A miR-30 Guided Molecular Profiling of Canine Osteosarcoma and Extraskeletal Osteosarcoma Reveals Non-Seed Regulatory Divergence
Abstract
1. Introduction
2. Materials and Methods
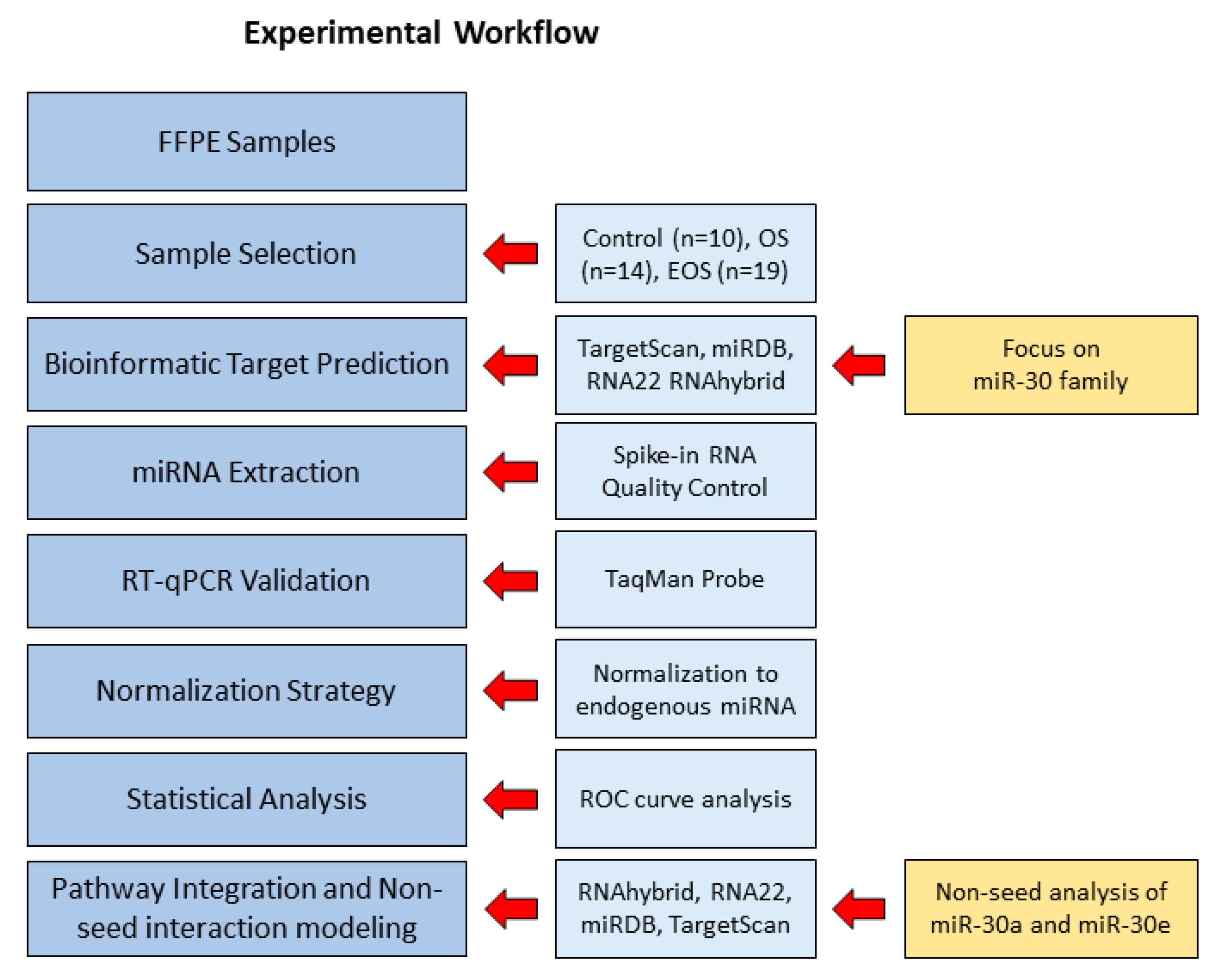
2.1. Experimental Workflow
2.2. Case Selection and Inclusion Criteria
2.3. Bioinformatics-Based Selection of RUNX2, KPNA2, and SATB2 miRNA-Target Interactions
2.4. Seed Sequences of miR-30 Family
2.5. RNA Spike-In Quality Assessment
2.6. MiRNA Isolation and Purification
2.7. RT-qPCR Analysis of miRNA Expression
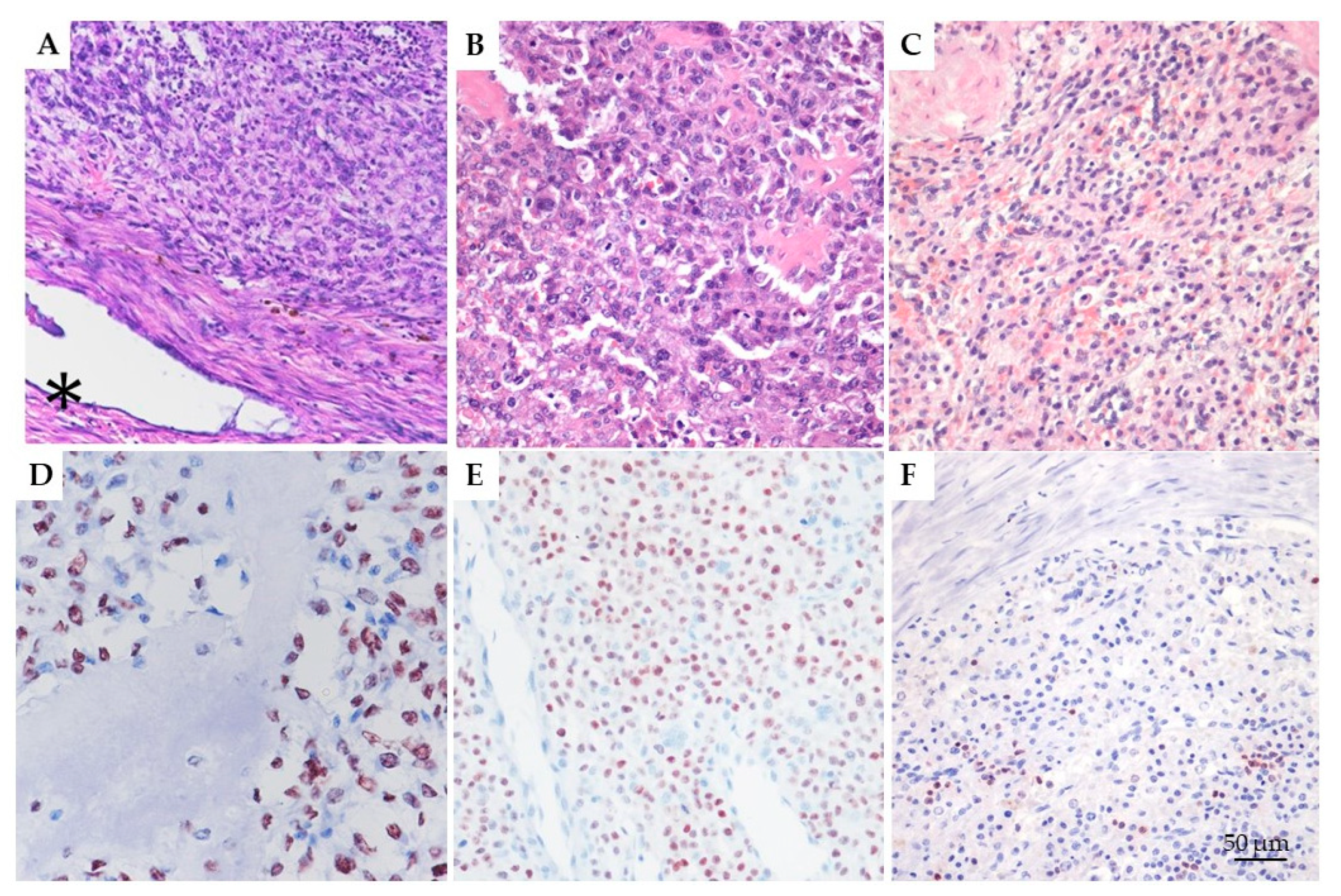
2.8. RUNX2 Immunohistochemistry in OS, EOS, and CTRL Tissue
2.9. Statistical Analysis
3. Results
3.1. Histological Diagnosis and IHC Evaluation of RUNX2
3.2. RNA Evaluation and Reference Gene Selection
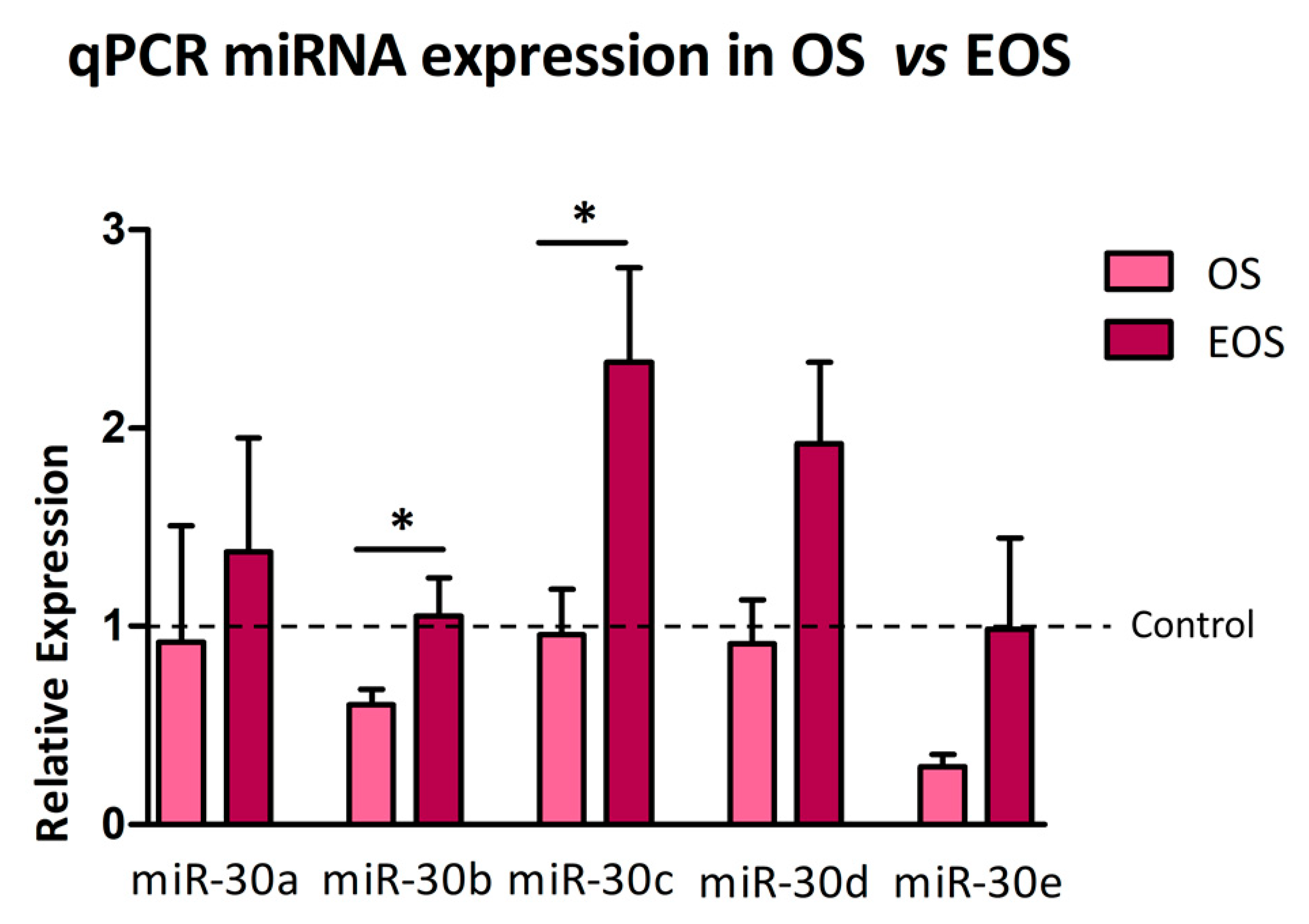
3.3. Expression Profiling of miR-30 Family Members in CTRL, OS, and EOS Samples
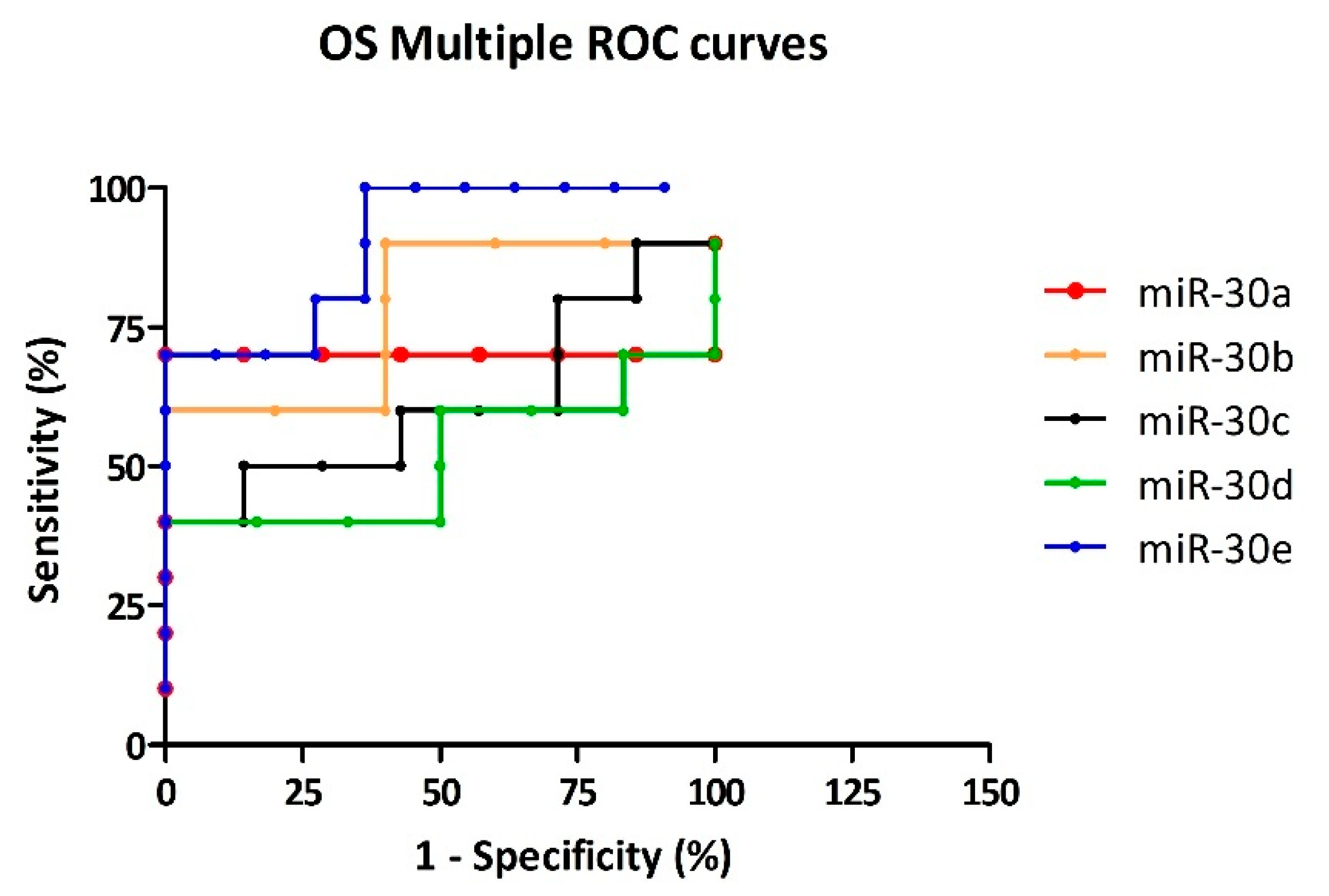
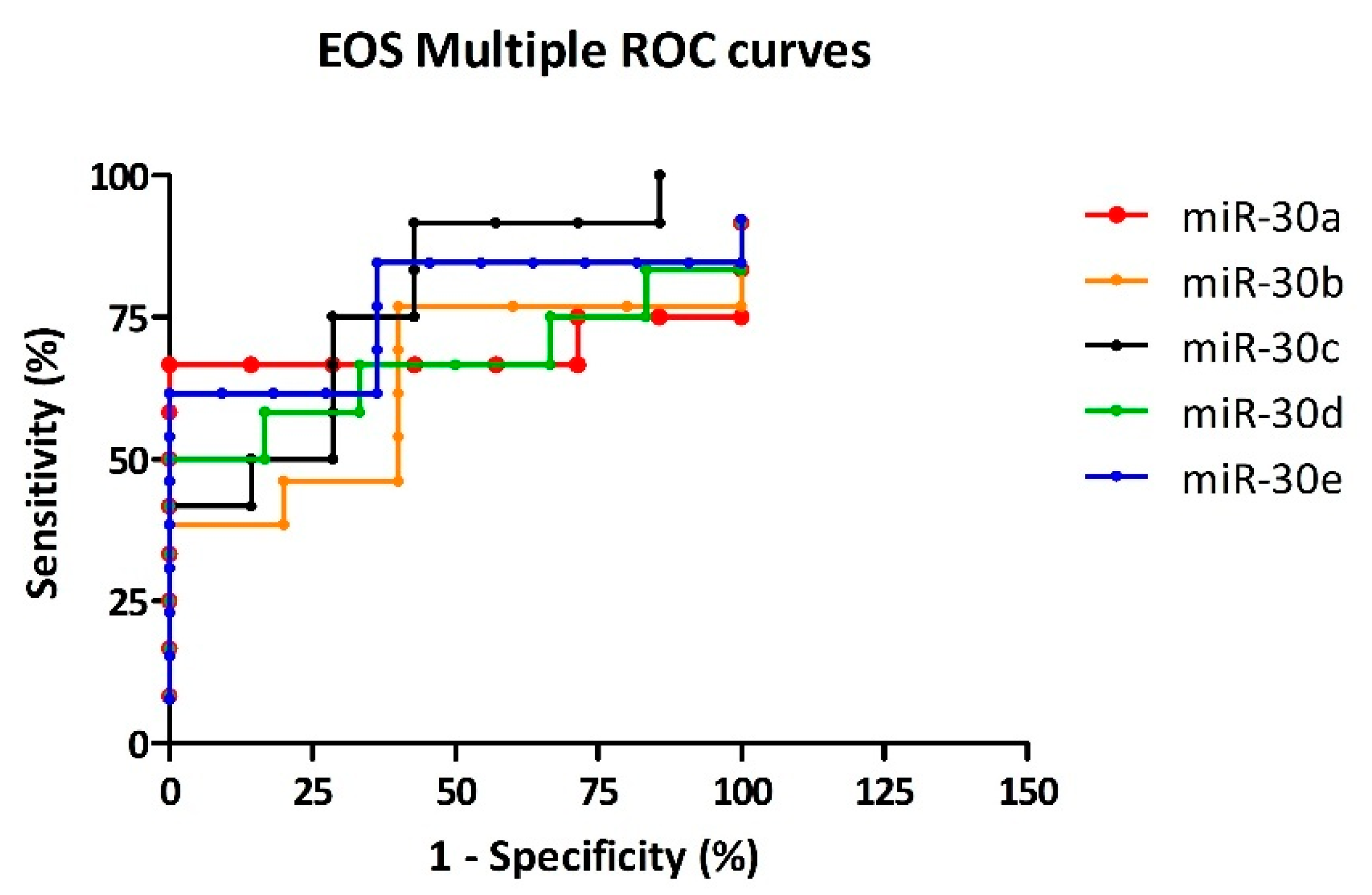
3.4. Cutoff Selection for Diagnostic Specificity
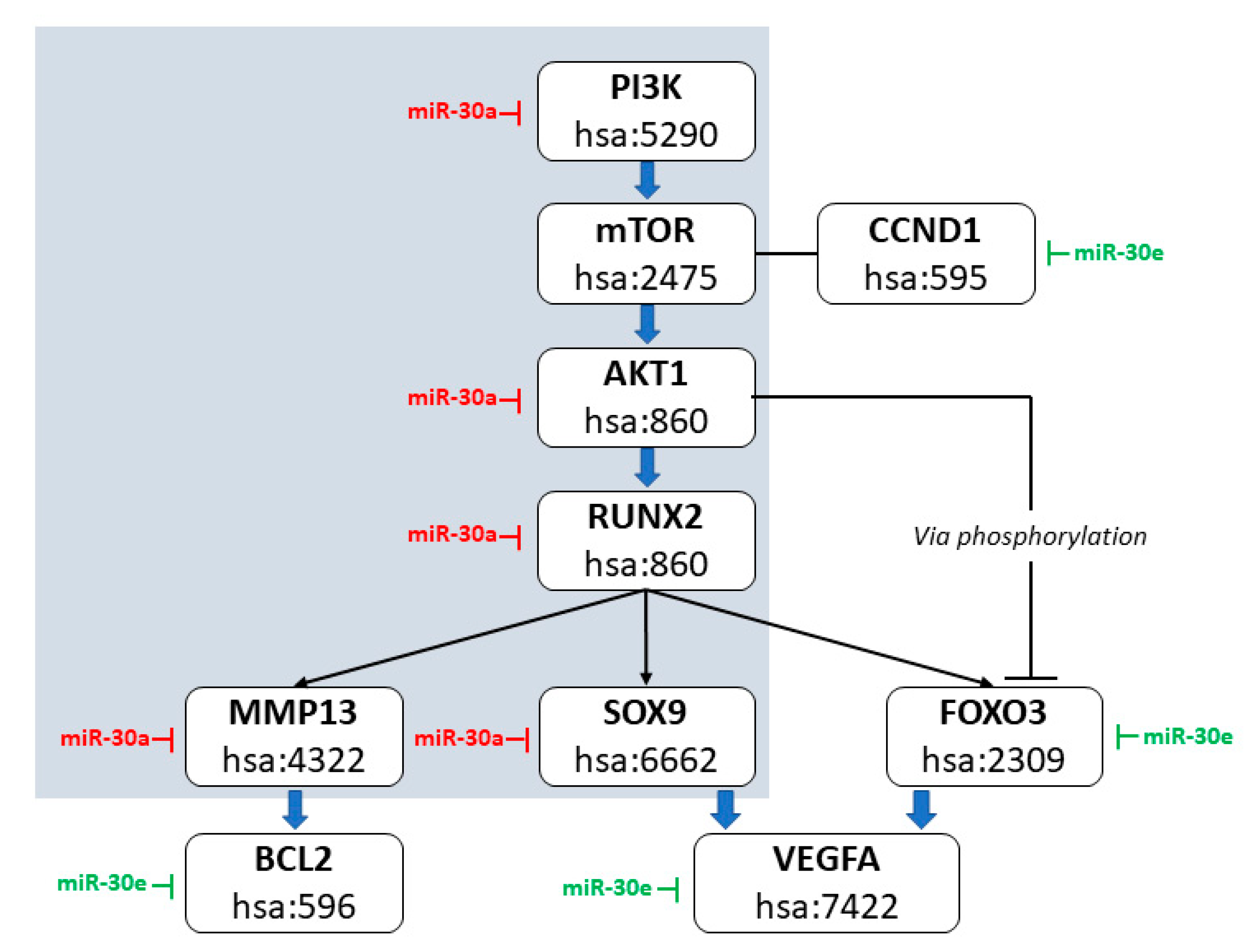
3.5. Non-Seed Bioinformatic Mapping of miR-30a and 30e in Canine OS and EOS Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Habeeb, O.; Weigelt, M.A.; Goldblum, J.R.; Ko, J.S.; Habermehl, G.; Rubin, B.P.; Billings, S.D. Primary Cutaneous Extraskeletal Osteosarcoma: A Series of 16 Cases. Pathology 2023, 55, 315–323. [Google Scholar] [CrossRef]
- Yenwongfai, L.N.; Liu, J.; Wang, C.; Bocklage, T.J. Extraskeletal Osteosarcoma and Its Histological Mimics. Hum. Pathol. Rep. 2022, 28, 300639. [Google Scholar] [CrossRef]
- Nance, R.L.; Sajib, A.M.; Smith, B.F. Canine Models of Human Cancer: Bridging the Gap to Improve Precision Medicine. Prog. Mol. Biol. Transl. Sci. 2022, 189, 67–99. [Google Scholar] [CrossRef]
- Langenbach, A.; Anderson, M.; Dambach, D.; Sorenmo, K.; Shofer, F. Extraskeletal Osteosarcomas in Dogs: A Retrospective Study of 169 Cases (1986–1996). J. Am. Anim. Hosp. Assoc. 1998, 34, 113–120. [Google Scholar] [CrossRef]
- Leonardi, L.; Manuali, E.; Bufalari, A.; Porcellato, I. Canine Soft Tissue Sarcomas: The Expression of RUNX2 and Karyopherin Alpha-2 in Extraskeletal (Soft Tissues) and Skeletal Osteosarcomas. Front. Vet. Sci. 2024, 11, 1292852. [Google Scholar] [CrossRef] [PubMed]
- Tattersall, L.; Davison, Z.; Gartland, A. Osteosarcoma. In Encyclopedia of Bone Biology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 362–378. ISBN 978-0-12-814082-6. [Google Scholar]
- Brown, H.K.; Tellez-Gabriel, M.; Heymann, D. Cancer Stem Cells in Osteosarcoma. Cancer Lett. 2017, 386, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, C.P.; Kukekov, V.G.; Reith, J.D.; Tchigrinova, O.; Suslov, O.N.; Scott, E.W.; Ghivizzani, S.C.; Ignatova, T.N.; Steindler, D.A. Stem-Like Cells in Bone Sarcomas: Implications for Tumorigenesis. Neoplasia 2005, 7, 967–976. [Google Scholar] [CrossRef]
