmann, J. LEM2 is a novel MAN1-related inner nuclear membrane protein associated with A-type lamins. J. Cell Sci. 2005, 118, 5797–5810. [Google Scholar] [CrossRef] [PubMed]
- Snyers, L.; Erhart, R.; Laffer, S.; Pusch, O.; Weipoltshammer, K.; Schöfer, C. LEM4/ANKLE-2 deficiency impairs post-mitotic re-localization of BAF, LAP2α and LaminA to the nucleus, causes nuclear envelope instability in telophase and leads to hyperploidy in HeLa cells. Eur. J. Cell Biol. 2018, 97, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Olins, A.L.; Rhodes, G.; Welch, D.B.; Zwerger, M.; Olins, D.E. Lamin B receptor: Multi-tasking at the nuclear envelope. Nucleus 2010, 1, 53–70. [Google Scholar] [CrossRef]
- Zhou, Z.; Du, X.; Cai, Z.; Song, X.; Zhang, H.; Mizuno, T.; Suzuki, E.; Yee, M.R.; Berezov, A.; Murali, R.; et al. Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J. Biol. Chem. 2012, 287, 5317–5326. [Google Scholar] [CrossRef]
- Foisner, R. Inner nuclear membrane proteins and the nuclear lamina. J. Cell Sci. 2001, 114, 3791–3792. [Google Scholar] [CrossRef]
- Linder, M.I.; Köhler, M.; Boersema, P.; Weberruss, M.; Wandke, C.; Marino, J.; Ashiono, C.; Picotti, P.; Antonin, W.; Kutay, U. Mitotic Disassembly of Nuclear Pore Complexes Involves CDK1- and PLK1-Mediated Phosphorylation of Key Interconnecting Nucleoporins. Dev. Cell 2017, 43, 141–156.e7. [Google Scholar] [CrossRef]
- Vivante, A.; Shoval, I.; Garini, Y. The Dynamics of Lamin a During the Cell Cycle. Front. Mol. Biosci. 2021, 8, 705595. [Google Scholar] [CrossRef]
- Gerace, L.; Blobel, G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell 1980, 19, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Collas, P. Sequential PKC- and Cdc2-mediated phosphorylation events elicit zebrafish nuclear envelope disassembly. J. Cell Sci. 1999, 112 Pt 6, 977–987. [Google Scholar] [CrossRef] [PubMed]
- de Castro, I.J.; Gil, R.S.; Ligammari, L.; Di Giacinto, M.L.; Vagnarelli, P. CDK1 and PLK1 coordinate the disassembly and reassembly of the nuclear envelope in vertebrate mitosis. Oncotarget 2018, 9, 7763–7773. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, T.; Koujin, T.; Hayakawa, T.; Kaneda, T.; Tsutsumi, C.; Imamoto, N.; Akazawa, C.; Sukegawa, J.; Yoneda, Y.; Hiraoka, Y. Live fluorescence imaging reveals early recruitment of emerin, LBR, RanBP2, and Nup153 to reforming functional nuclear envelopes. J. Cell Sci. 2000, 113, 779–794. [Google Scholar] [CrossRef]
- von Appen, A.; LaJoie, D.; Johnson, I.E.; Trnka, M.J.; Pick, S.M.; Burlingame, A.L.; Ullman, K.S.; Frost, A. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature 2020, 582, 115–118. [Google Scholar] [CrossRef]
- Chaudhary, N.; Courvalin, J.C. Stepwise reassembly of the nuclear envelope at the end of mitosis. J. Cell Biol. 1993, 122, 295–306. [Google Scholar] [CrossRef]
- Samwer, M.; Schneider, M.W.G.; Hoefler, R.; Schmalhorst, P.S.; Jude, J.G.; Zuber, J.; Gerlich, D.W. DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell 2017, 170, 956–972.e3. [Google Scholar] [CrossRef]
