DNA and Histone Modifications Identify a Putative Controlling Element (CE) on the X Chromosome of Sciara coprophila
Abstract
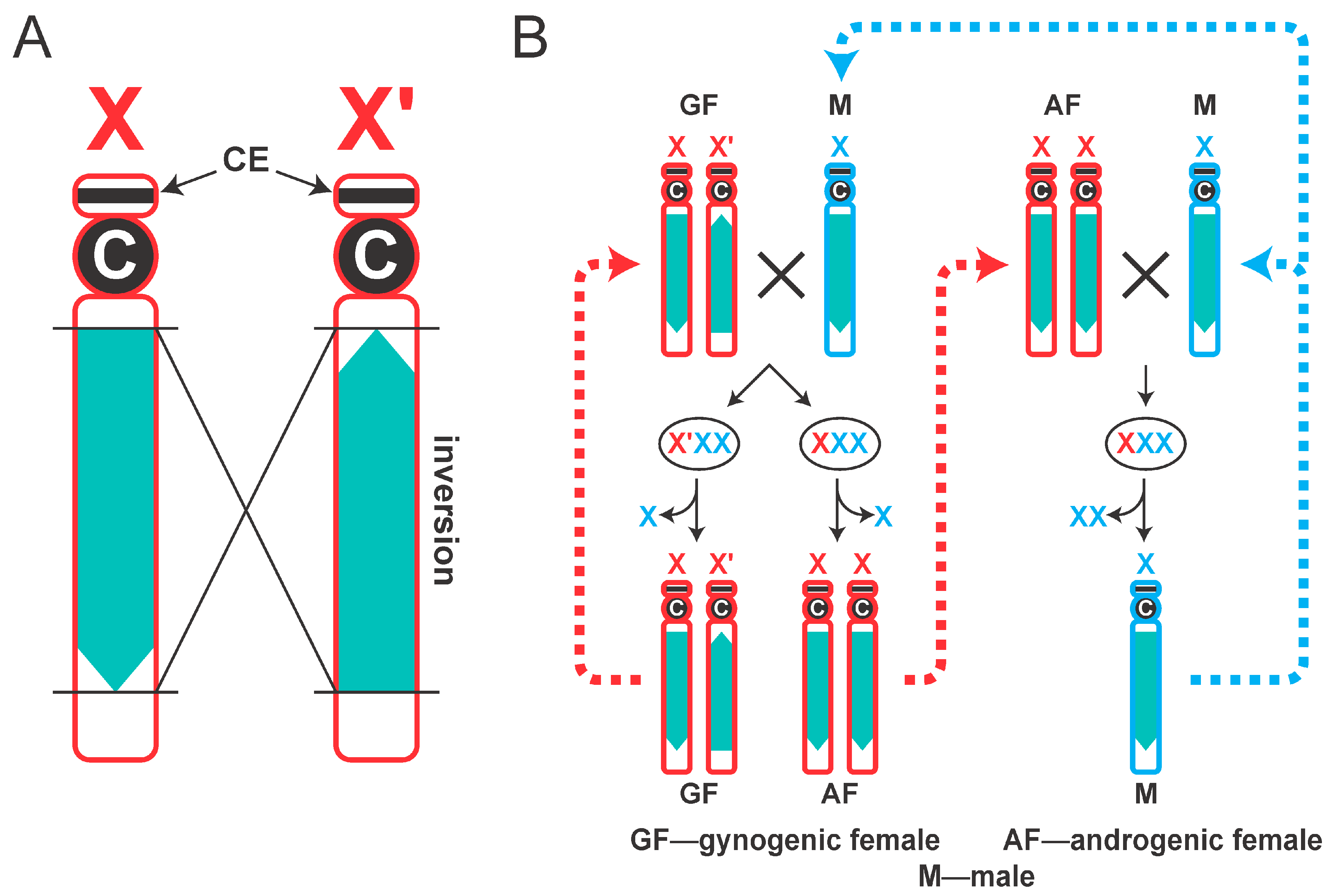
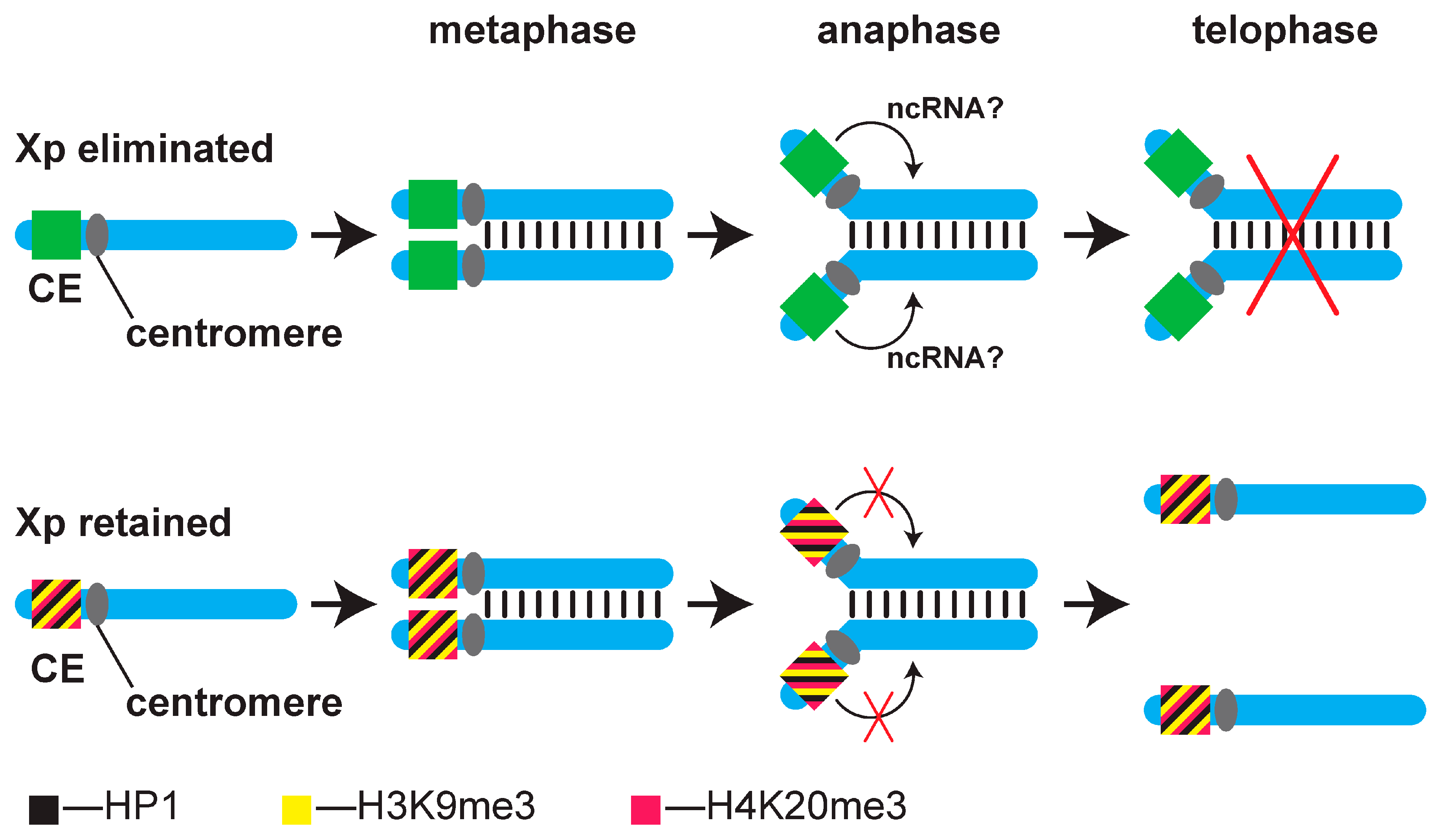
1. Introduction
2. Materials and Methods
2.1. Sciara coprophila Strains and Housbandry
2.2. Immunostaining of 5′ Methyl Cytosines in Polytene Chromosomes
2.3. Sciara coprophila Genome Sequencing and Assembly
2.4. Methylated DNA Immunoprecipitation (MeDIP)
2.5. ChIP-Seq
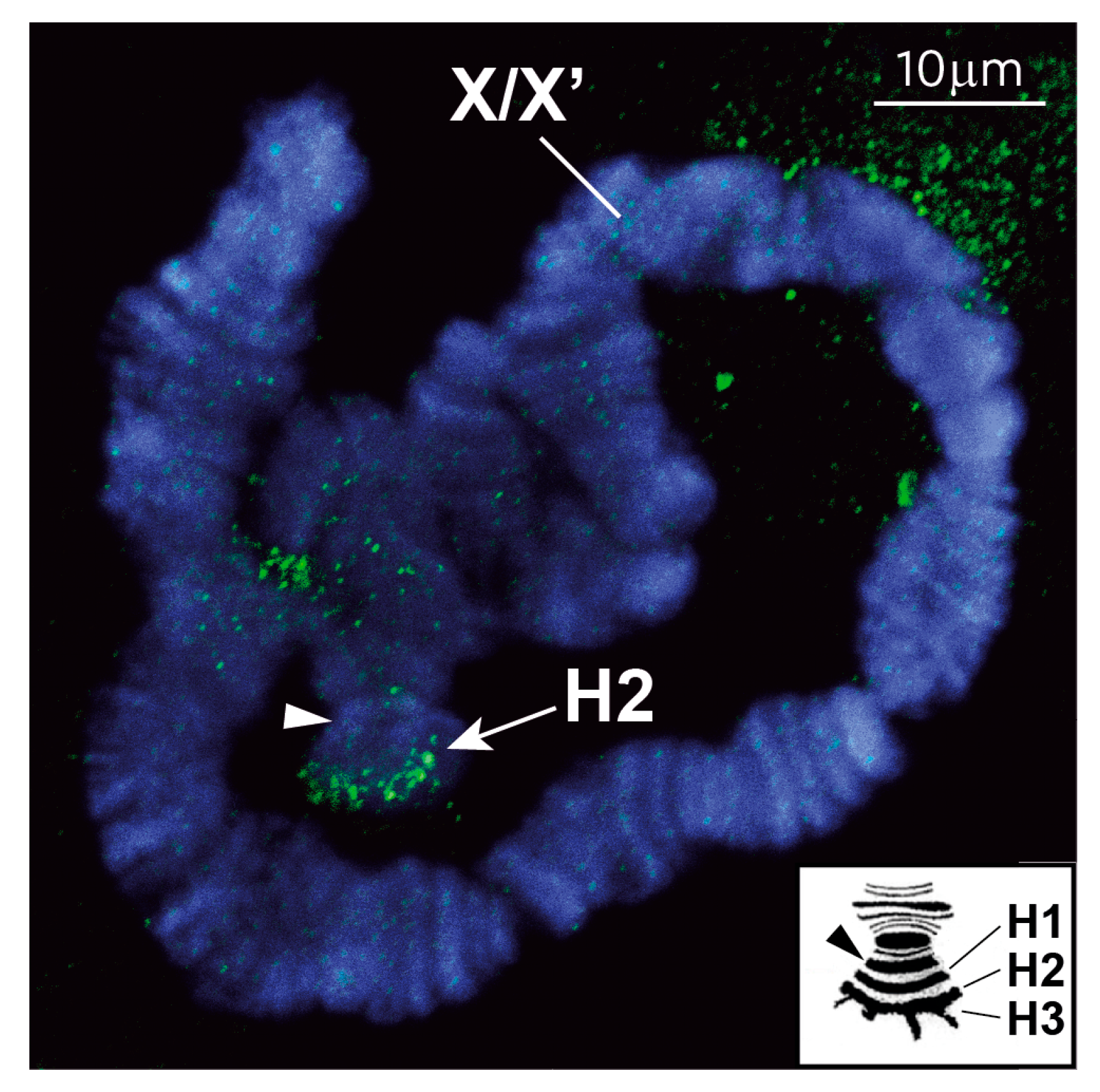
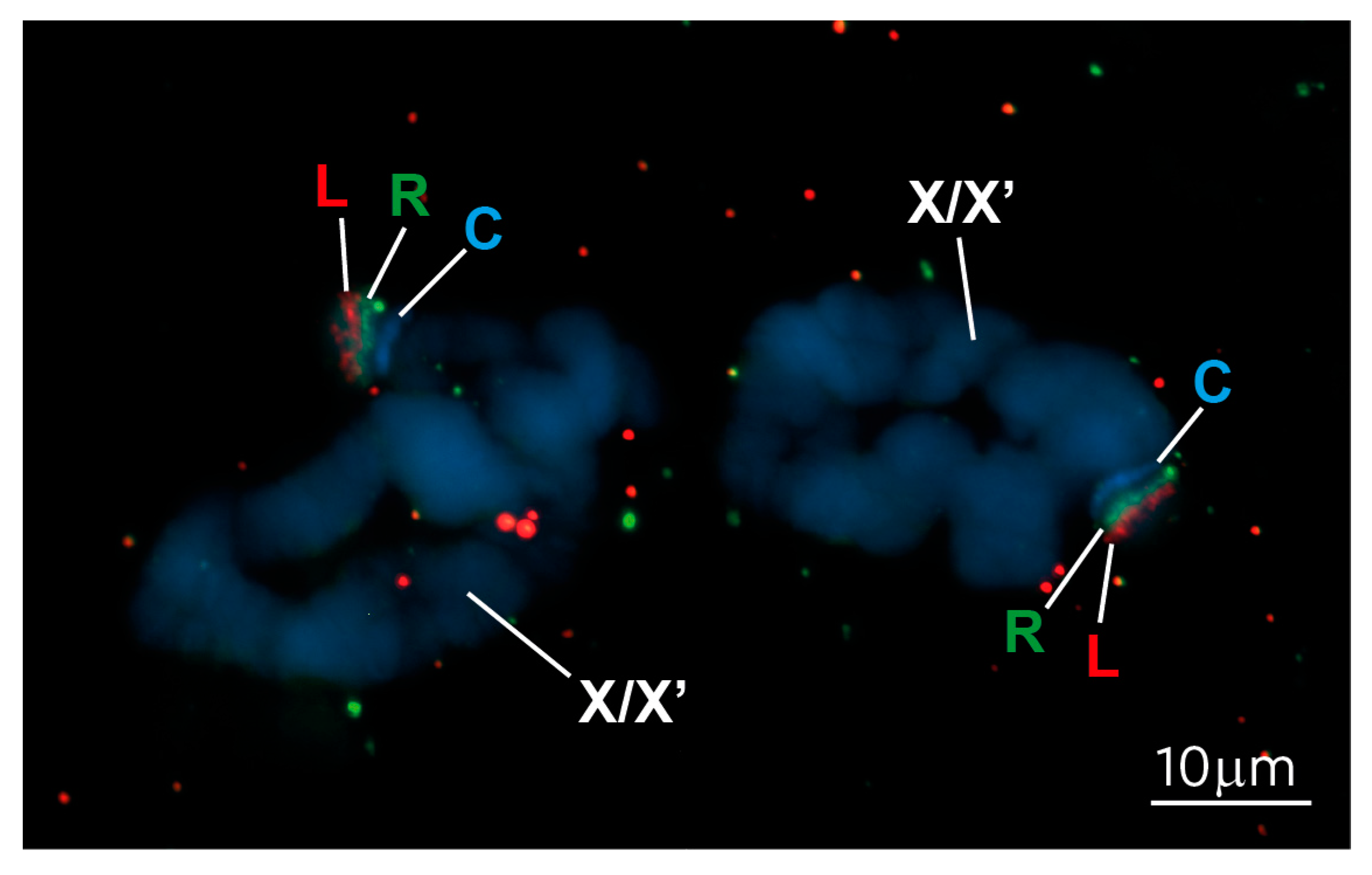
2.6. FISH on Polytene Chromosomes
- probe_L-for: 5′-GTCAGCGGATTAAAGCTCTCTAT-3′
- probe_L-rev: 5′-TTGGCGAACCACTTCTACTG-3′
- probe_R-for: 5′-CCATGCGTCGGTGAGTTATTA-3′
- probe_R-rev: 5′-GTGACCTGAAAGTACTGAGTGG-3′
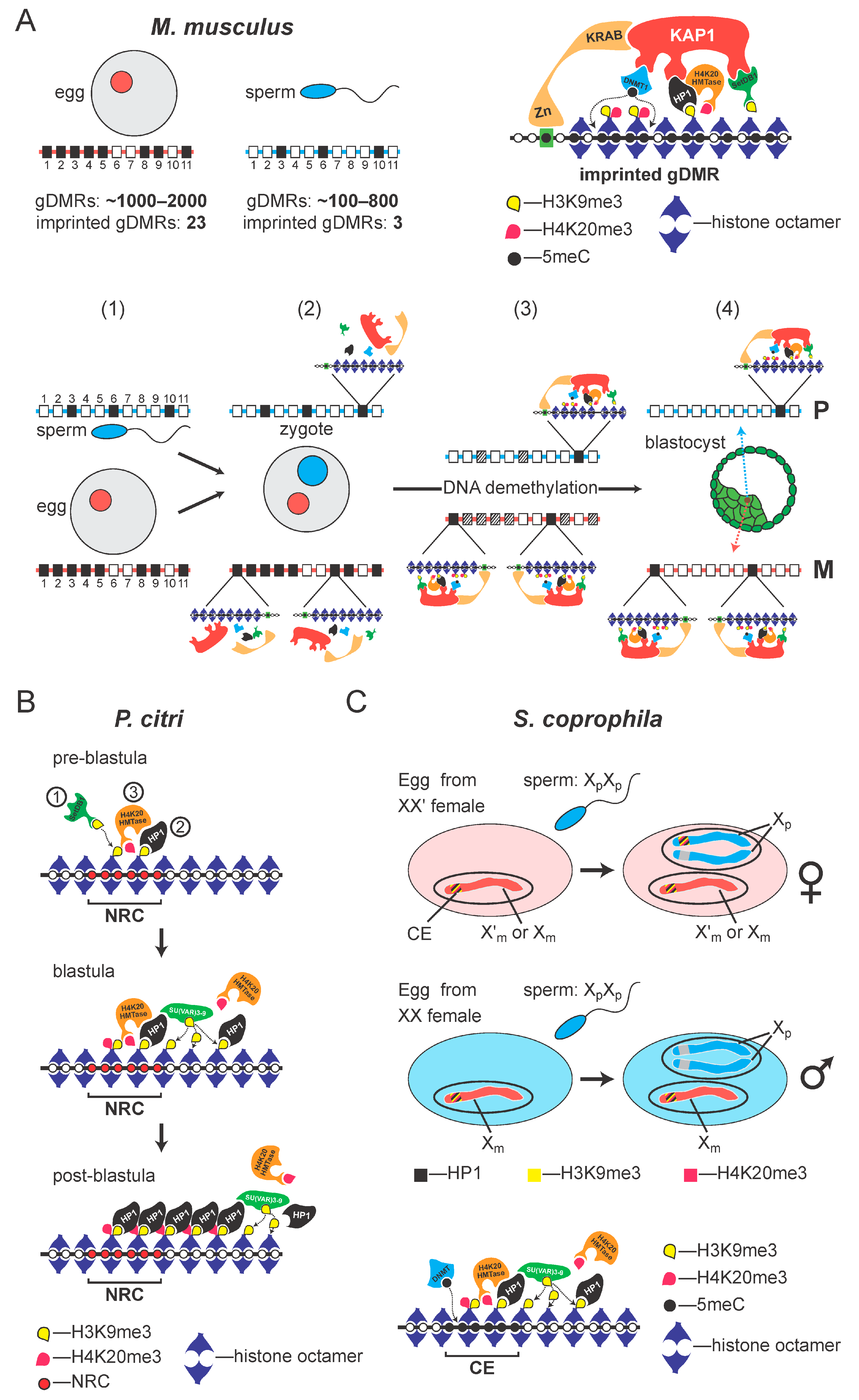
3. Results
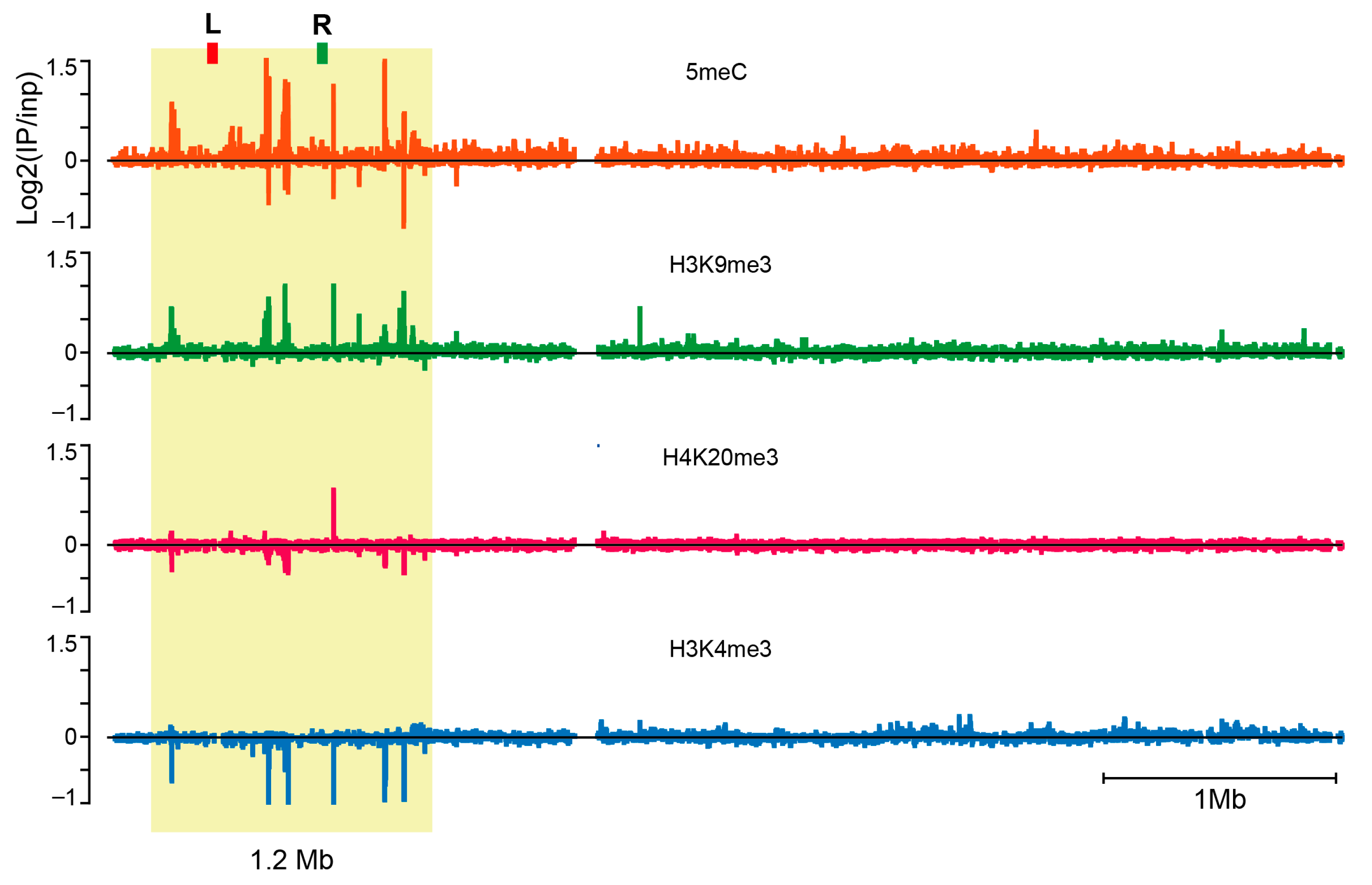
3.1. The Repressive Epigenetic DNA Modification 5′ Methyl Cytosine (5meC) Is Enriched at the CE
3.2. New S. coprophila Genome Assembly and Analysis of the Epigenetic Marks at the CE
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Metz, C.W. Chromosomes and Sex in Sciara. Science 1925, 61, 212–214. [Google Scholar] [CrossRef]
- Metz, C.W.C. Chromosome behavior, inheritance and sex determination in Sciara. Am. Nat. 1938, 72, 485–520. [Google Scholar] [CrossRef]
- Ferguson-Smith, A.C. Genomic imprinting: The emergence of an epigenetic paradigm. Nat. Rev. Genet. 2011, 12, 565–575. [Google Scholar] [CrossRef]
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a018382. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.B.; Shloma, V.V.; Belyakin, S.N. Maternal regulation of chromosomal imprinting in animals. Chromosoma 2019, 128, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Crouse, H.V. The Controlling Element in Sex Chromosome Behavior in Sciara. Genetics 1960, 45, 1429–1443. [Google Scholar] [CrossRef]
