Apolipoprotein L1 (APOL1): Consideration of Molecular Evolution, Interaction with APOL3, and Impact of Splice Isoforms Advances Understanding of Cellular and Molecular Mechanisms of Cell Injury
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. RNA Extraction and PCR Analysis
2.3. Plasmid Construction and Transfections
2.4. Quantification of APOL1 Efflux by ELISA
2.5. Immunoblotting Analysis
3. Results
3.1. Differential Splicing of Basal and IFNγ-Induced APOL1 Across Different Cell Types
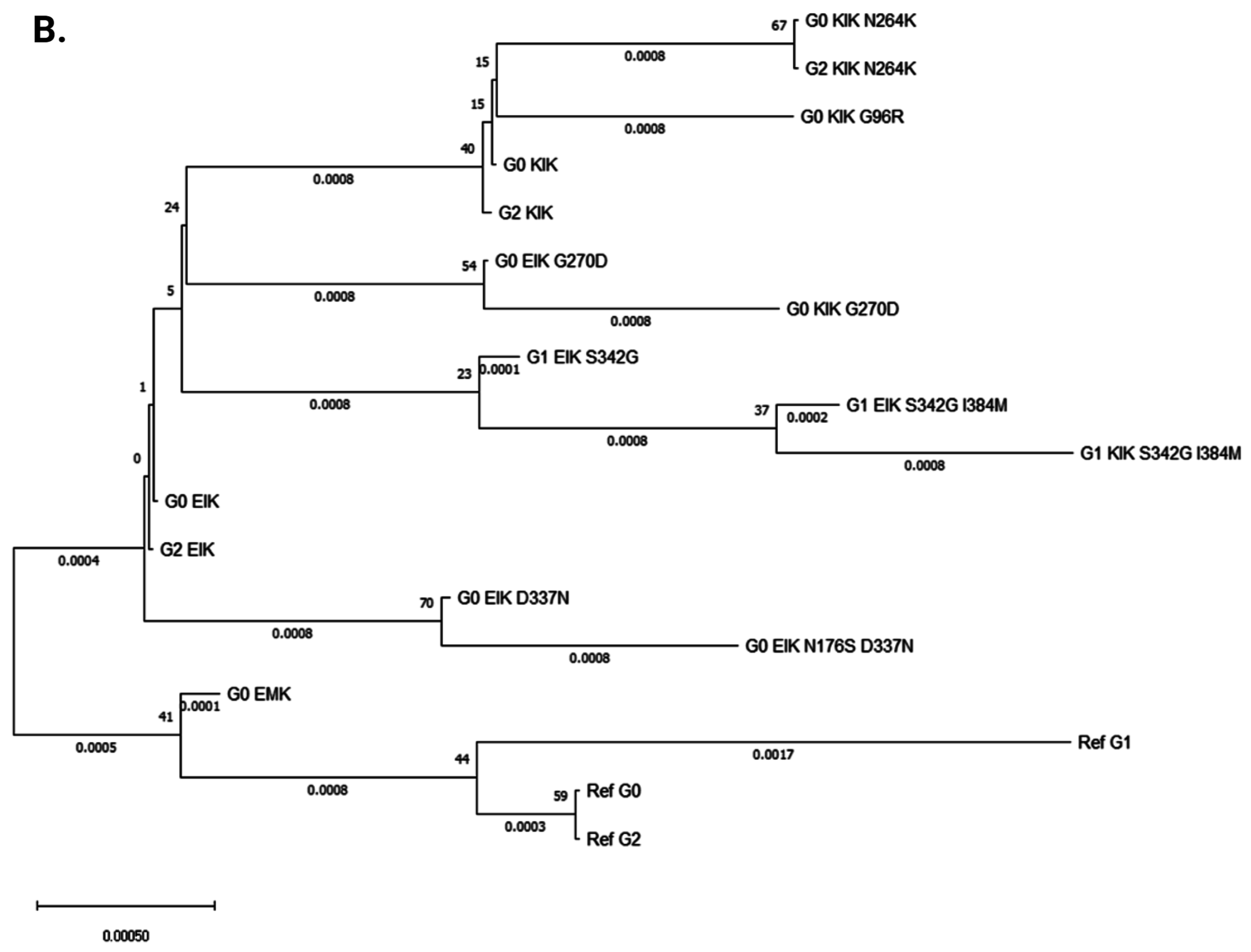
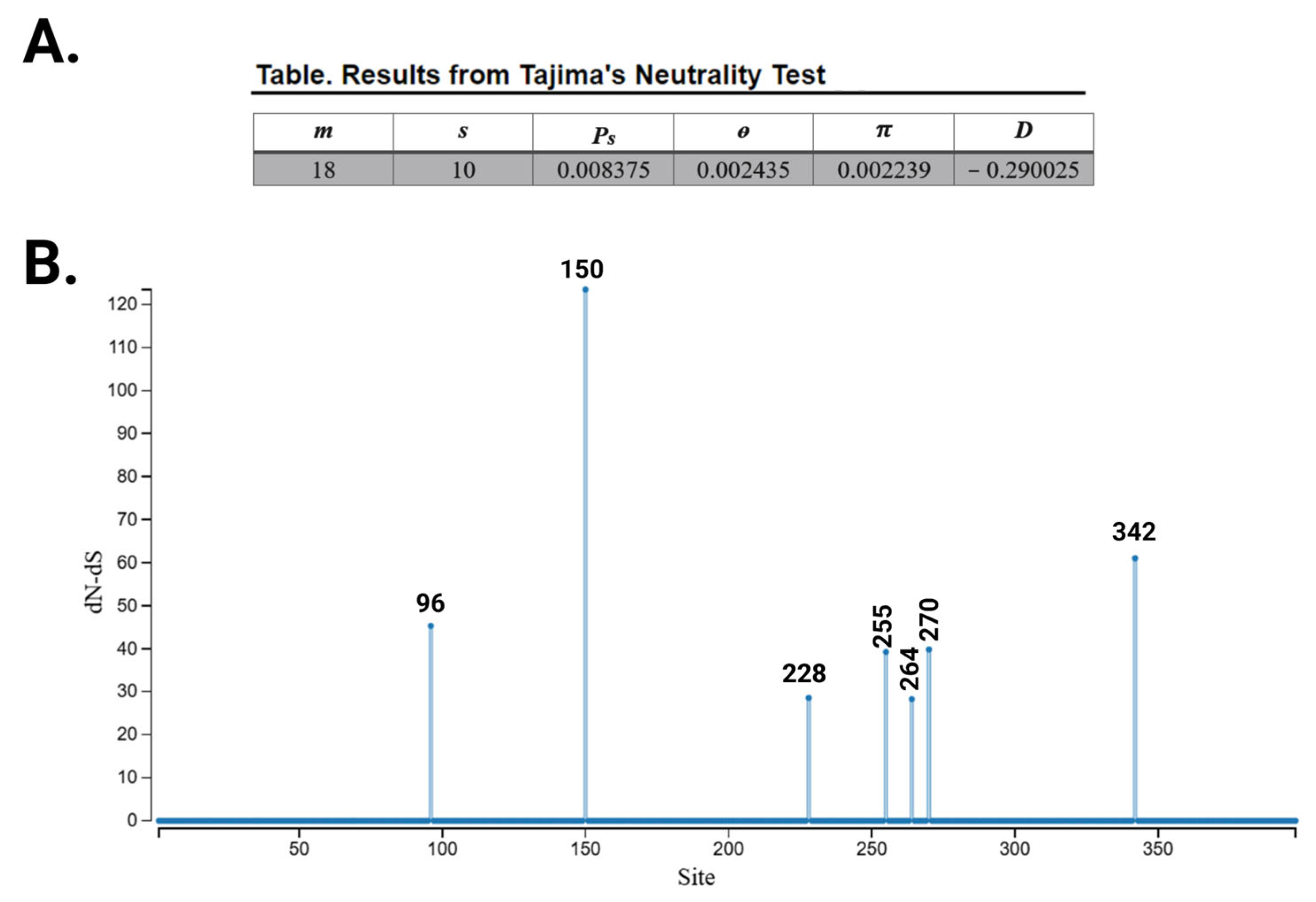
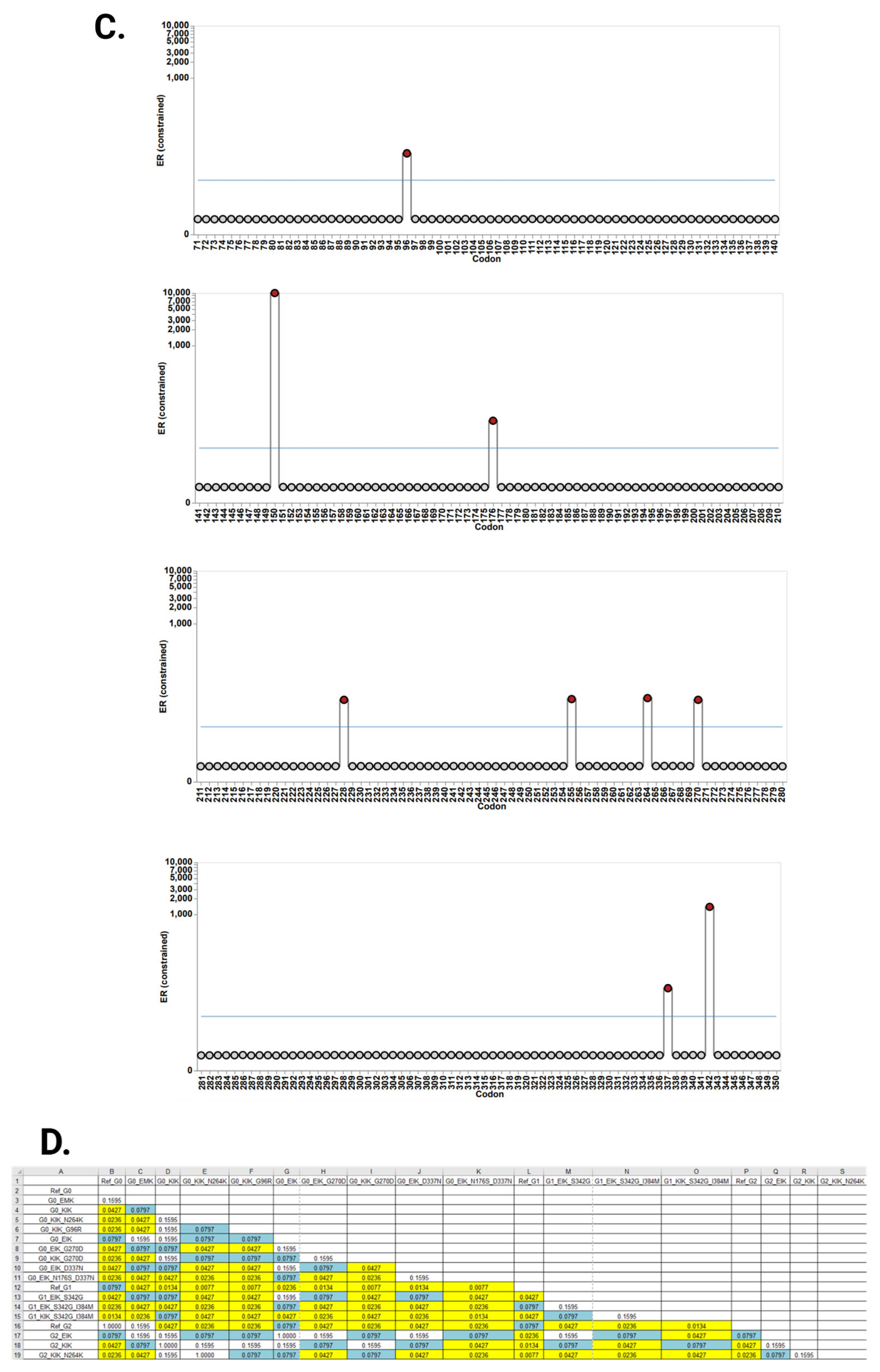
3.2. Evolutionary Chronology of APOL1 Haplotypes Reveals an Early Emergence of Toxic Gain-of-Function Variants and a Later Rise of Cell-Protective Ones
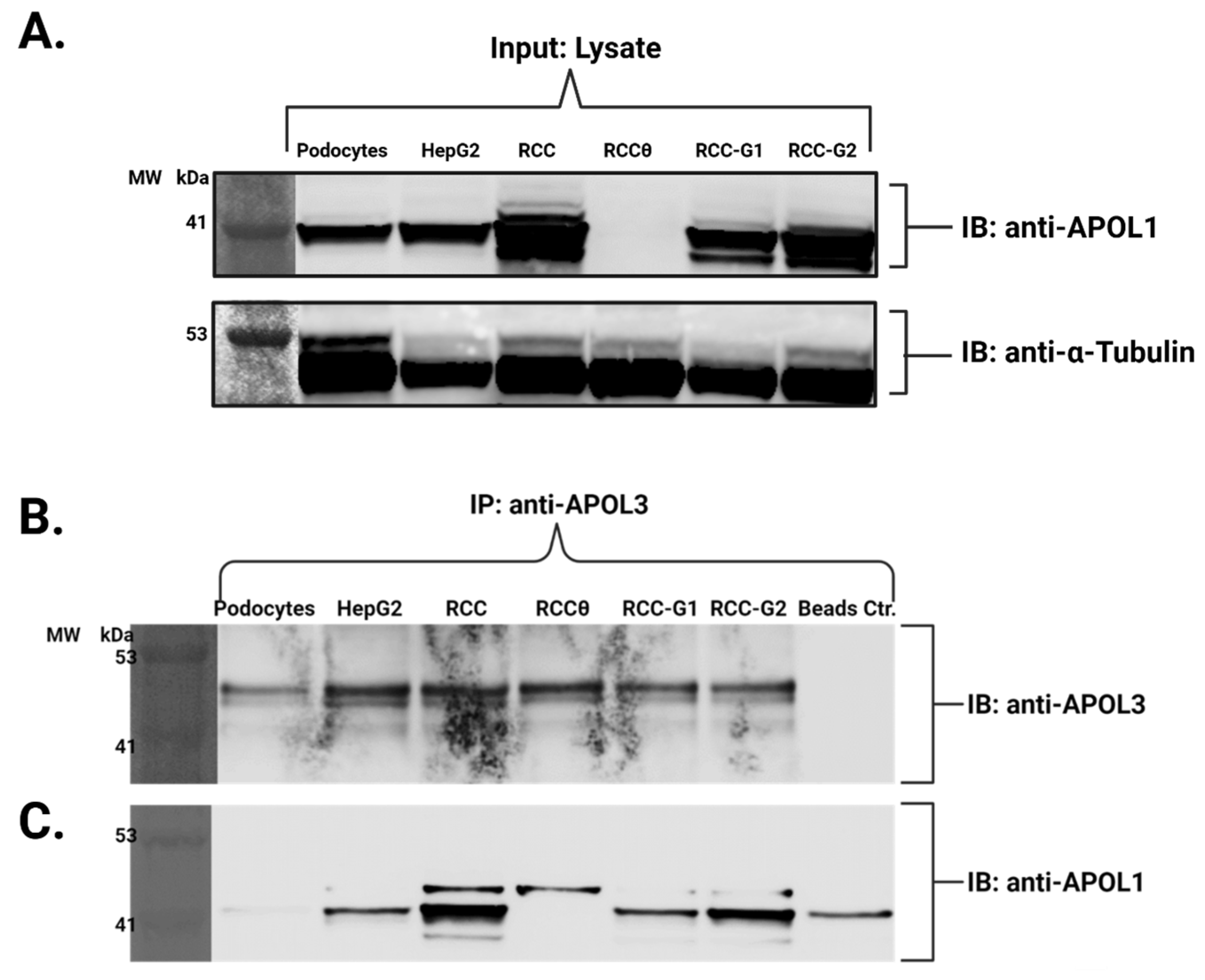
3.3. The Native Interaction Between APOL1 and APOL3 Is Modified by G1 and G2 Renal-Risk Variants
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMKD | APOL1 mediated kidney disease |
| ApoAI | Apolipoprotein AI |
| APOL1 | Apolipoprotein L1 (APOL1) |
| BH3 | Bcl-2 homology domain 3 |
| BUSTED | Branch-site unrestricted statistical test of episodic diversification |
| CKD | Chronic kidney disease |
| EHH | Extended haplotype homozygosity |
| ESKD | End-stage kidney disease |
| FSGS | Focal segmental glomerulosclerosis |
| HDL | High density lipoprotein |
| Hp | Haptoglobin |
| Hpr | Haptoglobin-related protein |
| HR | Hazard ratio |
| IFN-γ | Interferon gamma |
| ISG | Interferon-stimulated genes |
| MALD | Mapping by admixture linkage disequilibrium |
| SLAC | Single likelihood ancestor counting |
| SNP | Single-nucleotide polymorphism |
| SRA | Serum-resistance associated protein |
| TLF | Trypanosome lytic factor |
| VSG | Variant surface glycoprotein |
References
- Bruce, D. The Croonian Lectures on Trypanosomes Causing Disease in Man and Domestic Animals in Central Africa: Delivered Before the Royal College of Physicians of London. BMJ 1915, 2, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Stephens, J.W.W.; Fantham, H.B. On the peculiar morphology of a trypanosome from a case of sleeping sickness and the possibility of its being a new species (T. rhodesiense). Proc. R. Soc. Lond. Ser. B Contain. Pap. A Biol. Character 1910, 83, 28–33. [Google Scholar] [CrossRef]
- Hawking, F. The differentiation of Trypanosoma rhodesiense from T. brucei by means of human serum. Trans. R. Soc. Trop. Med. Hyg. 1973, 67, 517–527. [Google Scholar] [CrossRef]
- Rifkin, M.R. Identification of the trypanocidal factor in normal human serum: High density lipoprotein. Proc. Natl. Acad. Sci. USA 1978, 75, 3450–3454. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, S.L.; Moore, D.R.; Vasudevacharya, J.; Siqueira, H.; Torri, A.F.; Tytler, E.M.; Esko, J.D. Lysis of Trypanosoma brucei by a Toxic Subspecies of Human High Density Lipoprotein. J. Biol. Chem. 1989, 264, 5210–5217. [Google Scholar] [CrossRef]
- Barth, P. A new method for the isolation of the trypanocidal factor from normal human serum. Acta Trop. 1989, 46, 71–73. [Google Scholar] [CrossRef]
- Raper, J.; Nussenzweig, V.; Tomlinson, S. The main lytic factor of Trypanosoma brucei brucei in normal human serum is not high density lipoprotein. J. Exp. Med. 1996, 183, 1023–1029. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, S.; Muranjan, M.; Nussenzweig, V.; Raper, J. Haptoglobin-related protein and apolipoprotein AI are components of the two trypanolytic factors in human serum. Mol. Biochem. Parasitol. 1997, 86, 117–120. [Google Scholar] [CrossRef]