- Liu, S.; Pellman, D. The coordination of nuclear envelope assembly and chromosome segregation in metazoans. Nucleus 2020, 11, 35–52. [Google Scholar] [CrossRef]
- LaJoie, D.; Ullman, K.S. Coordinated events of nuclear assembly. Curr. Opin. Cell Biol. 2017, 46, 39–45. [Google Scholar] [CrossRef]
- Clever, M.; Mimura, Y.; Funakoshi, T.; Imamoto, N. Regulation and coordination of nuclear envelope and nuclear pore complex assembly. Nucleus 2013, 4, 105–114. [Google Scholar] [CrossRef]
- Holder, J.; Poser, E.; Barr, F.A. Getting out of mitosis: Spatial and temporal control of mitotic exit and cytokinesis by PP1 and PP2A. FEBS Lett. 2019, 593, 2908–2924. [Google Scholar] [CrossRef]
- Vagnarelli, P.; Hudson, D.F.; Ribeiro, S.A.; Trinkle-Mulcahy, L.; Spence, J.M.; Lai, F.; Farr, C.J.; Lamond, A.I.; Earnshaw, W.C. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat. Cell Biol. 2006, 8, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Petsalaki, E.; Dandoulaki, M.; Zachos, G. The ESCRT protein Chmp4c regulates mitotic spindle checkpoint signaling. J. Cell Biol. 2018, 217, 861–876. [Google Scholar] [CrossRef] [PubMed]
- Petsalaki, E.; Dandoulaki, M.; Zachos, G. Chmp4c is required for stable kinetochore-microtubule attachments. Chromosoma 2018, 127, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Sadler, J.B.A.; Wenzel, D.M.; Strohacker, L.K.; Guindo-Martínez, M.; Alam, S.L.; Mercader, J.M.; Torrents, D.; Ullman, K.S.; Sundquist, W.I.; Martin-Serrano, J. A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability. Proc. Natl. Acad. Sci. USA 2018, 115, E8900–E8908. [Google Scholar] [CrossRef]
- Haraguchi, T.; Kojidani, T.; Koujin, T.; Shimi, T.; Osakada, H.; Mori, C.; Yamamoto, A.; Hiraoka, Y. Live cell imaging and electron microscopy reveal dynamic processes of BAF-directed nuclear envelope assembly. J. Cell Sci. 2008, 121, 2540–2554. [Google Scholar] [CrossRef]
- Vlcek, S.; Dechat, T.; Foisner, R. Nuclear envelope and nuclear matrix: Interactions and dynamics. Cell Mol. Life Sci. 2001, 58, 1758–1765. [Google Scholar] [CrossRef]
- Pieper, G.H.; Sprenger, S.; Teis, D.; Oliferenko, S. ESCRT-III/Vps4 Controls Heterochromatin-Nuclear Envelope Attachments. Dev. Cell 2020, 53, 27–41.e6. [Google Scholar] [CrossRef]
- Burla, R.; Carcuro, M.; Torre, M.L.; Fratini, F.; Crescenzi, M.; D′Apice, M.R.; Spitalieri, P.; Raffa, G.D.; Astrologo, L.; Lattanzi, G.; et al. The telomeric protein AKTIP interacts with A- and B-type lamins and is involved in regulation of cellular senescence. Open Biol. 2016, 6, 160103, Erratum in Open Biol. 2024, 13 https://doi.org/10.1098/rsob.160103. [Google Scholar] [CrossRef]
- La Torre, M.; Merigliano, C.; Maccaroni, K.; Chojnowski, A.; Goh, W.I.; Giubettini, M.; Vernì, F.; Capanni, C.; Rhodes, D.; Wright, G.; et al. Combined alteration of lamin and nuclear morphology influences the localization of the tumor-associated factor AKTIP. J. Exp. Clin. Cancer Res. 2022, 41, 273. [Google Scholar] [CrossRef]
