Oligosaccharyltransferase Is Involved in Targeting to ER-Associated Degradation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Antibodies
2.3. Plasmids and Constructs
2.4. Cell Culture, Media and Transfections
2.5. BAP Construct Transfection and Probing
2.6. Metabolic Labeling
2.7. Immunoprecipitation, Crosslinking and Immunoblotting
2.8. Iodixanol Equilibrium Sedimentation Gradient
2.9. Immunofluorescence Microscopy
2.10. Proteomics
2.11. Bioinformatics Analysis
2.12. Statistical Analysis
3. Results
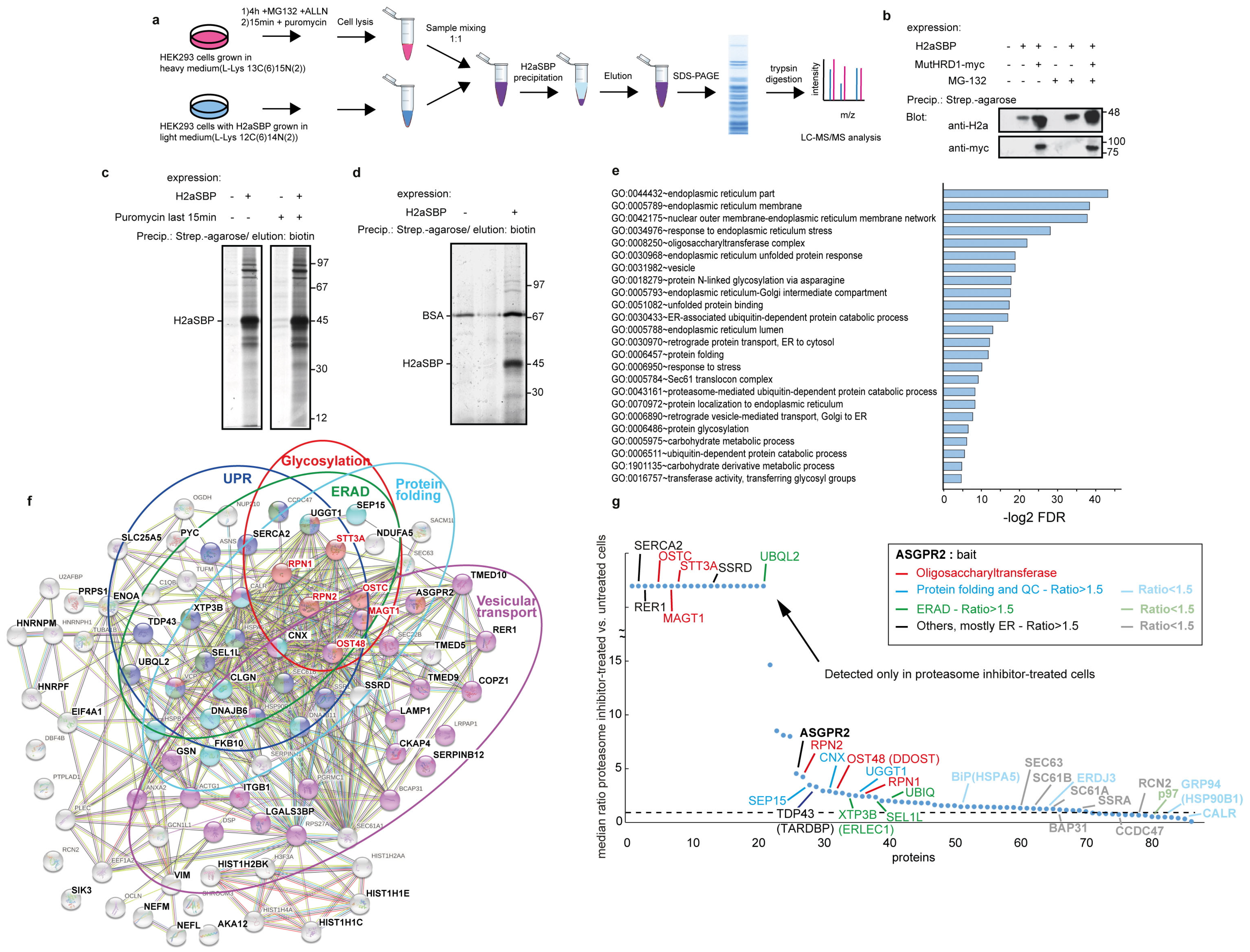
3.1. Identification of OST as ERAD Substrate Interactor by a SILAC Proteomics Approach
3.2. Shared Subcellular Localization and Increased Interaction of OST Subunits with the ERAD Substrate upon Proteasomal Inhibition
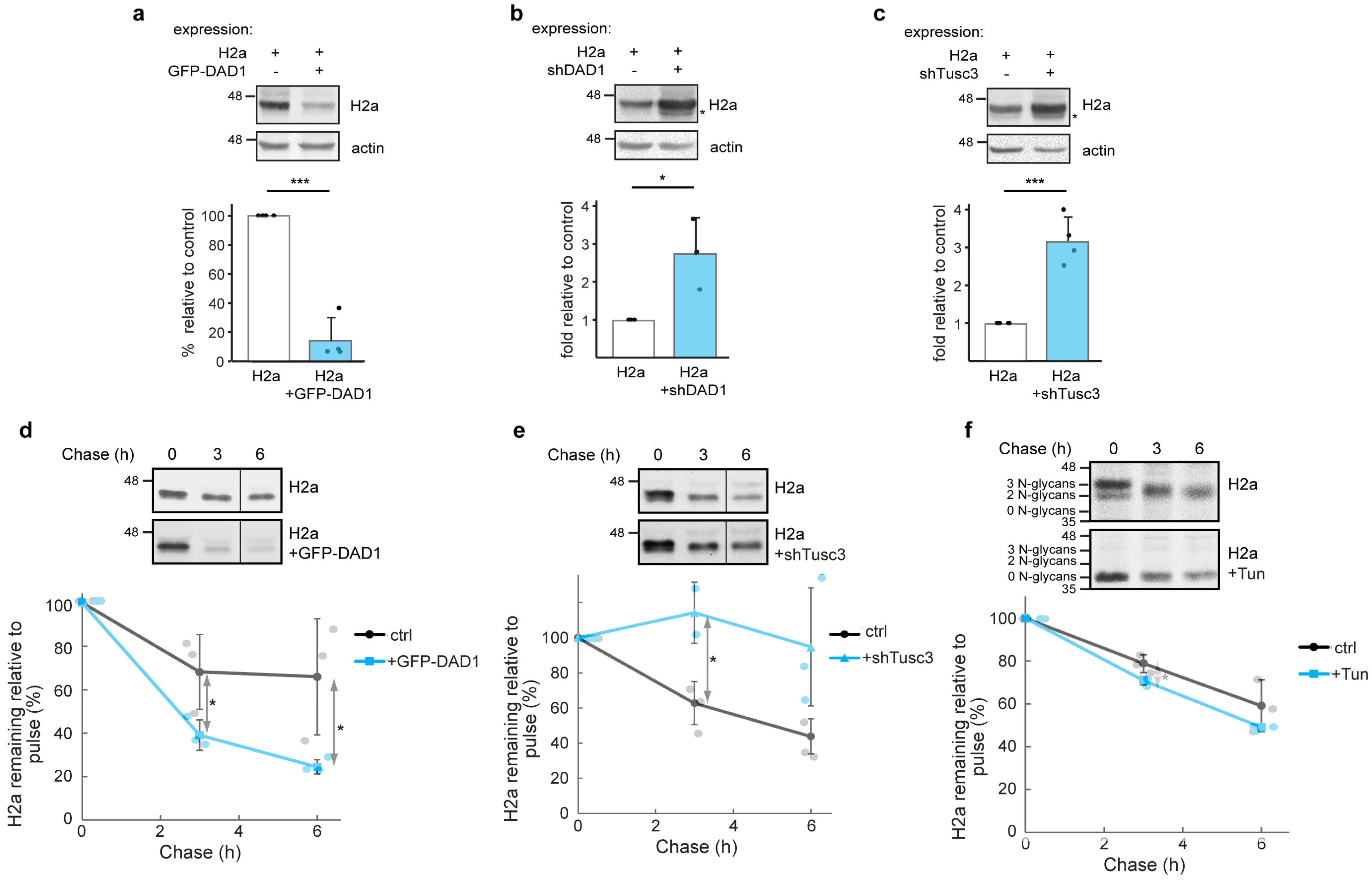
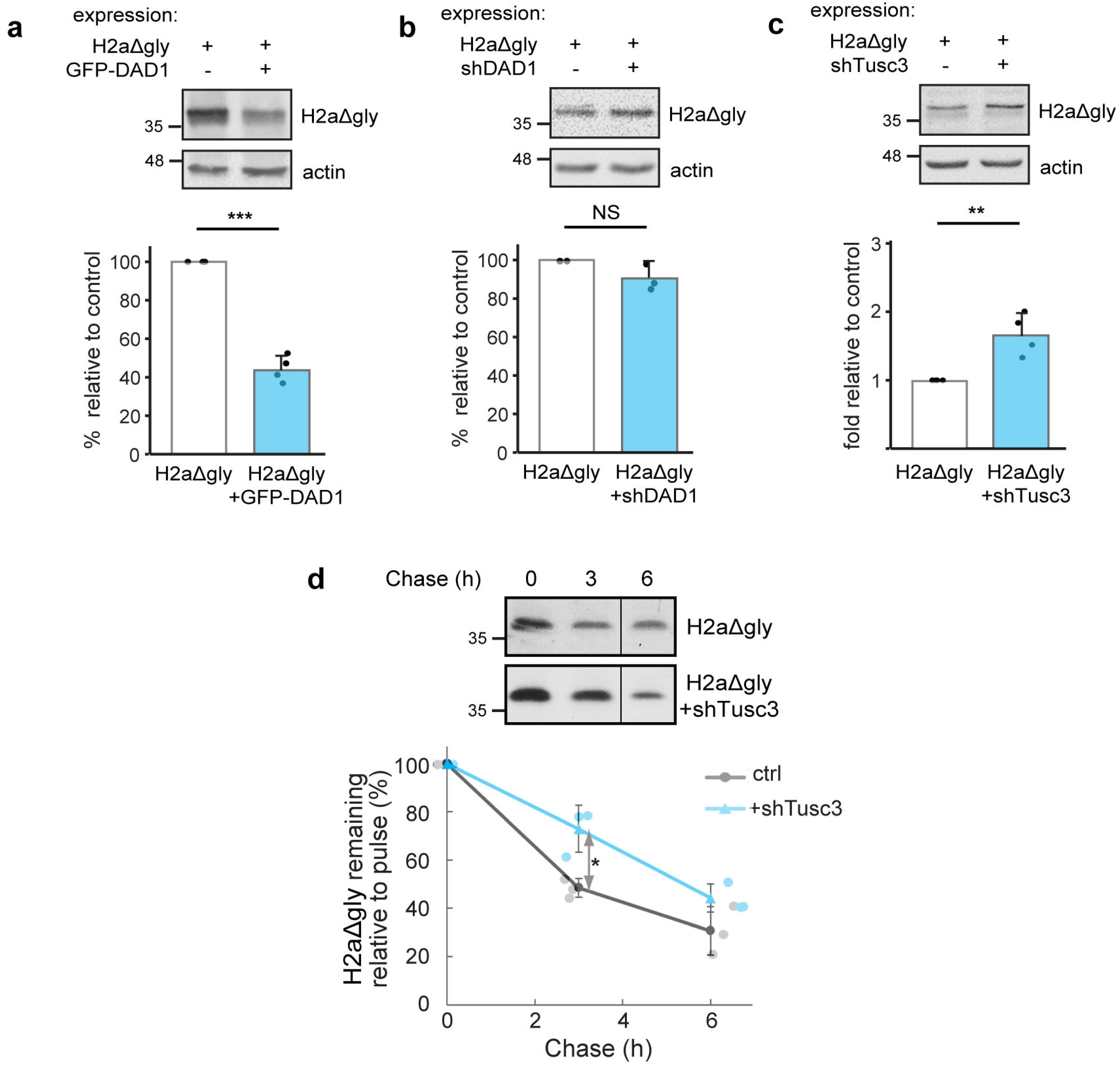
3.3. OST Subunits Promote the Degradation of Glycosylated H2a and to a Lesser Extent of Its Non-Glycosylated Mutant
3.4. OST Subunits Participate in ERAD of Other Membrane and Luminal Substrates
3.5. Interaction Between DAD1 and HRD1 and Promotion of Retrotranslocation by OST Subunits
3.6. Molecular Dynamics Simulation Suggest Membrane Thinning by OST
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALLN | N-acetyl-Leu-Leu-norleucinal |
| BACE476 | beta-secretase 1 mutant 476 |
| BAP | biotin-acceptor peptide |
| BirA | E. coli biotin ligase BirA |
| Bz | Bortezomib |
| CNX | Calnexin |
| DAD1 | Defender Against apoptotic cell Death 1 |
| ECL | enhanced chemiluminescence |
| ER | endoplasmic reticulum |
| ERAD | ER-associated degradation |
| ERQC | ER-derived quality control compartment |
| Fbs2 | Skp1-Cullin1-F-box protein 2 |
| Herp | Homocysteine-inducible Endoplasmic Reticulum (ER) Stress Protein |
| HRD1 | HMG-CoA Reductase Degradation Protein 1 |
| H2a | uncleaved precursor of human asialoglycoprotein receptor H2a. Gene: ASGPR2 |
| H2aΔgly | H2a mutated in its three glycosylation sites |
| H2aSBP | H2aG78R uncleavable mutant fused through its C-terminus to a 38-amino acid streptavidin binding peptide |
| Lac | Lactacystin |
| NHK | human α1-antitrypsin variant null Hong Kong |
| OS-9 | Amplified in Osteosarcoma 9 |
| OST | oligosaccharyltransferase |
| SEL1L | Suppressor of lin-12-like protein 1 |
| SEP15 | Selenoprotein 15 or Selenof |
| SERCA2 | Sarco(endo)plasmic reticulum calcium-ATPase 2 |