- Raper, J.; Fung, R.; Ghiso, J.; Nussenzweig, V.; Tomlinson, S. Characterization of a Novel Trypanosome Lytic Factor from Human Serum. Infect. Immun. 1999, 67, 1910–1916. [Google Scholar] [CrossRef][Green Version]
- Smith, A.B.; Esko, J.D.; Hajduk, S.L. Killing of trypanosomes by the human Haptoglobin-Related protein. Science 1995, 268, 284–286. [Google Scholar] [CrossRef]
- Fielding, C.J.; Fielding, P.E. Molecular physiology of reverse cholesterol transport. J. Lipid Res. 1995, 36, 211–228. [Google Scholar] [CrossRef] [PubMed]
- Duchateau, P.N.; Pullinger, C.R.; Orellana, R.E.; Kunitake, S.T.; Naya-Vigne, J.; O’Connor, P.M.; Malloy, M.J.; Kane, J.P. Apolipoprotein L, a new human high density lipoprotein apolipoprotein expressed by the pancreas. J. Biol. Chem. 1997, 272, 25576–25582. [Google Scholar] [CrossRef]
- Duchateau, P.N.; Pullinger, C.R.; Cho, M.H.; Eng, C.; Kane, J.P. Apolipoprotein L gene family: Tissue-specific expression, splicing, promoter regions; discovery of a new gene. J. Lipid Res. 2001, 42, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Page, N.M.; Butlin, D.J.; Lomthaisong, K.; Lowry, P.J. The Human Apolipoprotein L Gene Cluster: Identification, Classification, and sites of distribution. Genomics 2001, 74, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Vanhamme, L.; Paturiaux-Hanocq, F.; Poelvoorde, P.; Nolan, D.P.; Lins, L.; Van Den Abbeele, J.; Pays, A.; Tebabi, P.; Van Xong, H.; Jacquet, A.; et al. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 2003, 422, 83–87. [Google Scholar] [CrossRef]
- Hager, K.M.; Pierce, M.A.; Moore, D.R.; Tytler, E.M.; Esko, J.D.; Hajduk, S.L. Endocytosis of a cytotoxic human high density lipoprotein results in disruption of acidic intracellular vesicles and subsequent killing of African trypanosomes. J. Cell Biol. 1994, 126, 155–167. [Google Scholar] [CrossRef]
- Pereira, S.S.; Jackson, A.P.; Figueiredo, L.M. Evolution of the variant surface glycoprotein family in African trypanosomes. Trends Parasitol. 2021, 38, 23–36. [Google Scholar] [CrossRef]
- Green, H.P.; Del Pilar Molina Portela, M.; St Jean, E.N.; Lugli, E.B.; Raper, J. Evidence for a Trypanosoma brucei Lipoprotein Scavenger Receptor. J. Biol. Chem. 2002, 278, 422–427. [Google Scholar] [CrossRef]
- Vanhollebeke, B.; De Muylder, G.; Nielsen, M.J.; Pays, A.; Tebabi, P.; Dieu, M.; Raes, M.; Moestrup, S.K.; Pays, E. A Haptoglobin-Hemoglobin Receptor Conveys Innate Immunity to Trypanosoma brucei in Humans. Science 2008, 320, 677–681. [Google Scholar] [CrossRef]
- Weckerle, A.; Snipes, J.A.; Cheng, D.; Gebre, A.K.; Reisz, J.A.; Murea, M.; Shelness, G.S.; Hawkins, G.A.; Furdui, C.M.; Freedman, B.I.; et al. Characterization of circulating APOL1 protein complexes in African Americans. J. Lipid Res. 2016, 57, 120–130. [Google Scholar] [CrossRef]
- Pérez-Morga, D.; Vanhollebeke, B.; Paturiaux-Hanocq, F.; Nolan, D.P.; Lins, L.; Homble, F.; Vanhamme, L.; Tebabi, P.; Pays, A.; Poelvoorde, P.; et al. Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science 2005, 309, 469–472. [Google Scholar] [CrossRef] [PubMed]
- Kitiyakara, C.; Kopp, J.B.; Eggers, P. Trends in the epidemiology of focal segmental glomerulosclerosis. Semin. Nephrol. 2003, 23, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Kopp, J.B.; Winkler, C. HIV-associated nephropathy in African Americans11The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The publisher or recipient acknowledges right of the U.S. Government to retain a nonexclusive, royalty-free license in and to any copyright covering the article. Kidney Int. 2003, 63, S43–S49. [Google Scholar]
- Kao, W.H.L.; Klag, M.J.; Meoni, L.A.; Reich, D.; Berthier-Schaad, Y.; Li, M.; Coresh, J.; Patterson, N.; Tandon, A.; Powe, N.R.; et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet. 2008, 40, 1185–1192. [Google Scholar] [CrossRef]
