Dissecting the Immunological Microenvironment of Glioma Based on IDH Status: Implications for Immunotherapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Processing of Tumor Specimens
2.3. DNA and RNA Extraction
2.4. Molecular Genetic Diagnosis
2.5. RNA-Seq
2.6. Analysis of the Tumor Microenvironment
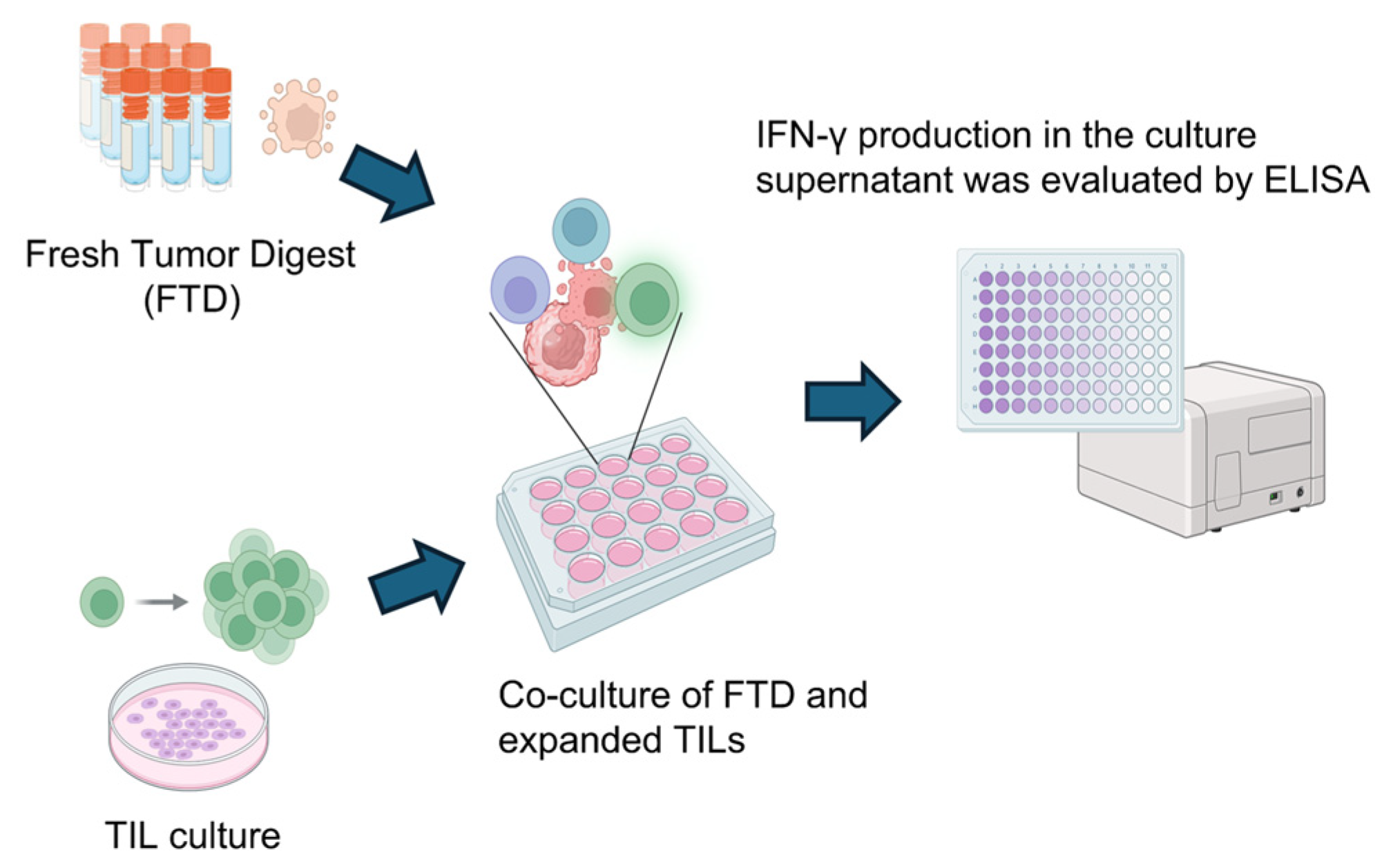
2.7. TIL Culture
2.8. Assessment of Tumor Reactivity
2.9. Statistical Analysis
3. Results
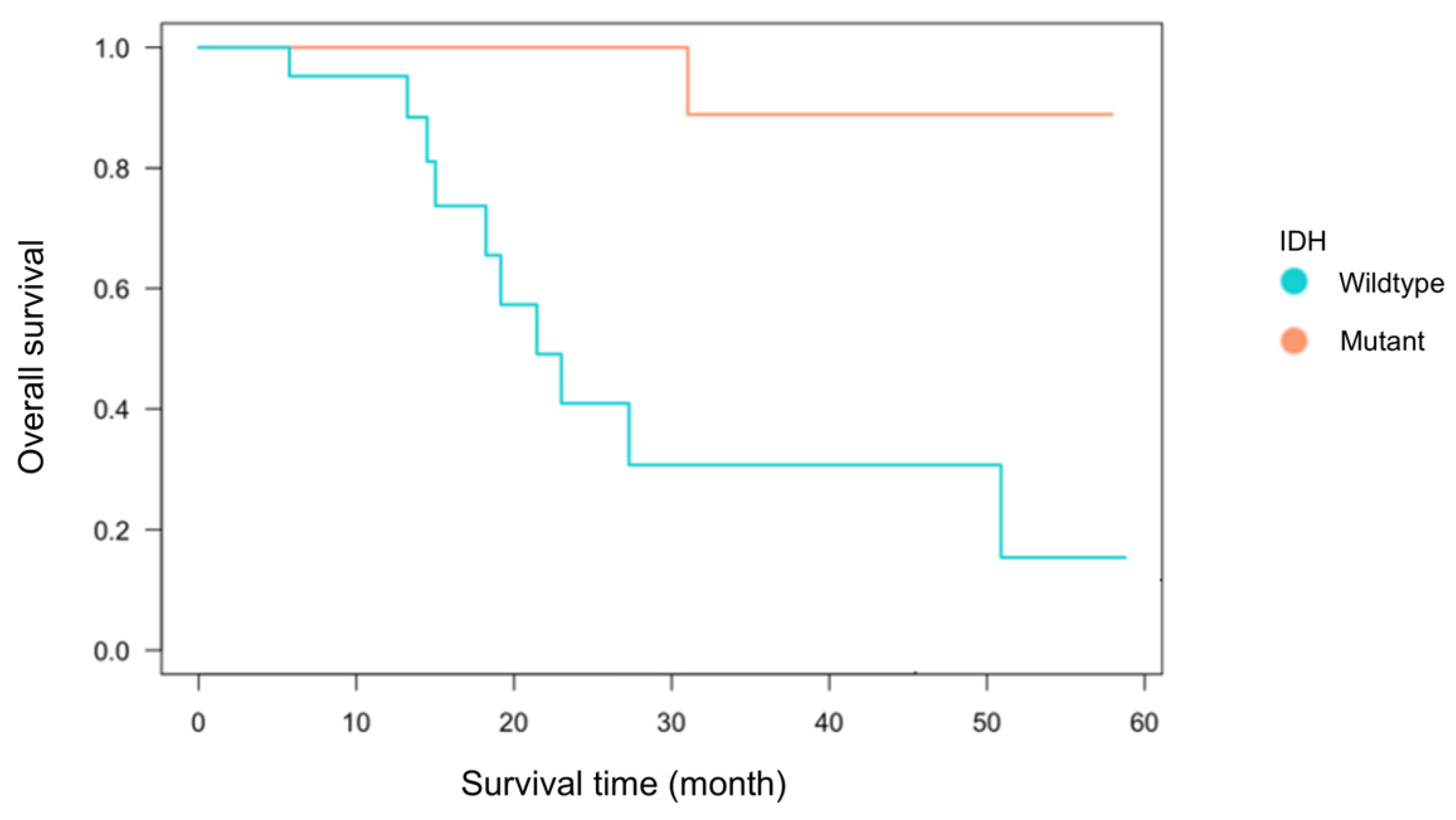
3.1. Patient Characteristics
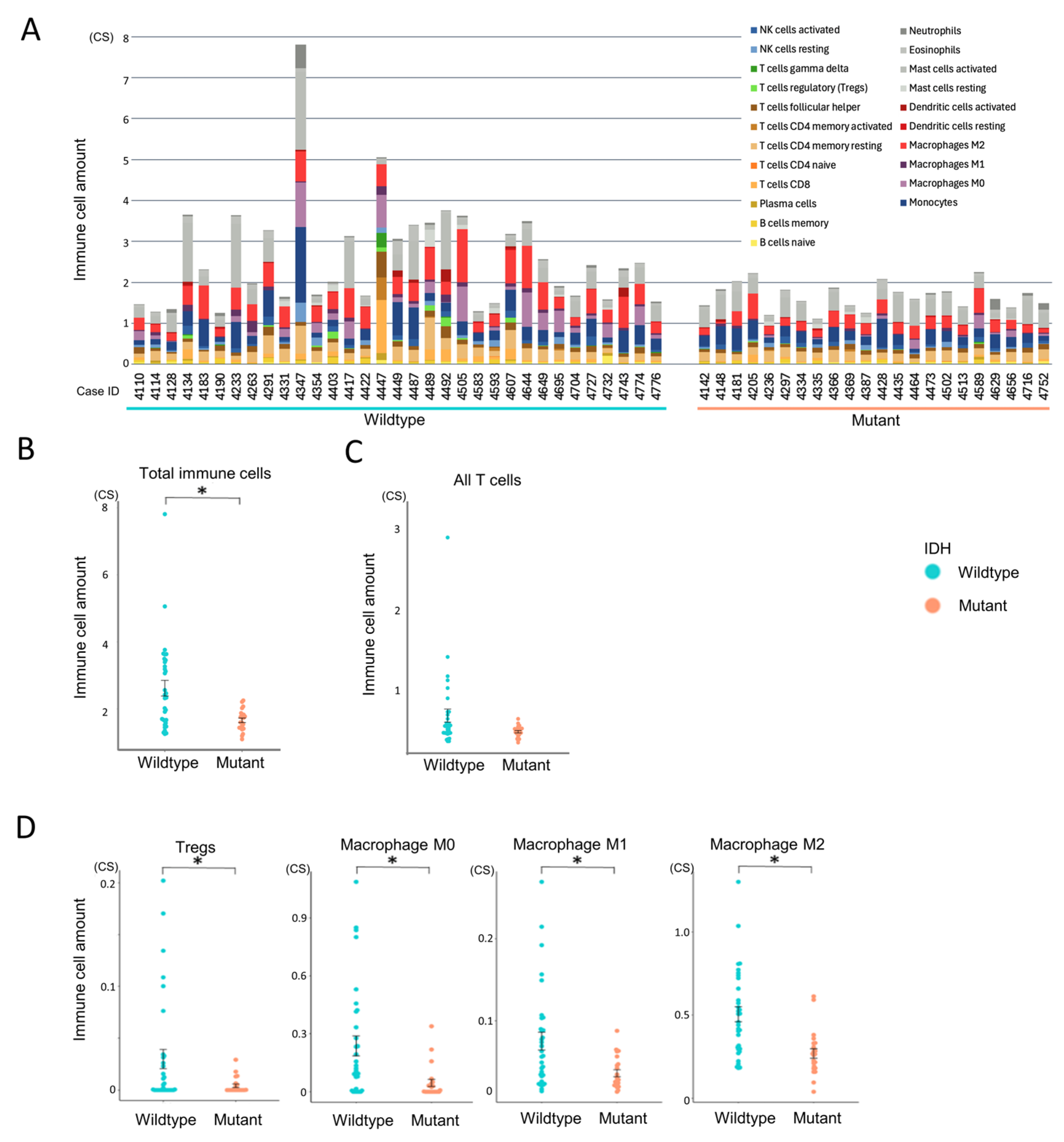
3.2. Immune Cell Infiltration According to IDH Status
3.3. Characterization of Immune Cell Functional States
3.4. Immune Cell State-Based Clustering of Gliomas
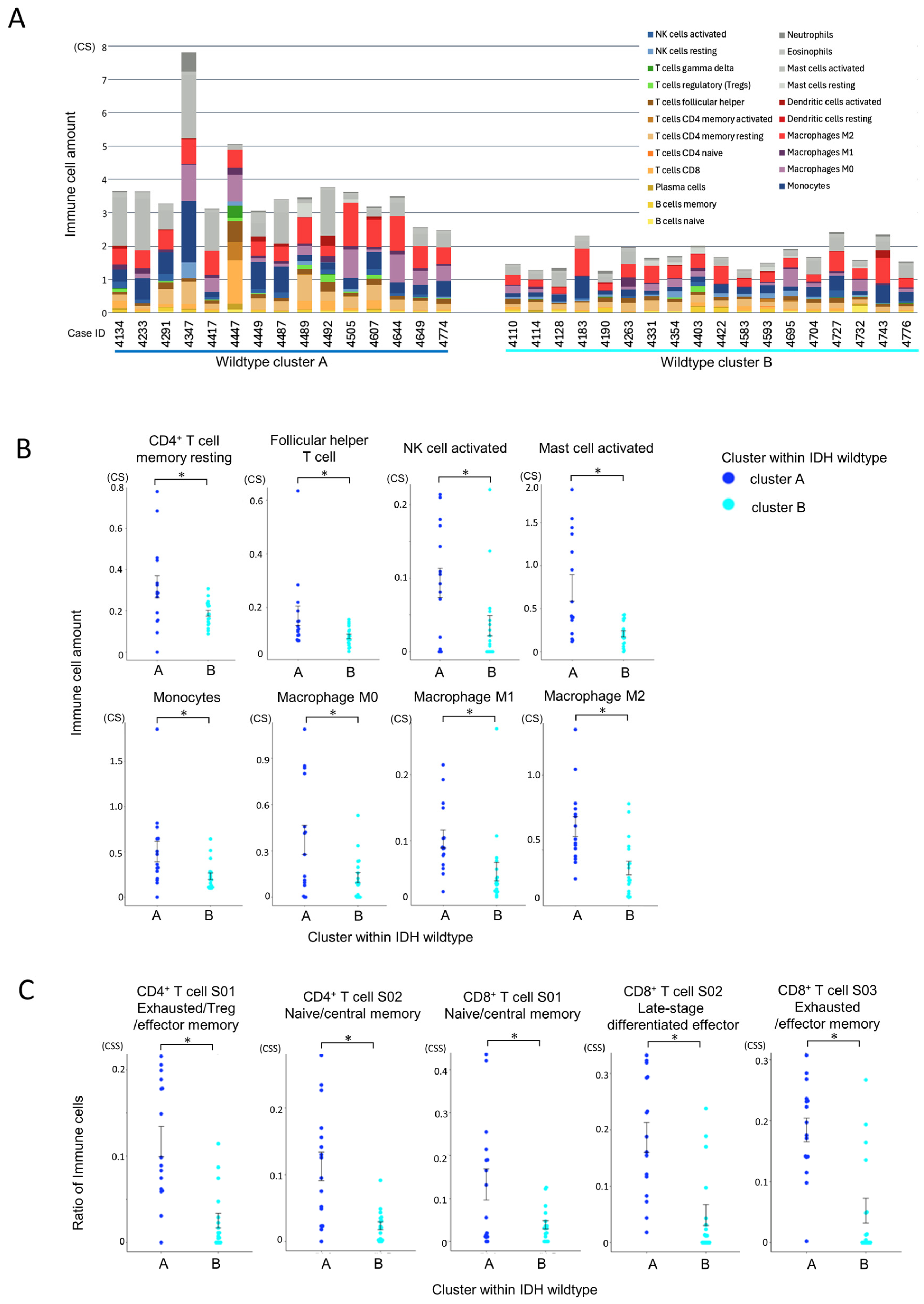
3.5. Immune Microenvironmental Divergence Within the IDH-Wildtype Group
3.5.1. Quantitative and Compositional Differences in Immune Infiltration
3.5.2. Functional States of Infiltrating Immune Cells
3.5.3. Pathway Enrichment Analysis Using Hallmark Gene Sets
3.6. Evaluation of Tumor Immune Microenvironment in the IDH-Wildtype Group via TIL Culture
3.6.1. TIL Culture Without Drug Addition
3.6.2. Effect of Anti-PD-1 Antibody on TIL Expansion
3.6.3. Effect of CSF1R Inhibitor on TIL Expansion
3.6.4. Effect of STAT3 Inhibitor on TIL Expansion
3.6.5. Influence of Combined Immune Suppression Pathways on Drug Responsiveness
3.7. Evaluation of Tumor Reactivity of Cultured TILs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CIBERSORTx | Cell type identification by estimating relative subsets of RNA transcripts x |
| CSF1 | Colony stimulating factor 1 |
| CSF1R | Colony stimulating factor 1 receptor |
| EGFR | Epidermal growth factor receptor |
| ELISA | Enzyme-linked immuno-sorbent assay |
| FTD | Fresh tumor digest |
| GBM | Glioblastoma |
| ICI | Immune checkpoint inhibitor |
| IDH | Isocitrate dehydrogenase |
| IFN-γ | Interferon γ |
| IL-10 | Interleukin-10 |
| IL-2 | Interleukin-2 |
| MGMT | O-6-methylguanine-DNA methyltransferase |
| NK cell | Natural killer cell |
| OS | Overall survival |
| PD-1 | Programmed death 1 |
| PD-L1 | Programmed death ligand 1 |