| SILAC | stable isotope labeling by amino acids in cell culture |
| STT3A | STT3 oligosaccharyltransferase complex catalytic subunit A |
| STT3B | STT3 oligosaccharyltransferase complex catalytic subunit B |
| TDP43 | TAR DNA-binding protein 43 |
| TMD | transmembrane domain |
| Tusc3 | tumor suppressor candidate 3 |
| Tun | Tunicamycin |
| Ubxd8 | ubiquitin regulatory X domain-containing protein 8 |
| UGGT1 | UDP-glucose glycoprotein glucosyltransferase 1 |
| XTP3-B | XTP3-transactivated gene B protein or ER lectin 1 |
References
- Sun, Z.; Brodsky, J.L. Protein quality control in the secretory pathway. J. Cell Biol. 2019, 218, 3171–3187. [Google Scholar] [CrossRef]
- Wu, X.; Rapoport, T.A. Mechanistic insights into ER-associated protein degradation. Curr. Opin. Cell Biol. 2018, 53, 22–28. [Google Scholar] [CrossRef]
- Salomo-Coll, C.; Jimenez-Moreno, N.; Wilkinson, S. Lysosomal Degradation of ER Client Proteins by ER-phagy and Related Pathways. J. Mol. Biol. 2025, 437, 169035. [Google Scholar] [CrossRef]
- Fregno, I.; Fasana, E.; Bergmann, T.J.; Raimondi, A.; Loi, M.; Soldà, T.; Galli, C.; D’Antuono, R.; Morone, D.; Danieli, A.; et al. ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 2018, 37, e99259. [Google Scholar] [CrossRef]
- Mochida, K.; Nakatogawa, H. ER-phagy: Selective autophagy of the endoplasmic reticulum. EMBO Rep. 2022, 23, e55192. [Google Scholar] [CrossRef]
- Ward, C.L.; Omura, S.; Kopito, R.R. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 1995, 83, 121–127. [Google Scholar] [CrossRef]
- Werner, E.D.; Brodsky, J.L.; McCracken, A.A. Proteasome-dependent endoplasmic reticulum-associated protein degradation: An unconventional route to a familiar fate. Proc. Natl. Acad. Sci. USA 1996, 93, 13797–13801. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.C.; Jarosch, E.; Sommer, T. Mechanisms of substrate processing during ER-associated protein degradation. Nat. Rev. Mol. Cell Biol. 2023, 24, 777–796. [Google Scholar] [CrossRef]
- Lopata, A.; Kniss, A.; Löhr, F.; Rogov, V.V.; Dötsch, V. Ubiquitination in the ERAD Process. Int. J. Mol. Sci. 2020, 21, 5369. [Google Scholar] [CrossRef] [PubMed]
- Berner, N.; Reutter, K.R.; Wolf, D.H. Protein Quality Control of the Endoplasmic Reticulum and Ubiquitin-Proteasome-Triggered Degradation of Aberrant Proteins: Yeast Pioneers the Path. Annu. Rev. Biochem. 2018, 87, 751–782. [Google Scholar] [CrossRef] [PubMed]