- Merigliano, C.; Burla, R.; La Torre, M.; Del Giudice, S.; Teo, H.; Liew, C.W.; Chojnowski, A.; Goh, W.I.; Olmos, Y.; Maccaroni, K.; et al. AKTIP interacts with ESCRT I and is needed for the recruitment of ESCRT III subunits to the midbody. PLoS Genet. 2021, 17, e1009757. [Google Scholar] [CrossRef] [PubMed]
- Burla, R.; Carcuro, M.; Raffa, G.D.; Galati, A.; Raimondo, D.; Rizzo, A.; La Torre, M.; Micheli, E.; Ciapponi, L.; Cenci, G.; et al. AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance. PLoS Genet. 2015, 11, e1005167. [Google Scholar] [CrossRef] [PubMed]
- Shankar, R.; Lettman, M.M.; Whisler, W.; Frankel, E.B.; Audhya, A. The ESCRT machinery directs quality control over inner nuclear membrane architecture. Cell Rep. 2022, 38, 110263. [Google Scholar] [CrossRef] [PubMed]
- Webster, B.M.; Colombi, P.; Jäger, J.; Lusk, C.P. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell 2014, 159, 388–401. [Google Scholar] [CrossRef]
- Borah, S.; Thaller, D.J.; Hakhverdyan, Z.; Rodriguez, E.C.; Isenhour, A.W.; Rout, M.P.; King, M.C.; Lusk, C.P. Heh2/Man1 may be an evolutionarily conserved sensor of NPC assembly state. Mol. Biol. Cell 2021, 32, 1359–1373. [Google Scholar] [CrossRef]
- Webster, B.M.; Lusk, C.P. ESCRTs breach the nuclear border. Nucleus 2015, 6, 197–202. [Google Scholar] [CrossRef][Green Version]
- Radulovic, M.; Schink, K.O.; Wenzel, E.M.; Nahse, V.; Bongiovanni, A.; Lafont, F.; Stenmark, H. ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival. EMBO J. 2018, 37, e99753. [Google Scholar] [CrossRef]
- Vrancx, C.; Annaert, W. Lysosome repair fails in ageing and Alzheimer′s disease. Nat. Cell Biol. 2025, 27, 553–555. [Google Scholar] [CrossRef]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2020, 21, 101–118. [Google Scholar] [CrossRef]
- Nowosad, A.; Besson, A. Lysosomes at the Crossroads of Cell Metabolism, Cell Cycle, and Stemness. Int. J. Mol. Sci. 2022, 23, 2290. [Google Scholar] [CrossRef] [PubMed]
- Stahl-Meyer, J.; Holland, L.K.K.; Liu, B.; Maeda, K.; Jaattela, M. Lysosomal Changes in Mitosis. Cells 2022, 11, 875. [Google Scholar] [CrossRef] [PubMed]
- Hamalisto, S.; Stahl, J.L.; Favaro, E.; Yang, Q.; Liu, B.; Christoffersen, L.; Loos, B.; Guasch Boldu, C.; Joyce, J.A.; Reinheckel, T.; et al. Spatially and temporally defined lysosomal leakage facilitates mitotic chromosome segregation. Nat. Commun. 2020, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Arii, J.; Maeda, F.; Maruzuru, Y.; Koyanagi, N.; Kato, A.; Mori, Y.; Kawaguchi, Y. ESCRT-III controls nuclear envelope deformation induced by progerin. Sci. Rep. 2020, 10, 18877. [Google Scholar] [CrossRef]
- Denais, C.M.; Gilbert, R.M.; Isermann, P.; McGregor, A.L.; te Lindert, M.; Weigelin, B.; Davidson, P.M.; Friedl, P.; Wolf, K.; Lammerding, J. Nuclear envelope rupture and repair during cancer cell migration. Science 2016, 352, 353–358. [Google Scholar] [CrossRef]