| PFS | Progression-free survival |
| RNA-Seq | RNA sequencing |
| ssGSEA | Single sample gene set enrichment analysis |
| STAT3 | Signal transducers and activator of transcription 3 |
| TERT | Telomerase reverse transcriptase |
| TGF-β | Transforming growth factor β |
| TIL | Tumor-infiltrating lymphocyte |
| Treg | Regulatory T cell |
| VEGF | Vascular endothelial growth factor |
| WHO | World Health Organization |
References
- Ostrom, Q.T.; Gittleman, H.; Xu, J.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2009–2013. Neuro Oncol. 2016, 18, v1–v75. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Committee of the Brain Tumor Registry of Japan, supported by the Japan Neurosurgical Society. Brain Tumor Registry of Japan (2005–2008). Neurol. Med. Chir. 2017, 57, 9–102. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Gritsch, S.; Batchelor, T.T.; Gonzalez Castro, L.N. Diagnostic, therapeutic, and prognostic implications of the 2021 World Health Organization classification of tumors of the central nervous system. Cancer 2022, 128, 47–58. [Google Scholar] [CrossRef]
- Brat, D.J.; Aldape, K.; Colman, H.; Figrarella-Branger, D.; Fuller, G.N.; Giannini, C.; Holland, E.C.; Jenkins, R.B.; Kleinschmidt-DeMasters, B.; Komori, T.; et al. cIMPACT-NOW update 5: Recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol. 2020, 139, 603–608. [Google Scholar] [CrossRef]
- Eckel-Passow, J.E.; Lachance, D.H.; Molinaro, A.M.; Walsh, K.M.; Decker, P.A.; Sicotte, H.; Pekmezci, M.; Rice, T.; Kosel, M.L.; Smirnov, I.V.; et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N. Engl. J. Med. 2015, 372, 2499–2508. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Reitman, Z.J.; Jin, G.; Karoly, E.D.; Spasojevic, I.; Yang, J.; Kinzler, K.W.; He, Y.; Bigner, D.D.; Vogelstein, B.; Yan, H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. USA 2011, 108, 3270–3275. [Google Scholar] [CrossRef]
- Pirozzi, C.J.; Yan, H. The implications of IDH mutations for cancer development and therapy. Nat. Rev. Clin. Oncol. 2021, 18, 645–661. [Google Scholar] [CrossRef] [PubMed]