- Kikkert, M.; Doolman, R.; Dai, M.; Avner, R.; Hassink, G.; van Voorden, S.; Thanedar, S.; Roitelman, J.; Chau, V.; Wiertz, E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 3525–3534. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Biunno, I.; Sun, S.; Qi, L. SEL1L-HRD1-mediated ERAD in mammals. Nat. Cell Biol. 2025, 27, 1063–1073. [Google Scholar] [CrossRef]
- Schoebel, S.; Mi, W.; Stein, A.; Ovchinnikov, S.; Pavlovicz, R.; DiMaio, F.; Baker, D.; Chambers, M.G.; Su, H.; Li, D.; et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 2017, 548, 352–355. [Google Scholar] [CrossRef]
- Wu, X.; Siggel, M.; Ovchinnikov, S.; Mi, W.; Svetlov, V.; Nudler, E.; Liao, M.; Hummer, G.; Rapoport, T.A. Structural basis of ER-associated protein degradation mediated by the Hrd1 ubiquitin ligase complex. Science 2020, 368, eaaz2449. [Google Scholar] [CrossRef]
- Guo, L.; Liu, G.; He, J.; Jia, X.; He, Y.; Wang, Z.; Qian, H. Structural insights into the human HRD1 ubiquitin ligase complex. Nat. Commun. 2025, 16, 6007. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.G.; Glaser, M.L.; Rapoport, T.A.; Baldridge, R.D. Cycles of autoubiquitination and deubiquitination regulate the ERAD ubiquitin ligase Hrd1. Elife 2019, 8, e50903. [Google Scholar] [CrossRef]
- Vasic, V.; Denkert, N.; Schmidt, C.C.; Riedel, D.; Stein, A.; Meinecke, M. Hrd1 forms the retrotranslocation pore regulated by auto-ubiquitination and binding of misfolded proteins. Nat. Cell Biol. 2020, 22, 274–281. [Google Scholar] [CrossRef]
- Leitman, J.; Shenkman, M.; Gofman, Y.; Shtern, N.O.; Ben-Tal, N.; Hendershot, L.M.; Lederkremer, G.Z. Herp coordinates compartmentalization and recruitment of HRD1 and misfolded proteins for ERAD. Mol. Biol. Cell 2014, 25, 1050–1060. [Google Scholar] [CrossRef]
- Shenkman, M.; Lederkremer, G.Z. Compartmentalization and Selective Tagging for Disposal of Misfolded Glycoproteins. Trends Biochem. Sci. 2019, 44, 827–836. [Google Scholar] [CrossRef]
- Denic, V.; Quan, E.M.; Weissman, J.S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 2006, 126, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Mueller, B.; Klemm, E.J.; Spooner, E.; Claessen, J.H.; Ploegh, H.L. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc. Natl. Acad. Sci. USA 2008, 105, 12325–12330. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.C.; Olzmann, J.A.; Shaler, T.A.; Sowa, M.E.; Bennett, E.J.; Richter, C.M.; Tyler, R.E.; Greenblatt, E.J.; Wade Harper, J.; Kopito, R.R. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 2012, 14, 93–105. [Google Scholar] [CrossRef]