- Kopp, J.B.; Smith, M.W.; Nelson, G.W.; Johnson, R.C.; Freedman, B.I.; Bowden, D.W.; Oleksyk, T.; McKenzie, L.M.; Kajiyama, H.; Ahuja, T.S.; et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. 2008, 40, 1175–1184. [Google Scholar] [CrossRef]
- Shlush, L.I.; Bercovici, S.; Wasser, W.G.; Yudkovsky, G.; Templeton, A.; Geiger, D.; Skorecki, K. Admixture mapping of end stage kidney disease genetic susceptibility using estimated mutual information ancestry informative markers. BMC Med. Genom. 2010, 3, 47. [Google Scholar] [CrossRef]
- Behar, D.M.; Shlush, L.I.; Maor, C.; Lorber, M.; Skorecki, K. Absence of HIV-Associated nephropathy in Ethiopians. Am. J. Kidney Dis. 2005, 47, 88–94. [Google Scholar] [CrossRef]
- Tzur, S.; Rosset, S.; Shemer, R.; Yudkovsky, G.; Selig, S.; Tarekegn, A.; Bekele, E.; Bradman, N.; Wasser, W.G.; Behar, D.M.; et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum. Genet. 2010, 128, 345–350. [Google Scholar] [CrossRef]
- Genovese, G.; Friedman, D.J.; Ross, M.D.; Lecordier, L.; Uzureau, P.; Freedman, B.I.; Bowden, D.W.; Langefeld, C.D.; Oleksyk, T.K.; Knob, A.L.U.; et al. Association of Trypanolytic ApoL1 Variants with Kidney Disease in African Americans. Science 2010, 329, 841–845. [Google Scholar] [CrossRef]
- Vivante, A. Genetics of chronic kidney Disease. N. Engl. J. Med. 2024, 391, 627–639. [Google Scholar] [CrossRef]
- Vy, H.M.T.; Coca, S.G.; Sawant, A.; Sakhuja, A.; Gutierrez, O.M.; Cooper, R.; Loos, R.J.; Horowitz, C.R.; Do, R.; Nadkarni, G.N. Genome-Wide Polygenic Risk Score for CKD in Individuals with APOL1 High-Risk Genotypes. Clin. J. Am. Soc. Nephrol. 2023, 19, 374–376. [Google Scholar] [CrossRef] [PubMed]
- Kopp, J.B.; Nelson, G.W.; Sampath, K.; Johnson, R.C.; Genovese, G.; An, P.; Friedman, D.; Briggs, W.; Dart, R.; Korbet, S.; et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-Associated nephropathy. J. Am. Soc. Nephrol. 2011, 22, 2129–2137. [Google Scholar] [CrossRef] [PubMed]
- Gbadegesin, R.A.; Ulasi, I.; Ajayi, S.; Raji, Y.; Olanrewaju, T.; Osafo, C.; Ademola, A.D.; Asinobi, A.; Winkler, C.A.; Burke, D.; et al. APOL1 bi- and monoallelic variants and chronic kidney disease in West Africans. N. Engl. J. Med. 2024, 392, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, D.B.; Shegokar, V.; Nihalani, D.; Rathore, Y.S.; Mallik, L.; Ashish, N.; Zare, V.; Ikizler, H.O.; Powar, R.; Holzman, L.B. APOL1 Null Alleles from a Rural Village in India Do Not Correlate with Glomerulosclerosis. PLoS ONE 2012, 7, e51546. [Google Scholar] [CrossRef]
- Wan, G.; Zhaorigetu, S.; Liu, Z.; Kaini, R.; Jiang, Z.; Hu, C.A. Apolipoprotein L1, a novel BCL-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J. Biol. Chem. 2008, 283, 21540–21549. [Google Scholar] [CrossRef]
- Ultsch, M.; Holliday, M.J.; Gerhardy, S.; Moran, P.; Scales, S.J.; Gupta, N.; Oltrabella, F.; Chiu, C.; Fairbrother, W.; Eigenbrot, C.; et al. Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif. Commun. Biol. 2021, 4, 916. [Google Scholar] [CrossRef]
- Heneghan, J.F.; Vandorpe, D.H.; Shmukler, B.E.; Giovinazzo, J.A.; Raper, J.; Friedman, D.J.; Pollak, M.R.; Alper, S.L. BH3 domain-independent apolipoprotein L1 toxicity rescued by BCL2 prosurvival proteins. AJP Cell Physiol. 2015, 309, C332–C347. [Google Scholar] [CrossRef]
- Lan, X.; Jhaveri, A.; Cheng, K.; Wen, H.; Saleem, M.A.; Mathieson, P.W.; Mikulak, J.; Aviram, S.; Malhotra, A.; Skorecki, K.; et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. AJP Ren. Physiol. 2014, 307, F326–F336. [Google Scholar] [CrossRef]