- Gerbi, S.A. Unusual chromosome movements in sciarid flies. Results Probl. Cell Differ. 1986, 13, 71–104. [Google Scholar] [PubMed]
- Singh, P.B.; Belyakin, S.N. L Chromosome Behaviour and Chromosomal Imprinting in Sciara coprophila. Genes 2018, 9, 440. [Google Scholar] [CrossRef] [PubMed]
- Crouse, H.V. X heterochromatin subdivision and cytogenetic analysis in Sciara coprophila (Diptera, Sciaridae). Chromosoma 1979, 74, 219–239. [Google Scholar] [CrossRef]
- Crouse, H.V.; Gerbi, S.A.; Liang, C.M.; Magnus, L.; Mercer, I.M. Localization of ribosomal DNA within the proximal X heterochromatin of Sciara coprophila (Diptera, Sciaridae). Chromosoma 1977, 64, 305–318. [Google Scholar] [CrossRef] [PubMed]
- de Saint Phalle, B.; Sullivan, W. Incomplete sister chromatid separation is the mechanism of programmed chromosome elimination during early Sciara coprophila embryogenesis. Development 1996, 122, 3775–3784. [Google Scholar] [CrossRef]
- Berry, R.O. Chromosome behavior in the germ cells and development of the gonads in Sciara ocellaris. J. Morphol. 1941, 68, 547–583. [Google Scholar] [CrossRef]
- Rieffel, S.M.; Crouse, H.V. The elimination and differentiation of chromosomes in the germ line of sciara. Chromosoma 1966, 19, 231–276. [Google Scholar] [CrossRef] [PubMed]
- Crouse, H.V.; Brown, A.; Mumford, B.C. L-chromosome inheritance and the problem of chromosome “imprinting” in Sciara (Sciaridae, Diptera). Chromosoma 1971, 34, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Baird, R.B.; Urban, J.M.; Mongue, A.J.; Jaron, K.S.; Hodson, C.N.; Grewoldt, M.; Martin, S.H.; Ross, L. Recent Evolution of a Maternally Acting Sex-Determining Supergene in a Fly with Single-Sex Broods. Mol. Biol. Evol. 2023, 40, msad148. [Google Scholar] [CrossRef] [PubMed]
- Crouse, H.V. The nature of the influence of x-translocations on sex of progeny in Sciara coprophila. Chromosoma 1960, 11, 146–166. [Google Scholar] [CrossRef] [PubMed]