- Hwang, J.; Walczak, C.P.; Shaler, T.A.; Olzmann, J.A.; Zhang, L.; Elias, J.E.; Kopito, R.R. Characterization of protein complexes of the endoplasmic reticulum-associated degradation E3 ubiquitin ligase Hrd1. J. Biol. Chem. 2017, 292, 9104–9116. [Google Scholar] [CrossRef]
- Kny, M.; Standera, S.; Hartmann-Petersen, R.; Kloetzel, P.M.; Seeger, M. Herp regulates Hrd1-mediated ubiquitylation in a ubiquitin-like domain-dependent manner. J. Biol. Chem. 2011, 286, 5151–5156. [Google Scholar] [CrossRef]
- Cherepanova, N.; Shrimal, S.; Gilmore, R. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 2016, 41, 57–65. [Google Scholar] [CrossRef]
- Mohanty, S.; Chaudhary, B.P.; Zoetewey, D. Structural Insight into the Mechanism of N-Linked Glycosylation by Oligosaccharyltransferase. Biomolecules 2020, 10, 624. [Google Scholar] [CrossRef]
- Qin, S.Y.; Hu, D.; Matsumoto, K.; Takeda, K.; Matsumoto, N.; Yamaguchi, Y.; Yamamoto, K. Malectin forms a complex with ribophorin I for enhanced association with misfolded glycoproteins. J. Biol. Chem. 2012, 287, 38080–38089. [Google Scholar] [CrossRef]
- Wilson, C.M.; Kraft, C.; Duggan, C.; Ismail, N.; Crawshaw, S.G.; High, S. Ribophorin I associates with a subset of membrane proteins after their integration at the sec61 translocon. J. Biol. Chem. 2005, 280, 4195–4206. [Google Scholar] [CrossRef]
- Ron, E.; Shenkman, M.; Groisman, B.; Izenshtein, Y.; Leitman, J.; Lederkremer, G.Z. Bypass of glycan-dependent glycoprotein delivery to ERAD by up-regulated EDEM1. Mol. Biol. Cell 2011, 22, 3945–3954. [Google Scholar] [CrossRef] [PubMed]
- Kamhi-Nesher, S.; Shenkman, M.; Tolchinsky, S.; Fromm, S.V.; Ehrlich, R.; Lederkremer, G.Z. A novel quality control compartment derived from the endoplasmic reticulum. Mol. Biol. Cell 2001, 12, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Kondratyev, M.; Avezov, E.; Shenkman, M.; Groisman, B.; Lederkremer, G.Z. PERK-dependent compartmentalization of ERAD and unfolded protein response machineries during ER stress. Exp. Cell Res. 2007, 313, 3395–3407. [Google Scholar] [CrossRef]
- Ogen-Shtern, N.; Chang, C.; Saad, H.; Mazkereth, N.; Patel, C.; Shenkman, M.; Lederkremer, G.Z. COP I and II dependent trafficking controls ER-associated degradation in mammalian cells. iScience 2023, 26, 106232. [Google Scholar] [CrossRef]