- Ma, L.; Chou, J.W.; Snipes, J.A.; Bharadwaj, M.S.; Craddock, A.L.; Cheng, D.; Weckerle, A.; Petrovic, S.; Hicks, P.J.; Hemal, A.K.; et al. APOL1 Renal-Risk variants induce mitochondrial dysfunction. J. Am. Soc. Nephrol. 2016, 28, 1093–1105. [Google Scholar] [CrossRef]
- Ma, L.; Ainsworth, H.C.; Snipes, J.A.; Murea, M.; Choi, Y.A.; Langefeld, C.D.; Parks, J.S.; Bharadwaj, M.S.; Chou, J.W.; Hemal, A.K.; et al. APOL1 Kidney-Risk variants induce mitochondrial fission. Kidney Int. Rep. 2020, 5, 891–904. [Google Scholar] [CrossRef]
- Shah, S.S.; Lannon, H.; Dias, L.; Zhang, J.; Alper, S.L.; Pollak, M.R.; Friedman, D.J. APOL1 kidney risk variants induce cell death via mitochondrial translocation and opening of the mitochondrial permeability transition pore. J. Am. Soc. Nephrol. 2019, 30, 2355–2368. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, S.M.; O’Toole, J.F.; Konieczkowski, M.; Barisoni, L.; Thomas, D.B.; Ganesan, S.; Bruggeman, L.A.; Buck, M.; Sedor, J.R. APOL1 variants change C-terminal conformational dynamics and binding to SNARE protein VAMP8. JCI Insight 2017, 2, e92581. [Google Scholar] [CrossRef]
- Kruzel-Davila, E.; Shemer, R.; Ofir, A.; Bavli-Kertselli, I.; Darlyuk-Saadon, I.; Oren-Giladi, P.; Wasser, W.G.; Magen, D.; Zaknoun, E.; Schuldiner, M.; et al. APOL1–Mediated cell injury involves disruption of conserved trafficking processes. J. Am. Soc. Nephrol. 2016, 28, 1117–1130. [Google Scholar] [CrossRef]
- Granado, D.; Müller, D.; Krausel, V.; Kruzel-Davila, E.; Schuberth, C.; Eschborn, M.; Wedlich-Söldner, R.; Skorecki, K.; Pavenstädt, H.; Michgehl, U.; et al. Intracellular APOL1 risk variants cause cytotoxicity accompanied by energy depletion. J. Am. Soc. Nephrol. 2017, 28, 3227–3238. [Google Scholar] [CrossRef] [PubMed]
- Gerstner, L.; Chen, M.; Kampf, L.L.; Milosavljevic, J.; Lang, K.; Schneider, R.; Hildebrandt, F.; Helmstädter, M.; Walz, G.; Hermle, T. Inhibition of endoplasmic reticulum stress signaling rescues cytotoxicity of human apolipoprotein-L1 risk variants in Drosophila. Kidney Int. 2022, 101, 1216–1231. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Kumar, V.; Lan, X.; Shoshtari, S.S.M.; Eng, J.M.; Zhou, X.; Wang, F.; Wang, H.; Skorecki, K.; Xing, G.; et al. APOL1 risk variants cause podocytes injury through enhancing endoplasmic reticulum stress. Biosci. Rep. 2018, 38, BSR20171713. [Google Scholar] [CrossRef]
- Thomson, R.; Finkelstein, A. Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: Relevance to trypanosome lysis. Proc. Natl. Acad. Sci. USA 2015, 112, 2894–2899. [Google Scholar] [CrossRef]
- Olabisi, O.A.; Zhang, J.; VerPlank, L.; Zahler, N.; DiBartolo, S.; Heneghan, J.F.; Schlöndorff, J.S.; Suh, J.H.; Yan, P.; Alper, S.L.; et al. APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc. Natl. Acad. Sci. USA 2015, 113, 830–837. [Google Scholar] [CrossRef]
- Datta, S.; Kataria, R.; Zhang, J.; Moore, S.; Petitpas, K.; Mohamed, A.; Zahler, N.; Pollak, M.R.; Olabisi, O.A. Kidney Disease-Associated APOL1 variants have Dose-Dependent, dominant toxic Gain-of-Function. J. Am. Soc. Nephrol. 2020, 31, 2083–2096. [Google Scholar] [CrossRef]
- Datta, S.; Antonio, B.M.; Zahler, N.H.; Theile, J.W.; Krafte, D.; Zhang, H.; Rosenberg, P.B.; Chaves, A.B.; Muoio, D.M.; Zhang, G.; et al. APOL1-mediated monovalent cation transport contributes to APOL1-mediated podocytopathy in kidney disease. J. Clin. Investig. 2024, 134, e172262. [Google Scholar] [CrossRef]
- Zimmerman, B.; Dakin, L.A.; Fortier, A.; Nanou, E.; Blasio, A.; Mann, J.; Miller, H.; Fletcher, M.; Wang, T.; Nanthakumar, S.; et al. Small molecule APOL1 inhibitors as a precision medicine approach for APOL1-mediated kidney disease. Nat. Commun. 2025, 16, 167. [Google Scholar] [CrossRef] [PubMed]