- Mehani, B.; Asanigari, S.; Chung, H.J.; Dazelle, K.; Singh, A.; Hannenhalli, S.; Aldape, K. Immune cell gene expression signatures in diffuse glioma are associated with IDH mutation status, patient outcome and malignant cell state, and highlight the importance of specific cell subsets in glioma biology. Acta Neuropathol. Commun. 2022, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.; Xia, Y.; Bettegowda, C.; Weller, M. Current state of immunotherapy for glioblastoma. Nat. Rev. Clin. Oncol. 2018, 15, 422–442. [Google Scholar] [CrossRef] [PubMed]
- Omuro, A.; Brandes, A.A.; Carpentier, A.F.; Idbaih, A.; Reardon, D.A.; Cloughesy, T.; Sumrall, A.; Baehring, J.; van den Bent, M.; Bähr, O.; et al. Radiotherapy combined with nivolumab or temozolomide for newly diagnosed glioblastoma with unmethylated MGMT promoter: An international randomized phase III trial. Neuro Oncol. 2023, 25, 123–134. [Google Scholar] [CrossRef]
- Reardon, D.A.; Brandes, A.A.; Omuro, A.; Mulholland, P.; Lim, M.; Wick, A.; Baehring, J.; Ahluwalia, M.S.; Roth, P.; Bähr, O.; et al. Effect of Nivolumab vs Bevacizumab in Patients With Recurrent Glioblastoma: The CheckMate 143 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020, 6, 1003–1010. [Google Scholar] [CrossRef]
- Lim, M.; Weller, M.; Idbaih, A.; Steinbach, J.; Finocchiaro, G.; Raval, R.R.; Ansstas, G.; Baehring, J.; Taylor, J.W.; Honnorat, J.; et al. Phase III trial of chemoradiotherapy with temozolomide plus nivolumab or placebo for newly diagnosed glioblastoma with methylated MGMT promoter. Neuro Oncol. 2022, 24, 1935–1949. [Google Scholar] [CrossRef]
- Fridman, W.H.; Zitvogel, L.; Sautès-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef]
- Jackson, C.M.; Choi, J.; Lim, M. Mechanisms of immunotherapy resistance: Lessons from glioblastoma. Nat. Immunol. 2019, 20, 1100–1109. [Google Scholar] [CrossRef]
- Pombo Antunes, A.R.; Scheyltjens, I.; Duerinck, J.; Neyns, B.; Movahedi, K.; Van Ginderachter, J.A. Understanding the glioblastoma immune microenvironment as basis for the development of new immunotherapeutic strategies. eLife 2020, 9, e52176. [Google Scholar] [CrossRef]