- Groisman, B.; Shenkman, M.; Ron, E.; Lederkremer, G.Z. Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps. J. Biol. Chem. 2011, 286, 1292–1300. [Google Scholar] [CrossRef]
- Sharma, N.; Patel, C.; Shenkman, M.; Kessel, A.; Ben-Tal, N.; Lederkremer, G.Z. The Sigma-1 receptor is an ER-localized type II membrane protein. J. Biol. Chem. 2021, 297, 101299. [Google Scholar] [CrossRef]
- Petris, G.; Vecchi, L.; Bestagno, M.; Burrone, O.R. Efficient detection of proteins retro-translocated from the ER to the cytosol by in vivo biotinylation. PLoS ONE 2011, 6, e23712. [Google Scholar] [CrossRef]
- Call, M.E.; Pyrdol, J.; Wiedmann, M.; Wucherpfennig, K.W. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 2002, 111, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bandla, C.; Kundu, D.J.; Kamatchinathan, S.; Bai, J.; Hewapathirana, S.; John, N.S.; Prakash, A.; Walzer, M.; Wang, S.; et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 2025, 53, D543–D553. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Adelman, M.R.; Sabatini, D.D.; Blobel, G. Ribosome-membrane interaction. Nondestructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J. Cell Biol. 1973, 56, 206–229. [Google Scholar] [CrossRef]
- Shenkman, M.; Ayalon, M.; Lederkremer, G.Z. Endoplasmic reticulum quality control of asialoglycoprotein receptor H2a involves a determinant for retention and not retrieval. Proc. Natl. Acad. Sci. USA 1997, 94, 11363–11368. [Google Scholar] [CrossRef]
- Keefe, A.D.; Wilson, D.S.; Seelig, B.; Szostak, J.W. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 2001, 23, 440–446. [Google Scholar] [CrossRef]
- Pfeffer, S.; Dudek, J.; Gogala, M.; Schorr, S.; Linxweiler, J.; Lang, S.; Becker, T.; Beckmann, R.; Zimmermann, R.; Förster, F. Structure of the mammalian oligosaccharyl-transferase complex in the native ER protein translocon. Nat. Commun. 2014, 5, 3072. [Google Scholar] [CrossRef]
- Scotter, E.L.; Vance, C.; Nishimura, A.L.; Lee, Y.B.; Chen, H.J.; Urwin, H.; Sardone, V.; Mitchell, J.C.; Rogelj, B.; Rubinsztein, D.C.; et al. Differential roles of the ubiquitin proteasome system and autophagy in the clearance of soluble and aggregated TDP-43 species. J. Cell Sci. 2014, 127, 1263–1278. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, D.J.; Gilmore, R. DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc. Natl. Acad. Sci. USA 1997, 94, 4994–4999. [Google Scholar] [CrossRef]