- Crouse, H.V. An inducible change in state on the chromosomes of Sciara: Its effects on the genetic components of the X. Chromosoma 1966, 18, 230–253. [Google Scholar] [CrossRef]
- Chandra, H.S.; Brown, S.W. Chromosome imprinting and the mammalian X chromosome. Nature 1975, 253, 165–168. [Google Scholar] [CrossRef]
- Singh, P.B. Heterochromatin and the molecular mechanisms of ‘parent-of-origin’ effects in animals. J. Biosci. 2016, 41, 759–786. [Google Scholar] [CrossRef]
- Greciano, P.G.; Ruiz, M.F.; Kremer, L.; Goday, C. Two new chromodomain-containing proteins that associate with heterochromatin in Sciara coprophila chromosomes. Chromosoma 2009, 118, 361–376. [Google Scholar] [CrossRef] [PubMed]
- Pindyurin, A.V.; Boldyreva, L.V.; Shloma, V.V.; Kolesnikova, T.D.; Pokholkova, G.V.; Andreyeva, E.N.; Kozhevnikova, E.N.; Ivanoschuk, I.G.; Zarutskaya, E.A.; Demakov, S.A.; et al. Interaction between the Drosophila heterochromatin proteins SUUR and HP1. J. Cell Sci. 2008, 121, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Zhang, B.; Li, D.; Wang, T. Comprehensive Whole DNA Methylome Analysis by Integrating MeDIP-seq and MRE-seq. Methods Mol. Biol. 2018, 1708, 209–246. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.P.; Stromberg, M.P.; Ward, A.; Stewart, C.; Garrison, E.P.; Marth, G.T. MOSAIK: A hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS ONE 2014, 9, e90581. [Google Scholar] [CrossRef] [PubMed]
- Koryakov, D.E.; Domanitskaya, E.V.; Belyakin, S.N.; Zhimulev, I.F. Abnormal tissue-dependent polytenization of a block of chromosome 3 pericentric heterochromatin in Drosophila melanogaster. J. Cell Sci. 2003, 116, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Crouse, H.V. Translocations in Sciara: Their bearing on chromosome behavior and sex determination. Res. Bull. 1943, 379, 1–75. [Google Scholar]