- Chen, W.; Jin, W.; Hardegen, N.; Lei, K.J.; Li, L.; Marinos, N.; McGrady, G.; Wahl, S.M. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003, 198, 1875–1886. [Google Scholar] [CrossRef]
- Crane, C.A.; Han, S.J.; Barry, J.J.; Ahn, B.J.; Lanier, L.L.; Parsa, A.T. TGF-beta downregulates the activating receptor NKG2D on NK cells and CD8+ T cells in glioma patients. Neuro Oncol. 2010, 12, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Berghoff, A.S.; Kiesel, B.; Widhalm, G.; Rajky, O.; Ricken, G.; Wöhrer, A.; Dieckmann, K.; Filipits, M.; Brandstetter, A.; Weller, M.; et al. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015, 17, 1064–1075. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Sturm, G.; Finotello, F.; Petitprez, F.; Zhang, J.D.; Baumbach, J.; Fridman, W.H.; List, M.; Aneichyk, T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 2019, 35, i436–i445. [Google Scholar] [CrossRef]
- White, B.S.; De Reyniès, A.; Newman, A.M.; Waterfall, J.J.; Lamb, A.; Petitprez, F.; Lin, Y.; Yu, R.; Guerrero-Gimenez, M.E.; Domanskyi, S.; et al. Community assessment of methods to deconvolve cellular composition from bulk gene expression. Nat. Commun. 2024, 15, 7362. [Google Scholar] [CrossRef]
- Berghoff, A.S.; Kiesel, B.; Widhalm, G.; Wilhelm, D.; Rajky, O.; Kurscheid, S.; Kresl, P.; Wöhrer, A.; Marosi, C.; Hegi, M.E.; et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol. 2017, 19, 1460–1468. [Google Scholar] [CrossRef]
- Luca, B.A.; Steen, C.B.; Matusiak, M.; Azizi, A.; Varma, S.; Zhu, C.; Przybyl, J.; Espín-Pérez, A.; Diehn, M.; Alizadeh, A.A.; et al. Atlas of clinically distinct cell states and ecosystems across human solid tumors. Cell 2021, 184, e5482–e5496. [Google Scholar] [CrossRef]
- González-Tablas Pimenta, M.; Otero, Á.; Arandia Guzman, D.A.; Pascual-Argente, D.; Ruíz Martín, L.; Sousa-Casasnovas, P.; García-Martin, A.; Roa Montes de Oca, J.C.; Villaseñor-Ledezma, J.; Torres Carretero, L.; et al. Tumor cell and immune cell profiles in primary human glioblastoma: Impact on patient outcome. Brain Pathol. 2021, 31, 365–380. [Google Scholar] [CrossRef]