- Nikonov, A.V.; Snapp, E.; Lippincott-Schwartz, J.; Kreibich, G. Active translocon complexes labeled with GFP-Dad1 diffuse slowly as large polysome arrays in the endoplasmic reticulum. J. Cell Biol. 2002, 158, 497–506. [Google Scholar] [CrossRef]
- Pereira, F.; Rettel, M.; Stein, F.; Savitski, M.M.; Collinson, I.; Römisch, K. Effect of Sec61 interaction with Mpd1 on endoplasmic reticulum-associated degradation. PLoS ONE 2019, 14, e0211180. [Google Scholar] [CrossRef]
- Shenkman, M.; Groisman, B.; Ron, E.; Avezov, E.; Hendershot, L.M.; Lederkremer, G.Z. A Shared Endoplasmic Reticulum-associated Degradation Pathway Involving the EDEM1 Protein for Glycosylated and Nonglycosylated Proteins. J. Biol. Chem. 2013, 288, 2167–2178. [Google Scholar] [CrossRef] [PubMed]
- Lamriben, L.; Oster, M.E.; Tamura, T.; Tian, W.; Yang, Z.; Clausen, H.; Hebert, D.N. EDEM1’s mannosidase-like domain binds ERAD client proteins in a redox-sensitive manner and possesses catalytic activity. J. Biol. Chem. 2018, 293, 13932–13945. [Google Scholar] [CrossRef]
- Ramírez, A.S.; Kowal, J.; Locher, K.P. Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B. Science 2019, 366, 1372–1375. [Google Scholar] [CrossRef] [PubMed]
- Newport, T.D.; Sansom, M.S.P.; Stansfeld, P.J. The MemProtMD database: A resource for membrane-embedded protein structures and their lipid interactions. Nucleic Acids Res. 2019, 47, D390–D397. [Google Scholar] [CrossRef]
- Wu, X.; Rapoport, T.A. Translocation of Proteins through a Distorted Lipid Bilayer. Trends Cell Biol. 2021, 31, 473–484. [Google Scholar] [CrossRef]
- Reggio, A.; Buonomo, V.; Berkane, R.; Bhaskara, R.M.; Tellechea, M.; Peluso, I.; Polishchuk, E.; Di Lorenzo, G.; Cirillo, C.; Esposito, M.; et al. Role of FAM134 paralogues in endoplasmic reticulum remodeling, ER-phagy, and Collagen quality control. EMBO Rep. 2021, 22, e52289. [Google Scholar] [CrossRef]
- Shibatani, T.; David, L.L.; McCormack, A.L.; Frueh, K.; Skach, W.R. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry 2005, 44, 5982–5992. [Google Scholar] [CrossRef]
- Losfeld, M.E.; Ng, B.G.; Kircher, M.; Buckingham, K.J.; Turner, E.H.; Eroshkin, A.; Smith, J.D.; Shendure, J.; Nickerson, D.A.; Bamshad, M.J.; et al. A new congenital disorder of glycosylation caused by a mutation in SSR4, the signal sequence receptor 4 protein of the TRAP complex. Hum. Mol. Genet. 2014, 23, 1602–1605. [Google Scholar] [CrossRef]