- Barger, A.; Baker, K.; Driskell, E.; Sander, W.; Roady, P.; Berry, M.; Schnelle, A.; Fan, T.M. The Use of Alkaline Phosphatase and Runx2 to Distinguish Osteosarcoma from Other Common Malignant Primary Bone Tumors in Dogs. Vet. Pathol. 2022, 59, 427–432. [Google Scholar] [CrossRef]
- Yamashita, K.; Kohashi, K.; Yamada, Y.; Nishida, Y.; Urakawa, H.; Oda, Y.; Toyokuni, S. Primary Extraskeletal Osteosarcoma: A Clinicopathological Study of 18 Cases Focusing on MDM2 Amplification Status. Hum. Pathol. 2017, 63, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Bane, B.L.; Evans, H.L.; Ro, J.Y.; Carrasco, C.H.; Grignon, D.J.; Benjamin, R.S.; Ayala, A.G. Extraskeletal Osteosarcoma. A Clinicopathologic Review of 26 Cases. Cancer 1990, 65, 2762–2770. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.-S.; Park, S.H.; Yang, S.-K.; Ye, B.D.; Yang, D.-H.; Kim, K.-J.; Byeon, J.-S.; Yoon, Y.S.; Yu, C.S.; et al. Pathologic Features of Colorectal Carcinomas Associated with Crohn’s Disease in Korean Population. Pathol.-Res. Pract. 2017, 213, 250–255. [Google Scholar] [CrossRef]
- Machado, I.; Navarro, S.; Picci, P.; Llombart-Bosch, A. The Utility of SATB2 Immunohistochemical Expression in Distinguishing between Osteosarcomas and Their Malignant Bone Tumor Mimickers, Such as Ewing Sarcomas and Chondrosarcomas. Pathol.-Res. Pract. 2016, 212, 811–816. [Google Scholar] [CrossRef]
- Seong, B.K.A.; Lau, J.; Adderley, T.; Kee, L.; Chaukos, D.; Pienkowska, M.; Malkin, D.; Thorner, P.; Irwin, M.S. SATB2 Enhances Migration and Invasion in Osteosarcoma by Regulating Genes Involved in Cytoskeletal Organization. Oncogene 2015, 34, 3582–3592. [Google Scholar] [CrossRef]
- Conner, J.R.; Hornick, J.L. SATB 2 Is a Novel Marker of Osteoblastic Differentiation in Bone and Soft Tissue Tumours. Histopathology 2013, 63, 36–49. [Google Scholar] [CrossRef]
- Shank, A.M.M.; Snook, E.; Cavender, K.; McCoy, J.; Sorensen, N.; Siegrist, B.; Tabb, B. Special AT-Rich Sequence-Binding Protein 2 Immunohistochemistry in the Diagnosis of Osteosarcoma in Dogs. J. Comp. Pathol. 2024, 215, 14–29. [Google Scholar] [CrossRef]
- Pan, X.; Wang, H.-L.; Lin, S.-M.; Lin, J.-L.; Ruan, D.-D.; Zhang, J.-H.; Chen, T.; Luo, J.-W.; Fang, Z.-T. A Primary Extraskeletal Osteosarcoma of the Spleen: Rare Case Report. Front. Oncol. 2022, 12, 892943. [Google Scholar] [CrossRef]