- Brat, D.J.; Verhaak, R.G.; Aldape, K.D.; Yung, W.K.; Salama, S.R.; Cooper, L.A.; Rheinbay, E.; Miller, C.R.; Vitucci, M.; Morozova, O.; et al. Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas. N. Engl. J. Med. 2015, 372, 2481–2498. [Google Scholar] [CrossRef]
- Wei, J.; Barr, J.; Kong, L.Y.; Wang, Y.; Wu, A.; Sharma, A.K.; Gumin, J.; Henry, V.; Colman, H.; Priebe, W.; et al. Glioblastoma cancer-initiating cells inhibit T-cell proliferation and effector responses by the signal transducers and activators of transcription 3 pathway. Mol. Cancer Ther. 2010, 9, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, P.; Zhou, Y.; Landström, M. The Synergistic Cooperation between TGF-β and Hypoxia in Cancer and Fibrosis. Biomolecules 2022, 12, 635. [Google Scholar] [CrossRef]
- Xia, T.; Zhang, M.; Lei, W.; Yang, R.; Fu, S.; Fan, Z.; Yang, Y.; Zhang, T. Advances in the role of STAT3 in macrophage polarization. Front. Immunol. 2023, 14, 1160719. [Google Scholar] [CrossRef]
- Lee, A.H.; Sun, L.; Mochizuki, A.Y.; Reynoso, J.G.; Orpilla, J.; Chow, F.; Kienzler, J.C.; Everson, R.G.; Nathanson, D.A.; Bensinger, S.J.; et al. Neoadjuvant PD-1 blockade induces T cell and cDC1 activation but fails to overcome the immunosuppressive tumor associated macrophages in recurrent glioblastoma. Nat. Commun. 2021, 12, 6938. [Google Scholar] [CrossRef] [PubMed]
- Pyonteck, S.M.; Akkari, L.; Schuhmacher, A.J.; Bowman, R.L.; Sevenich, L.; Quail, D.F.; Olson, O.C.; Quick, M.L.; Huse, J.T.; Teijeiro, V.; et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013, 19, 1264–1272. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Pollard, J.W. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 2004, 4, 71–78. [Google Scholar] [CrossRef]
- Kortylewski, M.; Yu, H. Role of Stat3 in suppressing anti-tumor immunity. Curr. Opin. Immunol. 2008, 20, 228–233. [Google Scholar] [CrossRef]
- Ren, X.; Zhang, L.; Zhang, Y.; Li, Z.; Siemers, N.; Zhang, Z. Insights Gained from Single-Cell Analysis of Immune Cells in the Tumor Microenvironment. Annu. Rev. Immunol. 2021, 39, 583–609. [Google Scholar] [CrossRef]