- Raab, M.; Gentili, M.; de Belly, H.; Thiam, H.R.; Vargas, P.; Jimenez, A.J.; Lautenschlaeger, F.; Voituriez, R.; Lennon-Dumenil, A.M.; Manel, N.; et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016, 352, 359–362. [Google Scholar] [CrossRef]
- Maciejowski, J.; Hatch, E.M. Nuclear Membrane Rupture and Its Consequences. Annu. Rev. Cell Dev. Biol. 2020, 36, 85–114. [Google Scholar] [CrossRef]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; de Lange, T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef]
- Maciejowski, J.; Chatzipli, A.; Dananberg, A.; Chu, K.; Toufektchan, E.; Klimczak, L.J.; Gordenin, D.A.; Campbell, P.J.; de Lange, T. APOBEC3-dependent kataegis and TREX1-driven chromothripsis during telomere crisis. Nat. Genet. 2020, 52, 884–890. [Google Scholar] [CrossRef]
- Nader, G.P.F.; Agüera-Gonzalez, S.; Routet, F.; Gratia, M.; Maurin, M.; Cancila, V.; Cadart, C.; Palamidessi, A.; Ramos, R.N.; San Roman, M.; et al. Compromised nuclear envelope integrity drives TREX1-dependent DNA damage and tumor cell invasion. Cell 2021, 184, 5230–5246.e22. [Google Scholar] [CrossRef]
- Mackenzie, K.J.; Carroll, P.; Martin, C.A.; Murina, O.; Fluteau, A.; Simpson, D.J.; Olova, N.; Sutcliffe, H.; Rainger, J.K.; Leitch, A.; et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017, 548, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678, Erratum in Nature 2008, 456, 274. https://doi.org/10.1038/nature07317. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhuang, Z.; Li, J.; Feng, Z. Significance of the cGAS-STING Pathway in Health and Disease. Int. J. Mol. Sci. 2023, 24, 13316. [Google Scholar] [CrossRef] [PubMed]
- Shumaker, D.K.; Dechat, T.; Kohlmaier, A.; Adam, S.A.; Bozovsky, M.R.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Khuon, S.; Collins, F.S.; et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. USA 2006, 103, 8703–8708. [Google Scholar] [CrossRef]
- De Vos, W.H.; Houben, F.; Kamps, M.; Malhas, A.; Verheyen, F.; Cox, J.; Manders, E.M.; Verstraeten, V.L.; van Steensel, M.A.; Marcelis, C.L.; et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet. 2011, 20, 4175–4186. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T. Lamin A-dependent nuclear defects in human aging. Science 2006, 312, 1059–1063. [Google Scholar] [CrossRef]
- Young, A.M.; Gunn, A.L.; Hatch, E.M. BAF facilitates interphase nuclear membrane repair through recruitment of nuclear transmembrane proteins. Mol. Biol. Cell 2020, 31, 1551–1560. [Google Scholar] [CrossRef]
- Halfmann, C.T.; Sears, R.M.; Katiyar, A.; Busselman, B.W.; Aman, L.K.; Zhang, Q.; O′Bryan, C.S.; Angelini, T.E.; Lele, T.P.; Roux, K.J. Repair of nuclear ruptures requires barrier-to-autointegration factor. J. Cell Biol. 2019, 218, 2136–2149. [Google Scholar] [CrossRef]
- Guey, B.; Wischnewski, M.; Decout, A.; Makasheva, K.; Kaynak, M.; Sakar, M.S.; Fierz, B.; Ablasser, A. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 2020, 369, 823–828. [Google Scholar] [CrossRef]