- Sugawara, M.; Kato, N.; Tsuchiya, T.; Motoyama, T. RUNX2 Expression in Developing Human Bones and Various Bone Tumors. Pathol. Int. 2011, 61, 565–571. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, X.; Xu, J.; Zhang, Z.; Lin, T.; Zhang, X.; Kang, X. The Role of lncRNA and miRNA on the Effects of Occurrence and Development of Osteosarcoma. Int. Immunopharmacol. 2025, 144, 113726. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, L.; Watson, J.; Feng, Y.; Lewis, B.; Harvey, G.; Post, G.; Megquier, K.; White, M.E.; Lambert, L.; Miller, A.; et al. Shared Hotspot Mutations in Oncogenes Position Dogs as an Unparalleled Comparative Model for Precision Therapeutics. Sci. Rep. 2023, 13, 10935. [Google Scholar] [CrossRef] [PubMed]
- Varvil, M.S.; Dos Santos, A.P. A Review on microRNA Detection and Expression Studies in Dogs. Front. Vet. Sci. 2023, 10, 1261085. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, L.; Edson, M.; Treleaven, H.; Viloria-Petit, A.M.; Mutsaers, A.J.; Moorehead, R.; Foster, R.A.; Ali, A.; Wood, R.D.; Wood, G.A. Plasma microRNA Signatures Predict Prognosis in Canine Osteosarcoma Patients. PLoS ONE 2024, 19, e0311104. [Google Scholar] [CrossRef]
- Yu, Y.; Li, K.; Peng, Y.; Zhang, Z.; Pu, F.; Shao, Z.; Wu, W. Tumor Microenvironment in Osteosarcoma: From Cellular Mechanism to Clinical Therapy. Genes Dis. 2025, 12, 101569. [Google Scholar] [CrossRef] [PubMed]
- Berindan-Neagoe, I.; Calin, G.A. Molecular Pathways: microRNAs, Cancer Cells, and Microenvironment. Clin. Cancer Res. 2014, 20, 6247–6253. [Google Scholar] [CrossRef]
- Solé, C.; Lawrie, C.H. MicroRNAs in Metastasis and the Tumour Microenvironment. Int. J. Mol. Sci. 2021, 22, 4859. [Google Scholar] [CrossRef]
- Conti, I.; Varano, G.; Simioni, C.; Laface, I.; Milani, D.; Rimondi, E.; Neri, L.M. miRNAs as Influencers of Cell–Cell Communication in Tumor Microenvironment. Cells 2020, 9, 220. [Google Scholar] [CrossRef] [PubMed]
- Buhagiar, A.F.; Kleaveland, B. To Kill a microRNA: Emerging Concepts in Target-Directed microRNA Degradation. Nucleic Acids Res. 2024, 52, 1558–1574. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Kim, J.; Yao, F.; Xiao, Z.; Sun, Y.; Ma, L. MicroRNAs and Metastasis: Small RNAs Play Big Roles. Cancer Metastasis Rev. 2018, 37, 5–15. [Google Scholar] [CrossRef]
- Llobat, L.; Gourbault, O. Role of MicroRNAs in Human Osteosarcoma: Future Perspectives. Biomedicines 2021, 9, 463. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The Role of MicroRNAs in Human Cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as Oncogenes and Tumor Suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kent, O.A.; Mendell, J.T. A Small Piece in the Cancer Puzzle: microRNAs as Tumor Suppressors and Oncogenes. Oncogene 2006, 25, 6188–6196. [Google Scholar] [CrossRef]
- Jiang, C.; Wang, P.; Tan, Z.; Zhang, Y. Long Non-Coding RNAs in Bone Formation: Key Regulators and Therapeutic Prospects. Open Life Sci. 2024, 19, 20220908. [Google Scholar] [CrossRef]