- Chinen, T.; Kannan, A.K.; Levine, A.G.; Fan, X.; Klein, U.; Zheng, Y.; Gasteiger, G.; Feng, Y.; Fontenot, J.D.; Rudensky, A.Y. An essential role for the IL-2 receptor in T(reg) cell function. Nat. Immunol. 2016, 17, 1322–1333. [Google Scholar] [CrossRef]
- Wang, J.; Tao, X.; Zhu, J.; Dai, Z.; Du, Y.; Xie, Y.; Chu, X.; Fu, G.; Lei, Z. Tumor organoid-immune co-culture models: Exploring a new perspective of tumor immunity. Cell Death Discov. 2025, 11, 195. [Google Scholar] [CrossRef]
- Lemoine, C.; Da Veiga, M.A.; Rogister, B.; Piette, C.; Neirinckx, V. An integrated perspective on single-cell and spatial transcriptomic signatures in high-grade gliomas. npj Precis. Oncol. 2025, 9, 44. [Google Scholar] [CrossRef]





| Wildtype (N = 33) | Mutant (N = 22) | p-Value | Statistical Analysis | ||
|---|---|---|---|---|---|
| Age of onset | 59.1 | 44.0 | <0.001 | t-test | |
| Gender | Male | 16 | 14 | 0.12 | Fisher |
| Female | 17 | 8 | |||
| Primary or recurrent | Primary | 22 | 16 | 0.21 | Fisher |
| Recurrent | 11 | 6 | |||
| Overall survival (month) | 15.1 | 29.5 | 0.07 | Log-rank test | |
| Resection | GTR | 11 | 9 | 0.22 | Fisher |
| PR | 16 | 11 | |||
| Unknown | 6 | 2 |
| ID | IDH | Cluster | Result of TIL Proliferation | Condition of Inhibition | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Anti-PD-1 Antibody | CSF1R Inhibitor | STAT3 Inhibitor | PD-1 (FPKM) | CSF1R (FPKM) | STAT3 (ssGSEA) | Number of Suppressing Pathways | Category | |||
| 4134 | wild | A | × | × | × | ▲ | ▲ | ▲ | 3 | ‡ |
| 4233 | wild | A | ○ | ○ | × | ▲ | ▲ | ▲ | 3 | † |
| 4291 | wild | A | × | NA | NA | ▲ | ▲ | Δ | 2 | ‡ |
| 4347 | wild | A | × | × | NA | Δ | Δ | ▲ | 1 | * |
| 4417 | wild | A | × | NA | NA | ▲ | ▲ | Δ | 2 | ‡ |
| 4447 | wild | A | × | NA | × | ▲ | Δ | Δ | 1 | ‡ |
| 4449 | wild | A | × | NA | × | Δ | Δ | ▲ | 1 | ‡ |
| 4487 | wild | A | × | NA | NA | Δ | ▲ | ▲ | 2 | * |
| 4489 | wild | A | × | NA | × | ▲ | Δ | ▲ | 2 | ‡ |
| 4492 | wild | A | × | NA | NA | ▲ | Δ | ▲ | 2 | ‡ |
| 4505 | wild | A | × | NA | NA | ▲ | ▲ | ▲ | 3 | ‡ |
| 4644 | wild | A | NA | × | × | ▲ | ▲ | ▲ | 3 | ‡ |
| 4649 | wild | A | NA | × | × | Δ | ▲ | ▲ | 2 | ‡ |
| 4774 | wild | A | NA | × | × | Δ | Δ | Δ | 0 | * |