- Bolderson, E.; Burgess, J.T.; Li, J.; Gandhi, N.S.; Boucher, D.; Croft, L.V.; Beard, S.; Plowman, J.J.; Suraweera, A.; Adams, M.N.; et al. Barrier-to-autointegration factor 1 (Banf1) regulates poly [ADP-ribose] polymerase 1 (PARP1) activity following oxidative DNA damage. Nat. Commun. 2019, 10, 5501. [Google Scholar] [CrossRef]
- Kono, Y.; Shimi, T. Crosstalk between mitotic reassembly and repair of the nuclear envelope. Nucleus 2024, 15, 2352203. [Google Scholar] [CrossRef] [PubMed]
- Kono, Y.; Adam, S.A.; Sato, Y.; Reddy, K.L.; Zheng, Y.; Medalia, O.; Goldman, R.D.; Kimura, H.; Shimi, T. Nucleoplasmic lamin C rapidly accumulates at sites of nuclear envelope rupture with BAF and cGAS. J. Cell Biol. 2022, 221, e202201024. [Google Scholar] [CrossRef] [PubMed]
- Young, T.W.; Mei, F.C.; Rosen, D.G.; Yang, G.; Li, N.; Liu, J.; Cheng, X. Up-regulation of tumor susceptibility gene 101 protein in ovarian carcinomas revealed by proteomics analyses. Mol. Cell Proteom. 2007, 6, 294–304. [Google Scholar] [CrossRef] [PubMed]
- Ferraiuolo, R.M.; Manthey, K.C.; Stanton, M.J.; Triplett, A.A.; Wagner, K.U. The Multifaceted Roles of the Tumor Susceptibility Gene 101 (TSG101) in Normal Development and Disease. Cancers 2020, 12, 450. [Google Scholar] [CrossRef]
- Liu, R.T.; Huang, C.C.; You, H.L.; Chou, F.F.; Hu, C.C.; Chao, F.P.; Chen, C.M.; Cheng, J.T. Overexpression of tumor susceptibility gene TSG101 in human papillary thyroid carcinomas. Oncogene 2002, 21, 4830–4837. [Google Scholar] [CrossRef]
- Oh, K.B.; Stanton, M.J.; West, W.W.; Todd, G.L.; Wagner, K.U. Tsg101 is upregulated in a subset of invasive human breast cancers and its targeted overexpression in transgenic mice reveals weak oncogenic properties for mammary cancer initiation. Oncogene 2007, 26, 5950–5959. [Google Scholar] [CrossRef]
- Liu, F.; Yu, Y.; Jin, Y.; Fu, S. TSG101, identified by screening a cancer cDNA library and soft agar assay, promotes cell proliferation in human lung cancer. Mol. Biol. Rep. 2010, 37, 2829–2838. [Google Scholar] [CrossRef]
- Xu, Z.; Liang, L.; Wang, H.; Li, T.; Zhao, M. HCRP1, a novel gene that is downregulated in hepatocellular carcinoma, encodes a growth-inhibitory protein. Biochem. Biophys. Res. Commun. 2003, 311, 1057–1066. [Google Scholar] [CrossRef]
- Wilson, E.M.; Oh, Y.; Hwa, V.; Rosenfeld, R.G. Interaction of IGF-binding protein-related protein 1 with a novel protein, neuroendocrine differentiation factor, results in neuroendocrine differentiation of prostate cancer cells. J. Clin. Endocrinol. Metab. 2001, 86, 4504–4511. [Google Scholar] [CrossRef][Green Version]
- Walker, G.E.; Antoniono, R.J.; Ross, H.J.; Paisley, T.E.; Oh, Y. Neuroendocrine-like differentiation of non-small cell lung carcinoma cells: Regulation by cAMP and the interaction of mac25/IGFBP-rP1 and 25. Oncogene 2006, 25, 1943–1954. [Google Scholar] [CrossRef]