- Papaioannou, G.; Mirzamohammadi, F.; Kobayashi, T. MicroRNAs Involved in Bone Formation. Cell. Mol. Life Sci. 2014, 71, 4747–4761. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, C.; Balmayor, E.R.; Van Griensven, M. miRNAs Related to Skeletal Diseases. Stem Cells Dev. 2016, 25, 1261–1281. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Liu, S.; Feng, J.; Kang, Y.; Zhou, Y.; Guo, S. Current Status of MicroRNAs That Target the Wnt Signaling Pathway in Regulation of Osteogenesis and Bone Metabolism: A Review. Med. Sci. Monit. 2021, 27, e929510. [Google Scholar] [CrossRef] [PubMed]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR Signaling Transduction Pathway and Targeted Therapies in Cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective microRNA Target Sites in Mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An Online Database for Prediction of Functional microRNA Targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Loher, P.; Rigoutsos, I. Interactive Exploration of RNA22 microRNA Target Predictions. Bioinformatics 2012, 28, 3322–3323. [Google Scholar] [CrossRef]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: microRNA Target Prediction Easy, Fast and Flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef]
- Braschi, B.; Denny, P.; Gray, K.; Jones, T.; Seal, R.; Tweedie, S.; Yates, B.; Bruford, E. Genenames.Org: The HGNC and VGNC Resources in 2019. Nucleic Acids Res. 2019, 47, D786–D792. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Foiani, G.; Guelfi, G.; Mandara, M.T. MicroRNA Dysregulation in Canine Meningioma: RT-qPCR Analysis of Formalin-Fixed Paraffin-Embedded Samples. J. Neuropathol. Exp. Neurol. 2021, 80, 769–775. [Google Scholar] [CrossRef]
- Ragni, E.; Colombini, A.; De Luca, P.; Libonati, F.; Viganò, M.; Perucca Orfei, C.; Zagra, L.; De Girolamo, L. miR-103a-3p and miR-22-5p Are Reliable Reference Genes in Extracellular Vesicles From Cartilage, Adipose Tissue, and Bone Marrow Cells. Front. Bioeng. Biotechnol. 2021, 9, 632440. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Guelfi, G.; Capaccia, C.; Santoro, M.M.; Diverio, S. Identification of Appropriate Endogenous Controls for Circulating miRNA Quantification in Working Dogs under Physiological Stress Conditions. Animals 2023, 13, 576. [Google Scholar] [CrossRef]
- Guelfi, G.; Iaboni, M.; Sansone, A.; Capaccia, C.; Santoro, M.M.; Diverio, S. Extracellular Circulating miRNAs as Stress-Related Signature to Search and Rescue Dogs. Sci. Rep. 2022, 12, 3213. [Google Scholar] [CrossRef] [PubMed]