| 4110 | wild | A | × | NA | NA | Δ | Δ | Δ | 0 | * |
| 4114 | wild | B | × | NA | NA | Δ | Δ | Δ | 0 | * |
| 4128 | wild | B | × | × | × | Δ | Δ | Δ | 0 | * |
| 4183 | wild | B | ○ | × | ○ | Δ | ▲ | Δ | 1 | § |
| 4190 | wild | B | × | NA | NA | Δ | Δ | Δ | 0 | * |
| 4263 | wild | B | × | × | ○ | Δ | Δ | Δ | 0 | § |
| 4331 | wild | B | × | NA | NA | Δ | Δ | Δ | 0 | * |
| 4354 | wild | B | × | NA | NA | Δ | Δ | Δ | 0 | * |
| 4403 | wild | B | × | × | × | Δ | Δ | Δ | 0 | * |
| 4422 | wild | B | × | × | NA | Δ | Δ | Δ | 0 | * |
| 4583 | wild | B | × | × | × | Δ | Δ | Δ | 0 | * |
| 4593 | wild | B | × | × | × | Δ | Δ | Δ | 0 | * |
| 4695 | wild | B | NA | × | × | Δ | Δ | Δ | 0 | * |
| 4704 | wild | B | NA | ○ | ○ | ▲ | Δ | Δ | 1 | § |
| 4727 | wild | B | NA | ○ | NA | Δ | ▲ | Δ | 1 | † |
| 4732 | wild | B | NA | × | NA | Δ | Δ | Δ | 0 | * |
| 4776 | wild | B | NA | ○ | × | Δ | Δ | Δ | 0 | § |
| ID | IDH | Cluster | Result of TIL Proliferation | Result of IFN-γ ELISA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No Additional Drug | Anti-PD-1 Antibody | CSF1R Inhibitor | STAT3 Inhibitor | No Additional Drug | Anti-PD-1 Antibody | CSF1R Inhibitor | STAT3 Inhibitor | Tumor Responsive TIL | |||
| 4233 | wild | A | ○ | ○ | ○ | × | - | + | - | - | + |
| 4644 | wild | A | ○ | NA | × | × | + | NA | + | + | + |
| 4649 | wild | A | ○ | NA | × | × | - | NA | - | - | - |
| 4774 | wild | A | ○ | NA | × | × | - | NA | - | + | + |
| 4183 | wild | B | ○ | ○ | × | ○ | - | - | - | - | + |
| 4263 | wild | B | ○ | × | × | ○ | - | NA | - | - | - |
| 4354 | wild | B | NA | × | NA | NA | NA | NA | NA | NA | - |
| 4403 | wild | B | ○ | × | × | × | + | NA | + | - | + |
| 4583 | wild | B | × | × | × | × | - | NA | - | - | - |
| 4593 | wild | B | ○ | × | × | × | - | NA | - | - | - |
| 4695 | wild | B | ○ | NA | × | × | - | NA | - | - | - |
| 4704 | wild | B | ○ | NA | ○ | ○ | - | NA | - | - | - |
| 4727 | wild | B | ○ | NA | ○ | NA | - | NA | - | NA | - |
| 4732 | wild | B | × | NA | × | × | + | NA | NA | NA | + |
| 4776 | wild | B | ○ | NA | ○ | × | - | NA | - | - | - |
| 4142 | mutant | B | NA | × | × | × | NA | NA | NA | NA | - |