- Sironi, M.; Cagliani, R.; Forni, D.; Clerici, M. Evolutionary insights into host–pathogen interactions from mammalian sequence data. Nat. Rev. Genet. 2015, 16, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Limou, S.; Dummer, P.D.; Nelson, G.W.; Kopp, J.B.; Winkler, C.A. APOL1 toxin, innate immunity, and kidney injury. Kidney Int. 2015, 88, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Juliar, B.A.; Stanaway, I.B.; Sano, F.; Fu, H.; Smith, K.D.; Akilesh, S.; Scales, S.J.; Saghir, J.E.; Bhatraju, P.K.; Liu, E.; et al. Interferon-γ induces combined pyroptotic angiopathy and APOL1 expression in human kidney disease. Cell Rep. 2024, 43, 114310. [Google Scholar] [CrossRef]
- Sidaway, P. Innate immunity—APOL1 interaction. Nat. Rev. Nephrol. 2014, 10, 543. [Google Scholar] [CrossRef]
- Lannon, H.; Shah, S.S.; Dias, L.; Blackler, D.; Alper, S.L.; Pollak, M.R.; Friedman, D.J. Apolipoprotein L1 (APOL1) risk variant toxicity depends on the haplotype background. Kidney Int. 2019, 96, 1303–1307. [Google Scholar] [CrossRef]
- Gupta, N.; Waas, B.; Austin, D.; De Mazière, A.M.; Kujala, P.; Stockwell, A.D.; Li, T.; Yaspan, B.L.; Klumperman, J.; Scales, S.J. Apolipoprotein L1 (APOL1) renal risk variant-mediated podocyte cytotoxicity depends on African haplotype and surface expression. Sci. Rep. 2024, 14, 3765. [Google Scholar] [CrossRef]
- Khatua, A.K.; Cheatham, A.M.; Kruzel, E.D.; Singhal, P.C.; Skorecki, K.; Popik, W. Exon 4-encoded sequence is a major determinant of cytotoxicity of apolipoprotein L1. AJP Cell Physiol. 2015, 309, C22–C37. [Google Scholar] [CrossRef]
- Kruzel-Davila, E.; Bavli-Kertselli, I.; Ofir, A.; Cheatham, A.M.; Shemer, R.; Zaknoun, E.; Chornyy, S.; Tabachnikov, O.; Davis, S.E.; Khatua, A.K.; et al. Endoplasmic reticulum-translocation is essential for APOL1 cellular toxicity. iScience 2021, 25, 103717. [Google Scholar] [CrossRef]
- Uzureau, S.; Lecordier, L.; Uzureau, P.; Hennig, D.; Graversen, J.H.; Homblé, F.; Mfutu, P.E.; Arcolino, F.O.; Ramos, A.R.; La Rovere, R.M.; et al. APOL1 C-Terminal Variants May Trigger Kidney Disease through Interference with APOL3 Control of Actomyosin. Cell Rep. 2020, 30, 3821–3836.e13. [Google Scholar] [CrossRef]
- Lecordier, L.; Heo, P.; Graversen, J.H.; Hennig, D.; Skytthe, M.K.; D’Elzius, A.C.; Pincet, F.; Pérez-Morga, D.; Pays, E. Apolipoproteins L1 and L3 control mitochondrial membrane dynamics. Cell Rep. 2023, 42, 113528. [Google Scholar] [CrossRef]
- Keren, H.; Lev-Maor, G.; Ast, G. Alternative splicing and evolution: Diversification, exon definition and function. Nat. Rev. Genet. 2010, 11, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Wakashin, H.; Heymann, J.; Roshanravan, H.; Daneshpajouhnejad, P.; Rosenberg, A.; Shin, M.K.; Hoek, M.; Kopp, J.B. APOL1 renal risk variants exacerbate podocyte injury by increasing inflammatory stress. BMC Nephrol. 2020, 21, 371. [Google Scholar] [CrossRef]
- Müller, D.; Schmitz, J.; Fischer, K.; Granado, D.; Groh, A.; Krausel, V.; Lüttgenau, S.M.; Amelung, T.M.; Pavenstädt, H.; Weide, T. Evolution of Renal-Disease factor APOL1 results in cis and trans orientations at the endoplasmic reticulum that both show cytotoxic effects. Mol. Biol. Evol. 2021, 38, 4962–4976. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.; Orchard, S.; Magrane, M.; Adesina, A.; Ahmad, S.; Bowler-Barnett, E.H.; Bye-A-Jee, H.; Carpentier, D.; Denny, P.; et al. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2024, 53, D609–D617. [Google Scholar]