- Olesin, E.; Nayar, R.; Saikumar-Lakshmi, P.; Berg, L.J. The Transcription Factor Runx2 Is Required for Long-Term Persistence of Antiviral CD8+ Memory T Cells. Immunohorizons 2018, 2, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper--Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Deeks, J.J.; Altman, D.G. Diagnostic Tests 4: Likelihood Ratios. BMJ 2004, 329, 168–169. [Google Scholar] [CrossRef]
- Grimes, D.A.; Schulz, K.F. Refining Clinical Diagnosis with Likelihood Ratios. Lancet 2005, 365, 1500–1505. [Google Scholar] [CrossRef] [PubMed]
- McGee, S. Simplifying Likelihood Ratios. J. Gen. Intern. Med. 2002, 17, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Rotwein, P. Selective Signaling by Akt1 Controls Osteoblast Differentiation and Osteoblast-Mediated Osteoclast Development. Mol. Cell. Biol. 2012, 32, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Solal, K.A.; Boregowda, R.K.; Lasfar, A. RUNX2 and the PI3K/AKT Axis Reciprocal Activation as a Driving Force for Tumor Progression. Mol. Cancer 2015, 14, 137. [Google Scholar] [CrossRef] [PubMed]
- Gayatri, M.B.; Gajula, N.N.; Chava, S.; Reddy, A.B.M. High Glutamine Suppresses Osteogenesis through mTORC1-Mediated Inhibition of the mTORC2/AKT-473/RUNX2 Axis. Cell Death Discov. 2022, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Chen, T.; Zhao, K.; Yang, D. miR-30a Suppresses Osteosarcoma Proliferation and Metastasis by Downregulating MEF2D Expression. OTT 2018, 11, 2195–2202. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Lu, S.; Wang, S.; Zhang, S. Identification of Critical Genes Associated with Human Osteosarcoma Metastasis Based on Integrated Gene Expression Profiling. Mol. Med. Rep. 2019, 20, 915–930. [Google Scholar] [CrossRef]
- Gong, J.; Wu, Y.; Zhang, X.; Liao, Y.; Sibanda, V.L.; Liu, W.; Guo, A.-Y. Comprehensive Analysis of Human Small RNA Sequencing Data Provides Insights into Expression Profiles and miRNA Editing. RNA Biol. 2014, 11, 1375–1385. [Google Scholar] [CrossRef]
- Moore, M.J.; Scheel, T.K.H.; Luna, J.M.; Park, C.Y.; Fak, J.J.; Nishiuchi, E.; Rice, C.M.; Darnell, R.B. miRNA–Target Chimeras Reveal miRNA 3′-End Pairing as a Major Determinant of Argonaute Target Specificity. Nat. Commun. 2015, 6, 8864. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt Signal Transduction for Cancer Therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef]
- Chen, F.; Wang, S.; Wei, Y.; Wu, J.; Huang, G.; Chen, J.; Shi, J.; Xia, J. Norcantharidin Modulates the miR-30a/Metadherin/AKT Signaling Axis to Suppress Proliferation and Metastasis of Stromal Tumor Cells in Giant Cell Tumor of Bone. Biomed. Pharmacother. 2018, 103, 1092–1100. [Google Scholar] [CrossRef]