- Eastman, E.M.; Goodman, R.M.; Erlanger, B.F.; Miller, O.J. 5-methylcytosine in the DNA of the polytene chromosomes of the diptera Sciara coprophila, Drosophila melanogaster and D. persimilis. Chromosoma 1980, 79, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Eastman, E.M.; Goodman, R.M.; Erlanger, B.F.; Miller, O.J. The organization of DNA in the mitotic and polytene chromosomes of Sciara corprophila. Chromosoma 1980, 79, 293–314. [Google Scholar] [CrossRef]
- Escriba, M.C.; Greciano, P.G.; Mendez-Lago, M.; de Pablos, B.; Trifonov, V.A.; Ferguson-Smith, M.A.; Goday, C.; Villasante, A. Molecular and cytological characterization of repetitive DNA sequences from the centromeric heterochromatin of Sciara coprophila. Chromosoma 2011, 120, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Regha, K.; Sloane, M.A.; Huang, R.; Pauler, F.M.; Warczok, K.E.; Melikant, B.; Radolf, M.; Martens, J.H.; Schotta, G.; Jenuwein, T.; et al. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol. Cell 2007, 27, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Kharchenko, P.V.; Alekseyenko, A.A.; Schwartz, Y.B.; Minoda, A.; Riddle, N.C.; Ernst, J.; Sabo, P.J.; Larschan, E.; Gorchakov, A.A.; Gu, T.; et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 2011, 471, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.W.; Nur, U. Heterochromatic Chromosomes in the Coccids. Science 1964, 145, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Prantera, G.; Bongiorni, S. Mealybug chromosome cycle as a paradigm of epigenetics. Genet. Res. Int. 2012, 2012, 867390. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Kelsey, G. The specification of imprints in mammals. Heredity 2014, 113, 176–183. [Google Scholar] [CrossRef]
- Smallwood, S.A.; Tomizawa, S.; Krueger, F.; Ruf, N.; Carli, N.; Segonds-Pichon, A.; Sato, S.; Hata, K.; Andrews, S.R.; Kelsey, G. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011, 43, 811–814. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Sakurai, T.; Imai, M.; Takahashi, N.; Fukuda, A.; Yayoi, O.; Sato, S.; Nakabayashi, K.; Hata, K.; Sotomaru, Y.; et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012, 8, e1002440. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, J.; Duan, J.; Gao, X.; Zhu, W.; Lu, X.; Yang, L.; Zhang, J.; Li, G.; Ci, W.; et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 2014, 157, 979–991. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ito, M.; Zhou, F.; Youngson, N.; Zuo, X.; Leder, P.; Ferguson-Smith, A.C. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 2008, 15, 547–557. [Google Scholar] [CrossRef]
- Quenneville, S.; Verde, G.; Corsinotti, A.; Kapopoulou, A.; Jakobsson, J.; Offner, S.; Baglivo, I.; Pedone, P.V.; Grimaldi, G.; Riccio, A.; et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell 2011, 44, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Coluccio, A.; Thorball, C.W.; Planet, E.; Shi, H.; Offner, S.; Turelli, P.; Imbeault, M.; Ferguson-Smith, A.C.; Trono, D. ZNF445 is a primary regulator of genomic imprinting. Genes Dev. 2019, 33, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Anvar, Z.; Cammisa, M.; Riso, V.; Baglivo, I.; Kukreja, H.; Sparago, A.; Girardot, M.; Lad, S.; De Feis, I.; Cerrato, F.; et al. ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic Acids Res. 2016, 44, 1118–1132. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Toh, H.; Sasaki, H.; Zhang, X.; Cheng, X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012, 26, 2374–2379. [Google Scholar] [CrossRef]
- Zuo, X.; Sheng, J.; Lau, H.T.; McDonald, C.M.; Andrade, M.; Cullen, D.E.; Bell, F.T.; Iacovino, M.; Kyba, M.; Xu, G.; et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J. Biol. Chem. 2012, 287, 2107–2118. [Google Scholar] [CrossRef]
- Schotta, G.; Lachner, M.; Sarma, K.; Ebert, A.; Sengupta, R.; Reuter, G.; Reinberg, D.; Jenuwein, T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004, 18, 1251–1262. [Google Scholar] [CrossRef]