- Sauerer, T.; Lischer, C.; Weich, A.; Berking, C.; Vera, J.; Dörrie, J. Single-Molecule RNA sequencing reveals IFNΓ-Induced differential expression of immune escape genes in Merkel cell Polyomavirus–Positive MCC cell lines. Front. Microbiol. 2021, 12, 785662. [Google Scholar] [CrossRef]
- Warre-Cornish, K.; Perfect, L.; Nagy, R.; Duarte, R.R.R.; Reid, M.J.; Raval, P.; Mueller, A.; Evans, A.L.; Couch, A.; Ghevaert, C.; et al. Interferon-γ signaling in human iPSC–derived neurons recapitulates neurodevelopmental disorder phenotypes. Sci. Adv. 2020, 6, eaay9506. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Jang, I.K.; Lee, M.W.; Ko, Y.J.; Lee, D.; Lee, J.W.; Sung, K.W.; Koo, H.H.; Yoo, K.H. Enhanced immunosuppressive properties of human mesenchymal stem cells primed by interferon-Γ. EBioMedicine 2018, 28, 261–273. [Google Scholar] [CrossRef]
- Kim, Y.J.; Sheu, K.M.; Tsoi, J.; Abril-Rodriguez, G.; Medina, E.; Grasso, C.S.; Torrejon, D.Y.; Champhekar, A.S.; Litchfield, K.; Swanton, C.; et al. Melanoma dedifferentiation induced by IFN-γ epigenetic remodeling in response to anti–PD-1 therapy. J. Clin. Investig. 2021, 131, e145859. [Google Scholar] [CrossRef]
- Mikulak, J.; Oriolo, F.; Portale, F.; Tentorio, P.; Lan, X.; Saleem, M.A.; Skorecki, K.; Singhal, P.C.; Mavilio, D. Impact of APOL1 polymorphism and IL-1β priming in the entry and persistence of HIV-1 in human podocytes. Retrovirology 2016, 13, 63. [Google Scholar] [CrossRef]
- Cheatham, A.M.; Davis, S.E.; Khatua, A.K.; Popik, W. Blocking the 5′ splice site of exon 4 by a morpholino oligomer triggers APOL1 protein isoform switch. Sci. Rep. 2018, 8, 8739. [Google Scholar] [CrossRef]
- Cole, C.G.; McCann, O.T.; Collins, J.E.; Oliver, K.; Willey, D.; Gribble, S.M.; Yang, F.; McLaren, K.; Rogers, J.; Ning, Z.; et al. Finishing the finished human chromosome 22 sequence. Genome Biol. 2008, 9, R78. [Google Scholar] [CrossRef]
- Shukha, K.; Mueller, J.L.; Chung, R.T.; Curry, M.P.; Friedman, D.J.; Pollak, M.R.; Berg, A.H. Most APOL1 is secreted by the liver. J. Am. Soc. Nephrol. 2016, 28, 1079–1083. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. Biorxiv (Cold Spring Harb. Lab.) 2022. [Google Scholar] [CrossRef]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [PubMed]
- McCarthy, G.M.; Blasio, A.; Donovan, O.G.; Schaller, L.B.; Bock-Hughes, A.; Magraner, J.M.; Suh, J.H.; Tattersfield, C.F.; Stillman, I.E.; Shah, S.S.; et al. Recessive, gain-of-function toxicity in an APOL1 BAC transgenic mouse model mirrors human APOL1 kidney disease. Dis. Models Mech. 2021, 14, dmm048952. [Google Scholar] [CrossRef]
- Bruggeman, L.A.; Wu, Z.; Luo, L.; Madhavan, S.M.; Konieczkowski, M.; Drawz, P.E.; Thomas, D.B.; Barisoni, L.; Sedor, J.R.; O’Toole, J.F. APOL1-G0 or APOL1-G2 transgenic models develop preeclampsia but not kidney disease. J. Am. Soc. Nephrol. 2016, 27, 3600–3610. [Google Scholar] [CrossRef]
- Gupta, Y.; Friedman, D.J.; McNulty, M.T.; Khan, A.; Lane, B.; Wang, C.; Ke, J.; Jin, G.; Wooden, B.; Knob, A.L.; et al. Strong protective effect of the APOL1 p.N264K variant against G2-associated focal segmental glomerulosclerosis and kidney disease. Nat. Commun. 2023, 14, 7836. [Google Scholar] [CrossRef]
- Hung, A.M.; Assimon, V.A.; Chen, H.; Yu, Z.; Vlasschaert, C.; Triozzi, J.L.; Chan, H.; Wheless, L.; Wilson, O.; Shah, S.C.; et al. Genetic inhibition of APOL1 Pore-Forming function prevents APOL1-Mediated kidney disease. J. Am. Soc. Nephrol. 2023, 34, 1889–1899. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, B.; Lecordier, L.; Meehan, C.J.; Van Den Broeck, F.; Imamura, H.; Büscher, P.; Dujardin, J.; Laukens, K.; Schnaufer, A.; Dewar, C.; et al. Apolipoprotein L1 Variant Associated with Increased Susceptibility to Trypanosome Infection. mBio 2016, 7, e02198-15. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Reich, D.E.; Higgins, J.M.; Levine, H.Z.P.; Richter, D.J.; Schaffner, S.F.; Gabriel, S.B.; Platko, J.V.; Patterson, N.J.; McDonald, G.J.; et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 2002, 419, 832–837. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W. Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinform 2005, 21, 2531–2533. [Google Scholar] [CrossRef]