- Huang, J.; Li, Y.; Zhu, S.; Wang, L.; Yang, L.; He, C. MiR-30 Family: A Novel Avenue for Treating Bone and Joint Diseases? Int. J. Med. Sci. 2023, 20, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhang, H.; Tang, J. MiR-30a: A Novel Biomarker and Potential Therapeutic Target for Cancer. J. Oncol. 2018, 2018, 5167829. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, S.; Yu, M. Mechanism of Low Expression of miR-30a-5p on Epithelial–Mesenchymal Transition and Metastasis in Ovarian Cancer. DNA Cell Biol. 2019, 38, 341–351. [Google Scholar] [CrossRef]
- Ding, L.; Jiang, H.; Li, Q.; Li, Q.; Zhang, T.-T.; Shang, L.; Xie, B.; Zhu, Y.; Ding, K.; Shi, X.; et al. Ropivacaine as a Novel AKT1 Specific Inhibitor Regulates the Stemness of Breast Cancer. J. Exp. Clin. Cancer Res. 2024, 43, 90. [Google Scholar] [CrossRef]
- Meuten, T.K.; Dean, G.A.; Thamm, D.H. Review: The PI3K-AKT-mTOR Signal Transduction Pathway in Canine Cancer. Vet. Pathol. 2024, 61, 339–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, X.-H.; Yan, Y.-G.; Wang, C.; Wang, W.-J. PI3K/Akt Signaling in Osteosarcoma. Clin. Chim. Acta 2015, 444, 182–192. [Google Scholar] [CrossRef] [PubMed]


| Breed | Age | Sex | Tumor Localization | Histological Diagnosis |
|---|---|---|---|---|
| English Cocker Spaniel | 14 | M | Intestine | EOS-1 |
| Mixed breed | 8 | F | Mammary gland | EOS-2 |
| Mixed breed | 14 | F | Mammary gland | EOS-3 |
| Mixed breed | 10 | F | Mammary gland | EOS-4 |
| NA | 9 | M | Urinary bladder | EOS-5 |
| Mixed breed | 12 | F | Spleen | EOS-6 |
| Mixed breed | 13 | F | Mammary gland | EOS-7 |
| Hunting dog | 13 | F | Uterus | EOS-8 |
| Mixed breed | 8 | F | Mammary gland | EOS-9 |
| Mixed breed | 12 | F | Intestine | EOS-10 |
| Cirneco dell’Etna | 15 | F | Haired skin (thorax) | EOS-11 |
| English Cocker Spaniel | 12 | M | Haired skin | EOS-12 |
| Mixed breed | 10 | F | Mammary gland | EOS-13 |
| Maremma Sheepdog | 9 | F | Subcutis (shoulder area) | EOS-14 |
| Labrador Retriever | 10 | M | Thyroid gland | EOS-15 |
| Mixed breed | 11 | F | Mammary gland | EOS-16 |
| Epagneul Breton | 10 | F | Subcutis (dorsal area) | EOS-17 |
| Mixed breed | 13 | F | Mammary gland | EOS-18 |
| English Pointer | 11 | F | Mammary gland | EOS-19 |
| Rottweiler | 11 | F | Femur | OS-1 |
| Labrador Retriever | 12 | F | Humerus | OS-2 |
| Mixed breed | 10 | F | Radium | OS-3 |
| Mixed breed | 7 | M | Vertebrae | OS-4 |
| Mixed breed | 9 | M | Maxilla | OS-5 |
| American Staffordshire Terrier | 9 | M | Tibia | OS-6 |
| Pinscher | 4 | F | Femur | OS-7 |
| Pitbull | 6 | F | Scapula | OS-8 |
| American Stafford | 12 | F | Front limb | OS-9 |
| Mixed breed | 9 | M | Carpus | OS-10 |
| Belgian Sheperd | 12 | M | Mandibula | OS-11 |
| Greyhound | 6 | F | Humerus | OS-12 |
| German Shepherd | 10 | F | Forelimb | OS-13 |
| Saint Bernard | 9 | F | Humerus | OS-14 |
| Mixed breed | 9 | M | Mandibula | CTRL-1 |