- Zheng, Y.; Yang, S.; Dai, W.; Wang, J.; Bi, S.; Zhang, X.; Zheng, Z.; Sun, Y.; Wu, S.; Kong, J. CHMP3 promotes the progression of hepatocellular carcinoma by inhibiting caspase-1-dependent pyroptosis. Int. J. Oncol. 2024, 64, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Belogortseva, N.; Porter, D.; Park, M. Chmp1A functions as a novel tumor suppressor gene in human embryonic kidney and ductal pancreatic tumor cells. Cell Cycle 2008, 7, 2886–2893. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Xin, Y.; Liu, Y.; Sun, J.; Zhou, G.; Gao, H.; Xu, P.; Chen, Y.; Chen, G.; Zhang, L.; et al. Chmp1A acts as a tumor suppressor gene that inhibits proliferation of renal cell carcinoma. Cancer Lett. 2012, 319, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Wang, M.; Cao, Q.Q.; Li, Q. Chromatin modified protein 4C (CHMP4C) facilitates the malignant development of cervical cancer cells. FEBS Open Bio 2020, 10, 1295–1303. [Google Scholar] [CrossRef]
- Xu, C.; Liu, M.; Li, Y.; Peng, X.; Zhou, W.; Zhang, W.; Zhang, J.; Yu, B. The role and mechanism of CHMP4C in poor prognosis and drug sensitivity of lung adenocarcinoma. Discov. Oncol. 2025, 16, 270. [Google Scholar] [CrossRef]
- Yu, L.; Guo, Q.; Li, Y.; Mao, M.; Liu, Z.; Li, T.; Wang, L.; Zhang, X. CHMP4C promotes pancreatic cancer progression by inhibiting necroptosis via the RIPK1/RIPK3/MLKL pathway. J. Adv. Res. 2025. In Press. [Google Scholar] [CrossRef]
- Neggers, J.E.; Paolella, B.R.; Asfaw, A.; Rothberg, M.V.; Skipper, T.A.; Yang, A.; Kalekar, R.L.; Krill-Burger, J.M.; Dharia, N.V.; Kugener, G.; et al. Synthetic Lethal Interaction between the ESCRT Paralog Enzymes VPS4A and VPS4B in Cancers Harboring Loss of Chromosome 18q or 16q. Cell Rep. 2020, 33, 108493. [Google Scholar] [CrossRef]
- Szymańska, E.; Nowak, P.; Kolmus, K.; Cybulska, M.; Goryca, K.; Derezińska-Wołek, E.; Szumera-Ciećkiewicz, A.; Brewińska-Olchowik, M.; Grochowska, A.; Piwocka, K.; et al. Synthetic lethality between VPS4A and VPS4B triggers an inflammatory response in colorectal cancer. EMBO Mol. Med. 2020, 12, e10812. [Google Scholar] [CrossRef]
- Huang, L.J.; Zhan, S.T.; Pan, Y.Q.; Bao, W.; Yang, Y. The role of Vps4 in cancer development. Front. Oncol. 2023, 13, 1203359. [Google Scholar] [CrossRef]
- Filimonenko, M.; Stuffers, S.; Raiborg, C.; Yamamoto, A.; Malerød, L.; Fisher, E.M.; Isaacs, A.; Brech, A.; Stenmark, H.; Simonsen, A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007, 179, 485–500. [Google Scholar] [CrossRef]
- Lee, J.A.; Beigneux, A.; Ahmad, S.T.; Young, S.G.; Gao, F.B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007, 17, 1561–1567. [Google Scholar] [CrossRef] [PubMed]
- Shirk, A.J.; Anderson, S.K.; Hashemi, S.H.; Chance, P.F.; Bennett, C.L. SIMPLE interacts with NEDD4 and TSG101: Evidence for a role in lysosomal sorting and implications for Charcot-Marie-Tooth disease. J. Neurosci. Res. 2005, 82, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, N.; Ince, P.G.; Smith, M.O.; Highley, R.; Skibinski, G.; Andersen, P.M.; Morrison, K.E.; Pall, H.S.; Hardiman, O.; Collinge, J.; et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 2006, 67, 1074–1077. [Google Scholar] [CrossRef]