- Nagasawa, K.; Higashi, T.; Hosokawa, N.; Kaufman, R.J.; Nagata, K. Simultaneous induction of the four subunits of the TRAP complex by ER stress accelerates ER degradation. EMBO Rep. 2007, 8, 483–489. [Google Scholar] [CrossRef]
- Russo, A. Understanding the mammalian TRAP complex function(s). Open Biol. 2020, 10, 190244. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Yamazaki, T.; Komazaki, S.; Yamashita, T.; Osaki, M.; Matsubayashi, M.; Kidoya, H.; Takakura, N.; Yamazaki, D.; Kakizawa, S. Contribution of calumin to embryogenesis through participation in the endoplasmic reticulum-associated degradation activity. Dev. Biol. 2014, 393, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Kon, T.; Mori, F.; Tanji, K.; Miki, Y.; Nishijima, H.; Nakamura, T.; Kinoshita, I.; Suzuki, C.; Kurotaki, H.; Tomiyama, M.; et al. Accumulation of Nonfibrillar TDP-43 in the Rough Endoplasmic Reticulum Is the Early-Stage Pathology in Amyotrophic Lateral Sclerosis. J. Neuropathol. Exp. Neurol. 2022, 81, 271–281. [Google Scholar] [CrossRef]
- Lin, D.L.; Cherepanova, N.A.; Bozzacco, L.; MacDonald, M.R.; Gilmore, R.; Tai, A.W. Dengue Virus Hijacks a Noncanonical Oxidoreductase Function of a Cellular Oligosaccharyltransferase Complex. mBio 2017, 8, e00939-17. [Google Scholar] [CrossRef]
- Patel, C.; Saad, H.; Shenkman, M.; Lederkremer, G.Z. Oxidoreductases in Glycoprotein Glycosylation, Folding, and ERAD. Cells 2020, 9, 2138. [Google Scholar] [CrossRef] [PubMed]
- Cherepanova, N.A.; Shrimal, S.; Gilmore, R. Oxidoreductase activity is necessary for N-glycosylation of cysteine-proximal acceptor sites in glycoproteins. J. Cell Biol. 2014, 206, 525–539. [Google Scholar] [CrossRef]
- Lilley, B.N.; Ploegh, H.L. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 2004, 429, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Christianson, J.C.; Shaler, T.A.; Tyler, R.E.; Kopito, R.R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008, 10, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Ng, B.G.; Freeze, H.H.; Himmelreich, N.; Blau, N.; Ferreira, C.R. Clinical and biochemical footprints of congenital disorders of glycosylation: Proposed nosology. Mol. Genet. Metab. 2024, 142, 108476. [Google Scholar] [CrossRef]