| Rottweiller | 11 | F | Haired skin | CTRL-2 |
| Briquet Griffon | 6 | F | Spleen | CTRL-3 |
| Briquet Griffon | 6 | F | Urinary Bladder | CTRL-4 |
| Mixed breed | 1 | F | Uterus | CTRL-5 |
| Mixed breed | 8 | F | Spleen | CTRL-6 |
| Mixed breed | 8 | F | Humerus | CTRL-7 |
| Mixed breed | 15 | M | Rib | CTRL-8 |
| Mixed breed | 8 | F | Subcutis (mammary area) | CTRL-9 |
| Mixed breed | 8 | F | Intestine | CTRL-10 |
| miRNA | Mature Sequence (5′–3′) | Seed | Non-Seed (nt 9–22) |
|---|---|---|---|
| (nt 2–8) | |||
| cfa-miR-30a MIMAT0006604 | UGUAAACAUCCUCGACUGGAAG | GUAAACA | AUCCUCGACUGGAAG |
| hsa-miR-30b-5p MIMAT0000420 | UGUAAACAUCCUACACUCAGCU | GUAAACA | AUCCUACACUCAGCU |
| gga-miR-30c-5p MIMAT0001137 | UGUAAACAUCCUACACUCUCAGCU | GUAAACA | AUCCUACACUCUCAGCU |
| cfa-miR-30d MIMAT0006616 | UGUAAACAUCCCCGACUGGAAGCU | GUAAACA | AUCCCCGACUGGAAGCU |
| hsa-miR-30e-5p MIMAT0000692 | UGUAAACAUCCUUGACUGGAAG | GUAAACA | AUCCUUGACUGGAAG |
| MiRNA | Specie Name | miRBase Accession | Qiagen ID |
|---|---|---|---|
| cfa- miR-30a | Canis familiaris | MIMAT0006604 | YP02101182 |
| hsa- miR-30b-5p | Homo sapiens | MIMAT0000420 | YP00204765 |
| gga- miR-30c-5p | Gallus gallus | MIMAT0001137 | YP00205948 |
| cfa- miR-30d | Canis familiaris | MIMAT0006616 | YP02118689 |
| hsa- miR-30e-3p | Homo sapiens | MIMAT0000693 | YP00204410 |
| hsa- miR-26a-5p [46] | Homo sapiens | MIMAT0000082 | YP00206023 |
| hsa- miR-103a-3p [47] | Homo sapiens | MIMAT0000101 | YP00204063 |
| hsa- miR-186-5p [46] | Homo sapiens | MIMAT0000456 | YP00206053 |
| UniSp2 | All species | None | YP00203950 |
| UniSp4 | All species | None | YP00203953 |
| UniSp6 | All species | None | YP00203954 |
| Histological Diagnosis | % Labeled Cells | Score | Intensity |
|---|---|---|---|
| EOS-1 | 30 b | 2 b | 3 b |
| EOS-2 | 70 b | 3 b | 2 b |
| EOS-3 | 80 b | 4 b | 3 b |
| EOS-4 | 60 b | 3 b | 2 b |
| EOS-5 | 90 b | 4 b | 3 b |
| EOS-6 | 40 b | 2 b | 3 b |
| EOS-7 | 40 b | 2 b | 3 b |
| EOS-8 | 80 b | 4 b | 3 b |
| EOS-9 | 50 b | 3 b | 3 b |
| EOS-10 | 75 b | 3 b | 2 b |
| EOS-11 | 90 b | 4 b | 3 b |
| EOS-12 | 30 b | 2 b | 3 b |
| EOS-13 | 70 b | 3 b | 3 b |
| EOS-14 | 25 b | 2 b | 3 b |
| EOS-15 | 50 b | 3 b | 3 b |
| EOS-16 | 35 b | 2 b | 3 b |
| EOS-17 | 40 b | 2 b | 3 b |
| EOS-18 | 15 b | 1 b | 3 b |
| EOS-19 | 50 b | 3 b | 3 b |
| OS-1 | 30 b | 4 b | 3 b |
| OS-2 | 60 b | 3 b | 2 b |
| OS-3 | 60 b | 3 b | 2 b |
| OS-4 | 50 b | 3 b | 3 b |
| OS-5 | 80 b | 4 b | 3 b |
| OS-6 | 30 b | 2 b | 2 b |
| OS-7 | 70 b | 3 b | 3 b |
| OS-8 | 30 b | 2 b | 2 b |
| OS-9 | 40 b | 2 b | 2 b |
| OS-10 | 30 b | 3 b | 3 b |
| OS-11 | 60 b | 2 b | 2 b |
| OS-12 | 70 b | 3 b | 3 b |
| OS-13 | 40 b | 4 b | 3 b |
| OS-14 | 30 b | 4 b | 3 b |