| 4148 | mutant | B | × | × | × | × | - | - | NA | NA | - |
| 4181 | mutant | B | NA | ○ | × | × | - | - | NA | NA | - |
| 4205 | mutant | B | × | × | × | × | - | NA | NA | NA | - |
| 4236 | mutant | B | × | × | × | × | - | NA | NA | NA | - |
| 4297 | mutant | B | ○ | × | × | × | - | NA | - | - | - |
| 4366 | mutant | B | ○ | × | × | × | - | NA | - | - | - |
| 4369 | mutant | B | × | ○ | × | × | - | - | - | - | - |
| 4387 | mutant | B | × | × | × | ○ | NA | NA | - | - | - |
| 4435 | mutant | B | ○ | × | × | × | - | NA | - | - | - |
| 4502 | mutant | B | ○ | × | × | × | - | NA | - | - | - |
| 4589 | mutant | B | ○ | × | × | × | - | NA | - | - | - |
| 4656 | mutant | B | ○ | NA | × | × | - | NA | - | - | - |
| 4716 | mutant | B | ○ | NA | × | × | - | NA | - | - | - |
| 4752 | mutant | B | × | NA | × | × | - | NA | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kikuchi, M.; Takami, H.; Kobayashi, Y.; Nagaoka, K.; Kitagawa, Y.; Nomura, M.; Takayanagi, S.; Tanaka, S.; Saito, N.; Kakimi, K. Dissecting the Immunological Microenvironment of Glioma Based on IDH Status: Implications for Immunotherapy. Cells 2025, 14, 1035. https://doi.org/10.3390/cells14131035
Kikuchi M, Takami H, Kobayashi Y, Nagaoka K, Kitagawa Y, Nomura M, Takayanagi S, Tanaka S, Saito N, Kakimi K. Dissecting the Immunological Microenvironment of Glioma Based on IDH Status: Implications for Immunotherapy. Cells. 2025; 14(13):1035. https://doi.org/10.3390/cells14131035
Chicago/Turabian StyleKikuchi, Miyu, Hirokazu Takami, Yukari Kobayashi, Koji Nagaoka, Yosuke Kitagawa, Masashi Nomura, Shunsaku Takayanagi, Shota Tanaka, Nobuhito Saito, and Kazuhiro Kakimi. 2025. "Dissecting the Immunological Microenvironment of Glioma Based on IDH Status: Implications for Immunotherapy" Cells 14, no. 13: 1035. https://doi.org/10.3390/cells14131035
APA StyleKikuchi, M., Takami, H., Kobayashi, Y., Nagaoka, K., Kitagawa, Y., Nomura, M., Takayanagi, S., Tanaka, S., Saito, N., & Kakimi, K. (2025). Dissecting the Immunological Microenvironment of Glioma Based on IDH Status: Implications for Immunotherapy. Cells, 14(13), 1035. https://doi.org/10.3390/cells14131035