- Wilson, M.P.; Garanto, A.; Pinto e Vairo, F.; Ng, B.G.; Ranatunga, W.K.; Ventouratou, M.; Baerenfaenger, M.; Huijben, K.; Thiel, C.; Ashikov, A.; et al. Active site variants in STT3A cause a dominant type I congenital disorder of glycosylation with neuromusculoskeletal findings. Am. J. Hum. Genet. 2021, 108, 2130–2144. [Google Scholar] [CrossRef]
- Cherepanova, N.A.; Gilmore, R. Mammalian cells lacking either the cotranslational or posttranslocational oligosaccharyltransferase complex display substrate-dependent defects in asparagine linked glycosylation. Sci. Rep. 2016, 6, 20946. [Google Scholar] [CrossRef]
- Knopf, J.D.; Landscheidt, N.; Pegg, C.L.; Schulz, B.L.; Kühnle, N.; Chao, C.W.; Huck, S.; Lemberg, M.K. Intramembrane protease RHBDL4 cleaves oligosaccharyltransferase subunits to target them for ER-associated degradation. J. Cell Sci. 2020, 133, jcs243790. [Google Scholar] [CrossRef]
- Hosokawa, N.; Wada, I.; Nagasawa, K.; Moriyama, T.; Okawa, K.; Nagata, K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J. Biol. Chem. 2008, 283, 20914–20924. [Google Scholar] [CrossRef] [PubMed]
- Jaenicke, L.A.; Brendebach, H.; Selbach, M.; Hirsch, C. Yos9p assists in the degradation of certain nonglycosylated proteins from the endoplasmic reticulum. Mol. Biol. Cell 2011, 22, 2937–2945. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shenkman, M.; Ogen-Shtern, N.; Patel, C.; Saad, H.; Groisman, B.; Pasmanik-Chor, M.; Schermann, S.M.; Körner, R.; Lederkremer, G.Z. Oligosaccharyltransferase Is Involved in Targeting to ER-Associated Degradation. Cells 2025, 14, 1593. https://doi.org/10.3390/cells14201593
Shenkman M, Ogen-Shtern N, Patel C, Saad H, Groisman B, Pasmanik-Chor M, Schermann SM, Körner R, Lederkremer GZ. Oligosaccharyltransferase Is Involved in Targeting to ER-Associated Degradation. Cells. 2025; 14(20):1593. https://doi.org/10.3390/cells14201593
Chicago/Turabian StyleShenkman, Marina, Navit Ogen-Shtern, Chaitanya Patel, Haddas Saad, Bella Groisman, Metsada Pasmanik-Chor, Sonya M. Schermann, Roman Körner, and Gerardo Z. Lederkremer. 2025. "Oligosaccharyltransferase Is Involved in Targeting to ER-Associated Degradation" Cells 14, no. 20: 1593. https://doi.org/10.3390/cells14201593
APA StyleShenkman, M., Ogen-Shtern, N., Patel, C., Saad, H., Groisman, B., Pasmanik-Chor, M., Schermann, S. M., Körner, R., & Lederkremer, G. Z. (2025). Oligosaccharyltransferase Is Involved in Targeting to ER-Associated Degradation. Cells, 14(20), 1593. https://doi.org/10.3390/cells14201593