| CTRL-1 | 0 a | 0 a | 0 a |
| CTRL-2 | 0 a | 0 a | 0 a |
| CTRL-3 | 0 a | 0 a | 0 a |
| CTRL-4 | 0 a | 0 a | 0 a |
| CTRL-5 | 0 a | 0 a | 0 a |
| CTRL-6 | 0 a | 0 a | 0 a |
| CTRL-7 | 0 a | 0 a | 0 a |
| CTRL-8 | 0 a | 0 a | 0 a |
| CTRL-9 | 0 a | 0 a | 0 a |
| CTRL-10 | 0 a | 0 a | 0 a |
| Analysis Method | |||
|---|---|---|---|
| Delta CT | miR-103a-3p | miR-26a-5p | miR-186-5p |
| BestKeepeer | miR-103a-3p | miR-26a-5p | miR-186-5p |
| NormFinder | miR-103a-3p | miR-26a-5p | miR-186-5p |
| GeNorm | miR-26a-5p/miR-103a-3p | miR-186-5p | |
| RefFinder Ranking Order | miR-103a-3p | miR-26a-5p | miR-186-5p |
| Better | Good | Average |
| miRNA | AUC (IC 95%) | Cutoff | Sensitivity (%) | Specificity (%) | LR+ | Diagnostic Value | |
|---|---|---|---|---|---|---|---|
| CTRL vs. OS | miR-30a | 0.714 | <0.280 | 71.43 | 85.71 | 5 | Good |
| miR-30b | 0.780 | <1.184 | 60.00 | 80.00 | 3 | Acceptable | |
| miR-30c | 0.614 | <0.587 | 50.00 | 85.71 | 3.5 | Acceptable | |
| miR-30d | 0.518 | <0.419 | 40.00 | 83.33 | 2.4 | Acceptable | |
| miR-30e | 0.900 | <0.094 | 70.00 | 90.91 | 7.7 | Good | |
| CTRL vs. EOS | miR-30a | 0.691 | <0.280 | 66.67 | 85.71 | 4.7 | Acceptable |
| miR-30b | 0.631 | <1.184 | 46.15 | 80.00 | 2.3 | Acceptable | |
| miR-30c | 0.774 | >1.225 | 50.00 | 85.71 | 3.5 | Acceptable | |
| miR-30d | 0.667 | >0.614 | 58.33 | 83.33 | 3.5 | Acceptable | |
| miR-30e | 0.762 | <0.093 | 61.54 | 90.91 | 6.8 | Good |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guelfi, G.; Munteanu, P.; Capaccia, C.; Porcellato, I.; Manuali, E.; Maranesi, M.; Leonardi, L. A miR-30 Guided Molecular Profiling of Canine Osteosarcoma and Extraskeletal Osteosarcoma Reveals Non-Seed Regulatory Divergence. Cells 2025, 14, 1279. https://doi.org/10.3390/cells14161279
Guelfi G, Munteanu P, Capaccia C, Porcellato I, Manuali E, Maranesi M, Leonardi L. A miR-30 Guided Molecular Profiling of Canine Osteosarcoma and Extraskeletal Osteosarcoma Reveals Non-Seed Regulatory Divergence. Cells. 2025; 14(16):1279. https://doi.org/10.3390/cells14161279
Chicago/Turabian StyleGuelfi, Gabriella, Petronela Munteanu, Camilla Capaccia, Ilaria Porcellato, Elisabetta Manuali, Margherita Maranesi, and Leonardo Leonardi. 2025. "A miR-30 Guided Molecular Profiling of Canine Osteosarcoma and Extraskeletal Osteosarcoma Reveals Non-Seed Regulatory Divergence" Cells 14, no. 16: 1279. https://doi.org/10.3390/cells14161279
APA StyleGuelfi, G., Munteanu, P., Capaccia, C., Porcellato, I., Manuali, E., Maranesi, M., & Leonardi, L. (2025). A miR-30 Guided Molecular Profiling of Canine Osteosarcoma and Extraskeletal Osteosarcoma Reveals Non-Seed Regulatory Divergence. Cells, 14(16), 1279. https://doi.org/10.3390/cells14161279