- Murrell, B.; Weaver, S.; Smith, M.D.; Wertheim, J.O.; Murrell, S.; Aylward, A.; Eren, K.; Pollner, T.; Martin, D.P.; Smith, D.M.; et al. Gene-Wide identification of episodic selection. Mol. Biol. Evol. 2015, 32, 1365–1371. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar]
- Skorecki, K.L.; Lee, J.H.; Langefeld, C.D.; Rosset, S.; Tzur, S.; Wasser, W.G.; Shemer, R.; Hawkins, G.A.; Divers, J.; Parekh, R.S.; et al. A null variant in the apolipoprotein L3 gene is associated with non-diabetic nephropathy. Nephrol. Dial. Transpl. 2016, 33, 323–330. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Levin, M.G.; Duda, J.T.; Landry, L.G.; Witschey, W.R.; Damrauer, S.M.; Ritchie, M.D.; Rader, D.J. Protein-truncating variant in APOL3 increases chronic kidney disease risk in epistasis with APOL1 risk alleles. JCI Insight 2024, 9, e181238. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, R.G.; Zhu, S.; Halder, A.; Kim, B.; Bradfield, C.J.; Huang, S.; Xu, D.; Mamiñska, A.; Nguyen, T.N.; Lazarou, M.; et al. A human apolipoprotein L with detergent-like activity kills intracellular pathogens. Science 2021, 373, e181238. [Google Scholar] [CrossRef]
- Pays, E. The Mechanism of Kidney Disease due to APOL1 Risk Variants. J. Am. Soc. Nephrol. 2024, 35, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.A.; O’Hare, M.J.; Reiser, J.; Coward, R.J.; Inward, C.D.; Farren, T.; Xing, C.Y.; Ni, L.; Mathieson, P.W.; Mundel, P. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 2002, 13, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Tzukerman, M.; Shamai, Y.; Abramovich, I.; Gottlieb, E.; Selig, S.; Skorecki, K. Comparative analysis of the APOL1 variants in the genetic landscape of renal carcinoma cells. Cancers 2022, 14, 733. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Pays, E. Apolipoprotein-L1 (APOL1): From sleeping sickness to kidney disease. Cells 2024, 13, 1738. [Google Scholar] [CrossRef]
- Egbuna, O.; Zimmerman, B.; Manos, G.; Fortier, A.; Chirieac, M.C.; Dakin, L.A.; Friedman, D.J.; Bramham, K.; Campbell, K.; Knebelmann, B.; et al. Inaxaplin for Proteinuric Kidney Disease in Persons with Two APOL1 Variants. N. Engl. J. Med. 2023, 388, 969–979. [Google Scholar] [CrossRef]
- O’Toole, J.F.; Schilling, W.; Kunze, D.; Madhavan, S.M.; Konieczkowski, M.; Gu, Y.; Luo, L.; Wu, Z.; Bruggeman, L.A.; Sedor, J.R. APOL1 overexpression drives Variant-Independent cytotoxicity. J. Am. Soc. Nephrol. 2017, 29, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Simeone, C.A.; McNulty, M.T.; Gupta, Y.; Genovese, G.; Sampson, M.G.; Sanna-Cherchi, S.; Friedman, D.J.; Pollak, M.R. The APOL1 p.N264K variant is co-inherited with the G2 kidney disease risk variant through a proximity recombination event. G3 Genes Genomes Genet. 2024, 15, jkae290. [Google Scholar] [CrossRef]
- Cooper, A.; Ilboudo, H.; Alibu, V.P.; Ravel, S.; Enyaru, J.; Weir, W.; Noyes, H.; Capewell, P.; Camara, M.; Milet, J.; et al. APOL1 renal risk variants have contrasting resistance and susceptibility associations with African trypanosomiasis. eLife 2017, 6, e25461. [Google Scholar] [CrossRef]