- Brugger, M.; Lauri, A.; Zhen, Y.; Gramegna, L.L.; Zott, B.; Sekulić, N.; Fasano, G.; Kopajtich, R.; Cordeddu, V.; Radio, F.C.; et al. Bi-allelic variants in SNF8 cause a disease spectrum ranging from severe developmental and epileptic encephalopathy to syndromic optic atrophy. Am. J. Hum. Genet. 2024, 111, 594–613. [Google Scholar] [CrossRef]
- Skibinski, G.; Parkinson, N.J.; Brown, J.M.; Chakrabarti, L.; Lloyd, S.L.; Hummerich, H.; Nielsen, J.E.; Hodges, J.R.; Spillantini, M.G.; Thusgaard, T.; et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005, 37, 806–808. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Barak, E.; Danial-Farran, N.; Chervinsky, E.; Alimi-Kasem, O.; Zagairy, F.; Livneh, I.; Mawassi, B.; Hreish, M.; Khayat, M.; Lossos, A.; et al. A homozygous variant in CHMP3 is associated with complex hereditary spastic paraplegia. J. Med. Genet. 2023, 60, 233–240. [Google Scholar] [CrossRef]
- Coulter, M.E.; Dorobantu, C.M.; Lodewijk, G.A.; Delalande, F.; Cianferani, S.; Ganesh, V.S.; Smith, R.S.; Lim, E.T.; Xu, C.S.; Pang, S.; et al. The ESCRT-III Protein CHMP1A Mediates Secretion of Sonic Hedgehog on a Distinctive Subtype of Extracellular Vesicles. Cell Rep. 2018, 24, 973–986.e9. [Google Scholar] [CrossRef]
- Mochida, G.H.; Ganesh, V.S.; de Michelena, M.I.; Dias, H.; Atabay, K.D.; Kathrein, K.L.; Huang, H.T.; Hill, R.S.; Felie, J.M.; Rakiec, D.; et al. CHMP1A encodes an essential regulator of BMI1-INK4A in cerebellar development. Nat. Genet. 2012, 44, 1260–1264. [Google Scholar] [CrossRef]
- Sakamoto, M.; Shiiki, T.; Matsui, S.; Okamoto, N.; Koshimizu, E.; Tsuchida, N.; Uchiyama, Y.; Hamanaka, K.; Fujita, A.; Miyatake, S.; et al. A novel homozygous CHMP1A variant arising from segmental uniparental disomy causes pontocerebellar hypoplasia type 8. J. Hum. Genet. 2023, 68, 247–253, Erratum in J. Hum. Genet. 2023, 68, 299. https://doi.org/10.1038/s10038-022-01098-x. [Google Scholar] [CrossRef]
- Coyne, A.N.; Baskerville, V.; Zaepfel, B.L.; Dickson, D.W.; Rigo, F.; Bennett, F.; Lusk, C.P.; Rothstein, J.D. Nuclear accumulation of CHMP7 initiates nuclear pore complex injury and subsequent TDP-43 dysfunction in sporadic and familial ALS. Sci. Transl. Med. 2021, 13, eabe1923. [Google Scholar] [CrossRef]
- Seu, K.G.; Trump, L.R.; Emberesh, S.; Lorsbach, R.B.; Johnson, C.; Meznarich, J.; Underhill, H.R.; Chou, S.T.; Sakthivel, H.; Nassar, N.N.; et al. VPS4A Mutations in Humans Cause Syndromic Congenital Dyserythropoietic Anemia due to Cytokinesis and Trafficking Defects. Am. J. Hum. Genet. 2020, 107, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Rodger, C.; Flex, E.; Allison, R.J.; Sanchis-Juan, A.; Hasenahuer, M.A.; Cecchetti, S.; French, C.E.; Edgar, J.R.; Carpentieri, G.; Ciolfi, A.; et al. De Novo VPS4A Mutations Cause Multisystem Disease with Abnormal Neurodevelopment. Am. J. Hum. Genet. 2020, 107, 1129–1148. [Google Scholar] [CrossRef] [PubMed]