- Strogantsev, R.; Krueger, F.; Yamazawa, K.; Shi, H.; Gould, P.; Goldman-Roberts, M.; McEwen, K.; Sun, B.; Pedersen, R.; Ferguson-Smith, A.C. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 2015, 16, 112. [Google Scholar] [CrossRef]
- Bongiorni, S.; Pasqualini, B.; Taranta, M.; Singh, P.B.; Prantera, G. Epigenetic regulation of facultative heterochromatinisation in Planococcus citri via the Me(3)K9H3-HP1-Me(3)K20H4 pathway. J. Cell Sci. 2007, 120, 1072–1080. [Google Scholar] [CrossRef]
- Cowell, I.G.; Aucott, R.; Mahadevaiah, S.K.; Burgoyne, P.S.; Huskisson, N.; Bongiorni, S.; Prantera, G.; Fanti, L.; Pimpinelli, S.; Wu, R.; et al. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 2002, 111, 22–36. [Google Scholar] [CrossRef]
- Epstein, H.; James, T.C.; Singh, P.B. Cloning and expression of Drosophila HP1 homologs from a mealybug, Planococcus citri. J. Cell Sci. 1992, 101, 463–474. [Google Scholar] [CrossRef]
- Kourmouli, N.; Jeppesen, P.; Mahadevhaiah, S.; Burgoyne, P.; Wu, R.; Gilbert, D.M.; Bongiorni, S.; Prantera, G.; Fanti, L.; Pimpinelli, S.; et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 2004, 117, 2491–2501. [Google Scholar] [CrossRef]
- Nur, U. Meiotic parthenogenesis and heterochromatization in a soft scale, pulvinaria hydrangeae (Coccoidea: Homoptera). Chromosoma 1963, 14, 123–139. [Google Scholar] [CrossRef]
- Nur, U. Parthenogenesis in Coccids (Homoptera). Am. Zool. 1971, 11, 301–308. [Google Scholar] [CrossRef]
- Nur, U. Diploid arrhenotoky and automictic thelytoky in soft scale insects (Lecaniidae: Coccoidea: Homoptera). Chromosoma 1972, 39, 381–401. [Google Scholar] [CrossRef]
- Osipov, Y.A.; Posukh, O.V.; Kalashnikova, D.A.; Antoshina, P.A.; Laktionov, P.P.; Skrypnik, P.A.; Belyakin, S.N.; Singh, P.B. H3K9 and H4K20 methyltransferases are directly involved in the heterochromatinization of the paternal chromosomes in male Planococcus citri embryos. Chromosoma 2023, 132, 317–328. [Google Scholar] [CrossRef]