- Gonzales, G.A.; Huang, S.; Wilkinson, L.; Nguyen, J.A.; Sikdar, S.; Piot, C.; Naumenko, V.; Rajwani, J.; Wood, C.M.; Dinh, I.; et al. The pore-forming apolipoprotein APOL7C drives phagosomal rupture and antigen cross-presentation by dendritic cells. Sci. Immunol. 2024, 9, eadn2168. [Google Scholar] [CrossRef] [PubMed]
- Frith, M.C. Further varieties of ancient endogenous retrovirus in human DNA. Mob. DNA 2025, 16, 11. [Google Scholar] [CrossRef]
- Young, B.A.; Fullerton, S.M.; Wilson, J.G.; Cavanaugh, K.; Blacksher, E.; Spigner, C.; Himmelfarb, J.; Burke, W. Clinical Genetic Testing for APOL1: Are we There Yet? Semin. Nephrol. 2017, 37, 552–557. [Google Scholar] [CrossRef]

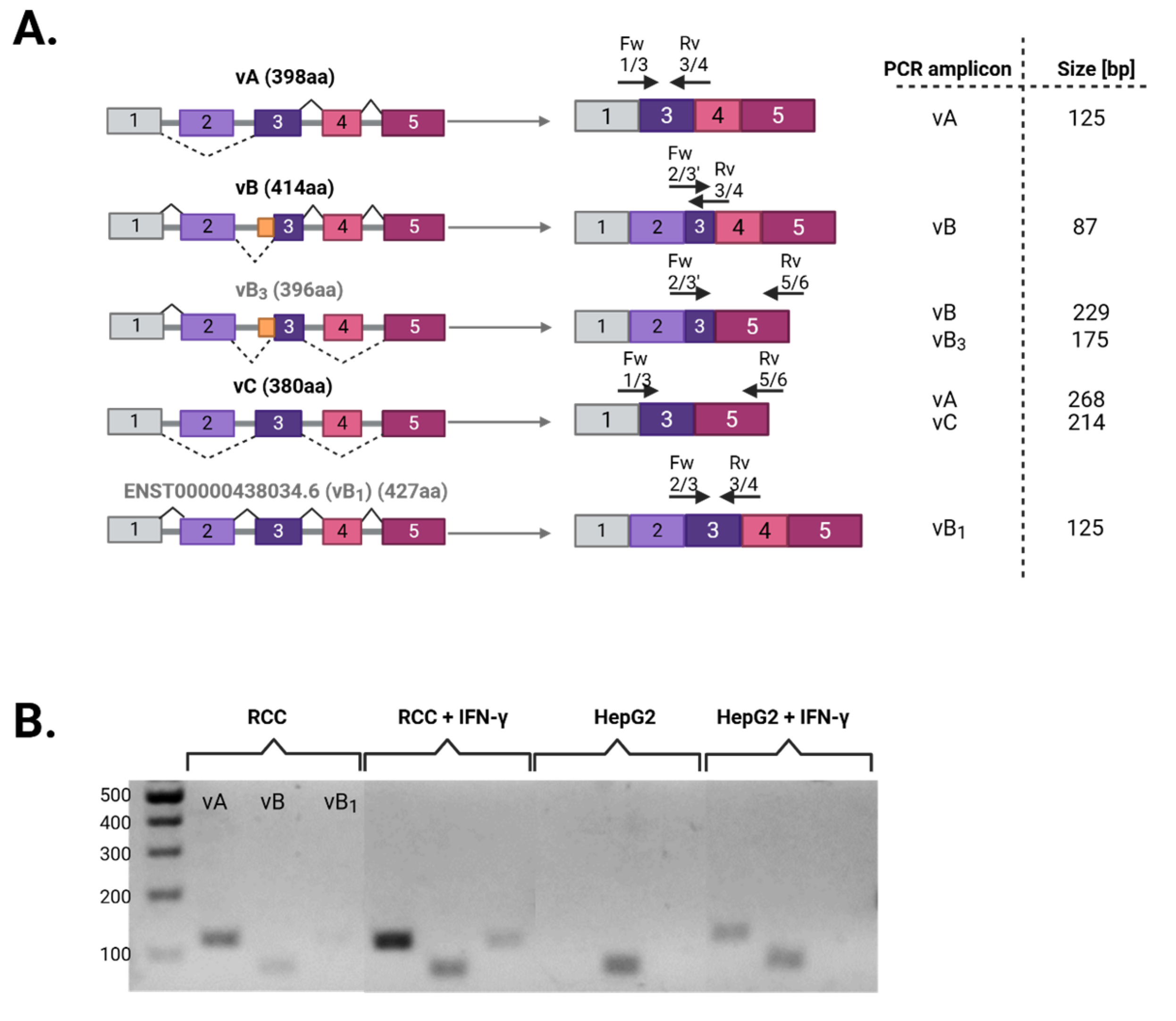
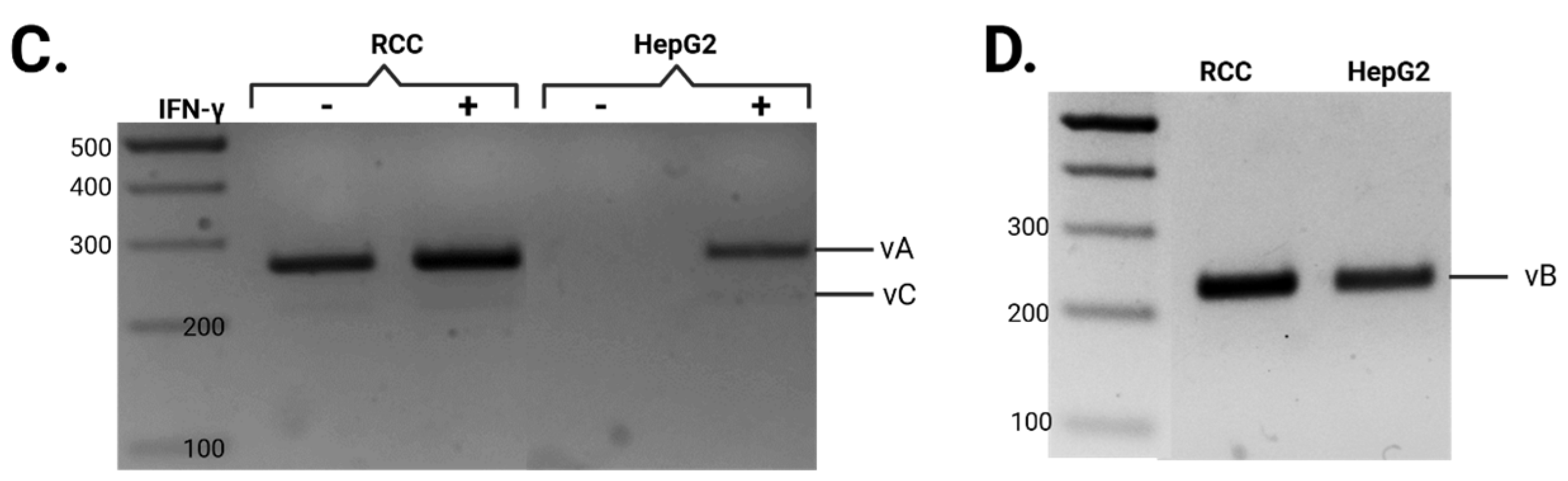





| # Primer | Sequence |
|---|---|
| APOL1 Fw 1/3 | GAAGATTCCTTGGAGGAGCACAC |
| APOL1 Fw 2/3′ | CACTGTGGAATTGAGGAGCACAC |
| APOL1 Fw 2/3 | CACTGTGGAATTGAGGAGGCCC |
| APOL1 Rv 3/4 | GTGCACTCATCCAGATGCAGAG |
| APOL1 Rv 5/6 | GATACTGCTCTCTGGGTCCATGG |
| APOL3 Fw 1/3 | GAGAAGCTTCCTTGAAAAGAAACGCTTTAC |
| APOL3 Rv 4 | CAAAACAAGACCAGCAAGGGAC |
| Amino Acid Position | 96 | 150 | 176 | 228 | 255 | 264 | 270 | 337 | 342 | 384 | 388–389 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Ref G0 | G | E | N | M | R | N | G | E | S | I | IN |
| 2 | G0 ‘EMK’ | G | E | N | M | K | N | G | E | S | I | IN |
| 3 | G0 ‘KIK’ | G | K | N | I | K | N | G | E | S | I | IN |
| 4 | G0 ‘KIK’ N264K | G | K | N | I | K | K | G | E | S | I | IN |
| 5 | G0 ‘KIK’ G96R | R | K | N | I | K | N | G | E | S | I | IN |
| 6 | G0 ‘EIK’ | G | E | N | I | K | N | G | E | S | I | IN |
| 7 | G0 ‘EIK’ G270D | G | E | N | I | K | N | D | E | S | I | IN |
| 8 | G0 ‘KIK’ G270D | G | K | N | I | K | N | D | E | S | I | IN |
| 9 | G0 ‘EIK’ D337N | G | E | N | I | K | N | G | N | S | I | IN |
| 10 | G0 ‘EIK’ N176S D337N | G | E | S | I | K | N | G | N | S | I | IN |
| 11 | Ref G1 | G | E | N | M | R | N | G | E | G | M | IN |
| 12 | G1 ‘EIK’ S342G | G | E | N | I | K | N | G | E | G | I | IN |
| 13 | G1 ‘EIK’ S342G I384M | G | E | N | I | K | N | G | E | G | M | IN |
| 14 | G1 ‘KIK’ S342G I384M | G | K | N | I | K | N | G | E | G | M | IN |
| 15 | Ref G2 | G | E | N | M | R | N | G | E | S | I | Del |
| 16 | G2 ‘EIK’ | G | E | N | I | K | N | G | E | S | I | Del |
| 17 | G2 ‘KIK’ N264K | G | K | N | I | K | K | G | E | S | I | Del |
| 18 | G2 ‘KIK’ | G | K | N | I | K | N | G | E | S | I | Del |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalaila, R.; Skorecki, K. Apolipoprotein L1 (APOL1): Consideration of Molecular Evolution, Interaction with APOL3, and Impact of Splice Isoforms Advances Understanding of Cellular and Molecular Mechanisms of Cell Injury. Cells 2025, 14, 1011. https://doi.org/10.3390/cells14131011
Khalaila R, Skorecki K. Apolipoprotein L1 (APOL1): Consideration of Molecular Evolution, Interaction with APOL3, and Impact of Splice Isoforms Advances Understanding of Cellular and Molecular Mechanisms of Cell Injury. Cells. 2025; 14(13):1011. https://doi.org/10.3390/cells14131011
Chicago/Turabian StyleKhalaila, Razi, and Karl Skorecki. 2025. "Apolipoprotein L1 (APOL1): Consideration of Molecular Evolution, Interaction with APOL3, and Impact of Splice Isoforms Advances Understanding of Cellular and Molecular Mechanisms of Cell Injury" Cells 14, no. 13: 1011. https://doi.org/10.3390/cells14131011
APA StyleKhalaila, R., & Skorecki, K. (2025). Apolipoprotein L1 (APOL1): Consideration of Molecular Evolution, Interaction with APOL3, and Impact of Splice Isoforms Advances Understanding of Cellular and Molecular Mechanisms of Cell Injury. Cells, 14(13), 1011. https://doi.org/10.3390/cells14131011