- Sekar, R.; Motzler, K.; Kwon, Y.; Novikoff, A.; Jülg, J.; Najafi, B.; Wang, S.; Warnke, A.L.; Seitz, S.; Hass, D.; et al. Vps37a regulates hepatic glucose production by controlling glucagon receptor localization to endosomes. Cell Metab. 2022, 34, 1824–1842.e29. [Google Scholar] [CrossRef] [PubMed]
- Shiels, A.; Bennett, T.M.; Knopf, H.L.; Yamada, K.; Yoshiura, K.; Niikawa, N.; Shim, S.; Hanson, P.I. CHMP4B, a novel gene for autosomal dominant cataracts linked to chromosome 20q. Am. J. Hum. Genet. 2007, 81, 596–606. [Google Scholar] [CrossRef]
- Sagona, A.P.; Nezis, I.P.; Stenmark, H. Association of CHMP4B and autophagy with micronuclei: Implications for cataract formation. Biomed. Res. Int. 2014, 2014, 974393. [Google Scholar] [CrossRef]
- Dai, J.; Feng, Y.; Liao, Y.; Tan, L.; Sun, Y.; Song, C.; Qiu, X.; Ding, C. ESCRT machinery and virus infection. Antivir. Res. 2024, 221, 105786. [Google Scholar] [CrossRef]
- Gordon, D.J.; Resio, B.; Pellman, D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012, 13, 189–203. [Google Scholar] [CrossRef]
- La Torre, M.; Merigliano, C.; Burla, R.; Mottini, C.; Zanetti, G.; Del Giudice, S.; Carcuro, M.; Virdia, I.; Bucciarelli, E.; Manni, I.; et al. Mice with reduced expression of the telomere-associated protein Ft1 develop p53-sensitive progeroid traits. Aging Cell 2018, 17, e12730. [Google Scholar] [CrossRef]
- Keeley, O.; Coyne, A.N. Nuclear and degradative functions of the ESCRT-III pathway: Implications for neurodegenerative disease. Nucleus 2024, 15, 2349085. [Google Scholar] [CrossRef]
- Lee, J.A.; Liu, L.; Gao, F.B. Autophagy defects contribute to neurodegeneration induced by dysfunctional ESCRT-III. Autophagy 2009, 5, 1070–1072. [Google Scholar] [CrossRef]
- Fare, C.M.; Rothstein, J.D. Nuclear pore dysfunction and disease: A complex opportunity. Nucleus 2024, 15, 2314297. [Google Scholar] [CrossRef]
- Megat, S.; Mora, N.; Sanogo, J.; Roman, O.; Catanese, A.; Alami, N.O.; Freischmidt, A.; Mingaj, X.; De Calbiac, H.; Muratet, F.; et al. Integrative genetic analysis illuminates ALS heritability and identifies risk genes. Nat. Commun. 2023, 14, 342, Erratum in Nat. Commun. 2023, 14, 8026. https://doi.org/10.1038/s41467-022-35724-1. [Google Scholar] [CrossRef]
- Van Bergen, N.J.; Bell, K.M.; Carey, K.; Gear, R.; Massey, S.; Murrell, E.K.; Gallacher, L.; Pope, K.; Lockhart, P.J.; Kornberg, A.; et al. Pathogenic variants in nucleoporin TPR (translocated promoter region, nuclear basket protein) cause severe intellectual disability in humans. Hum. Mol. Genet. 2022, 31, 362–375. [Google Scholar] [CrossRef]
- Nim, S.; O′Hara, D.M.; Corbi-Verge, C.; Perez-Riba, A.; Fujisawa, K.; Kapadia, M.; Chau, H.; Albanese, F.; Pawar, G.; De Snoo, M.L.; et al. Disrupting the alpha-synuclein-ESCRT interaction with a peptide inhibitor mitigates neurodegeneration in preclinical models of Parkinson′s disease. Nat. Commun. 2023, 14, 2150. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).