- Bongiorni, S.; Cintio, O.; Prantera, G. The relationship between DNA methylation and chromosome imprinting in the coccid Planococcus citri. Genetics 1999, 151, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.B.; Belyakin, S.N.; Laktionov, P.P. Biology and Physics of Heterochromatin-Like Domains/Complexes. Cells 2020, 9, 1881. [Google Scholar] [CrossRef] [PubMed]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Field, M.A.; Rosen, B.D.; Dudchenko, O.; Chan, E.K.F.; Minoche, A.E.; Edwards, R.J.; Barton, K.; Lyons, R.J.; Tuipulotu, D.E.; Hayes, V.M.; et al. Canfam_GSD: De novo chromosome-length genome assembly of the German Shepherd Dog (Canis lupus familiaris) using a combination of long reads, optical mapping, and Hi-C. GigaScience 2020, 9, giaa027. [Google Scholar] [CrossRef] [PubMed]
- Elbers, J.P.; Rogers, M.F.; Perelman, P.L.; Proskuryakova, A.A.; Serdyukova, N.A.; Johnson, W.E.; Horin, P.; Corander, J.; Murphy, D.; Burger, P.A. Improving Illumina assemblies with Hi-C and long reads: An example with the North African dromedary. Mol. Ecol. Resour. 2019, 19, 1015–1026. [Google Scholar] [CrossRef]
- Schopflin, R.; Melo, U.S.; Moeinzadeh, H.; Heller, D.; Laupert, V.; Hertzberg, J.; Holtgrewe, M.; Alavi, N.; Klever, M.K.; Jungnitsch, J.; et al. Integration of Hi-C with short and long-read genome sequencing reveals the structure of germline rearranged genomes. Nat. Commun. 2022, 13, 6470. [Google Scholar] [CrossRef]
- Harewood, L.; Kishore, K.; Eldridge, M.D.; Wingett, S.; Pearson, D.; Schoenfelder, S.; Collins, V.P.; Fraser, P. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol. 2017, 18, 125. [Google Scholar] [CrossRef]
- Posukh, O.V.; Maksimov, D.A.; Laktionov, P.P.; Koryakov, D.E.; Belyakin, S.N. Functional dissection of Drosophila melanogaster SUUR protein influence on H3K27me3 profile. Epigenet. Chromatin 2017, 10, 56. [Google Scholar] [CrossRef] [PubMed]
- Belaghzal, H.; Borrman, T.; Stephens, A.D.; Lafontaine, D.L.; Venev, S.V.; Weng, Z.; Marko, J.F.; Dekker, J. Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Nat. Genet. 2021, 53, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Durand, N.C.; Shamim, M.S.; Machol, I.; Rao, S.S.; Huntley, M.H.; Lander, E.S.; Aiden, E.L. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016, 3, 95–98. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Posukh, O.V.; Shloma, V.V.; Skrypnik, P.A.; Maksimov, D.A.; Antoshina, P.A.; Kalashnikova, D.A.; Nurislamov, A.; Lukyanchikova, V.A.; Torgunakov, N.; Battulin, N.R.; et al. DNA and Histone Modifications Identify a Putative Controlling Element (CE) on the X Chromosome of Sciara coprophila. Cells 2025, 14, 1243. https://doi.org/10.3390/cells14161243
Posukh OV, Shloma VV, Skrypnik PA, Maksimov DA, Antoshina PA, Kalashnikova DA, Nurislamov A, Lukyanchikova VA, Torgunakov N, Battulin NR, et al. DNA and Histone Modifications Identify a Putative Controlling Element (CE) on the X Chromosome of Sciara coprophila. Cells. 2025; 14(16):1243. https://doi.org/10.3390/cells14161243
Chicago/Turabian StylePosukh, Olga V., Victor V. Shloma, Polina A. Skrypnik, Daniil A. Maksimov, Polina A. Antoshina, Daria A. Kalashnikova, Artem Nurislamov, Varvara A. Lukyanchikova, Nikita Torgunakov, Nariman R. Battulin, and et al. 2025. "DNA and Histone Modifications Identify a Putative Controlling Element (CE) on the X Chromosome of Sciara coprophila" Cells 14, no. 16: 1243. https://doi.org/10.3390/cells14161243
APA StylePosukh, O. V., Shloma, V. V., Skrypnik, P. A., Maksimov, D. A., Antoshina, P. A., Kalashnikova, D. A., Nurislamov, A., Lukyanchikova, V. A., Torgunakov, N., Battulin, N. R., Fishman, V. S., Vyatkin, Y. V., Smelova, A. A., Romanov, S. E., Laktionov, P. P., Valishayev, D., Belyakin, S. N., & Singh, P. B. (2025). DNA and Histone Modifications Identify a Putative Controlling Element (CE) on the X Chromosome of Sciara coprophila. Cells, 14(16), 1243. https://doi.org/10.3390/cells14161243







