Defining Keypoints to Align H&E Images and Xenium DAPI-Stained Images Automatically
Abstract
1. Introduction
2. Materials and Methods
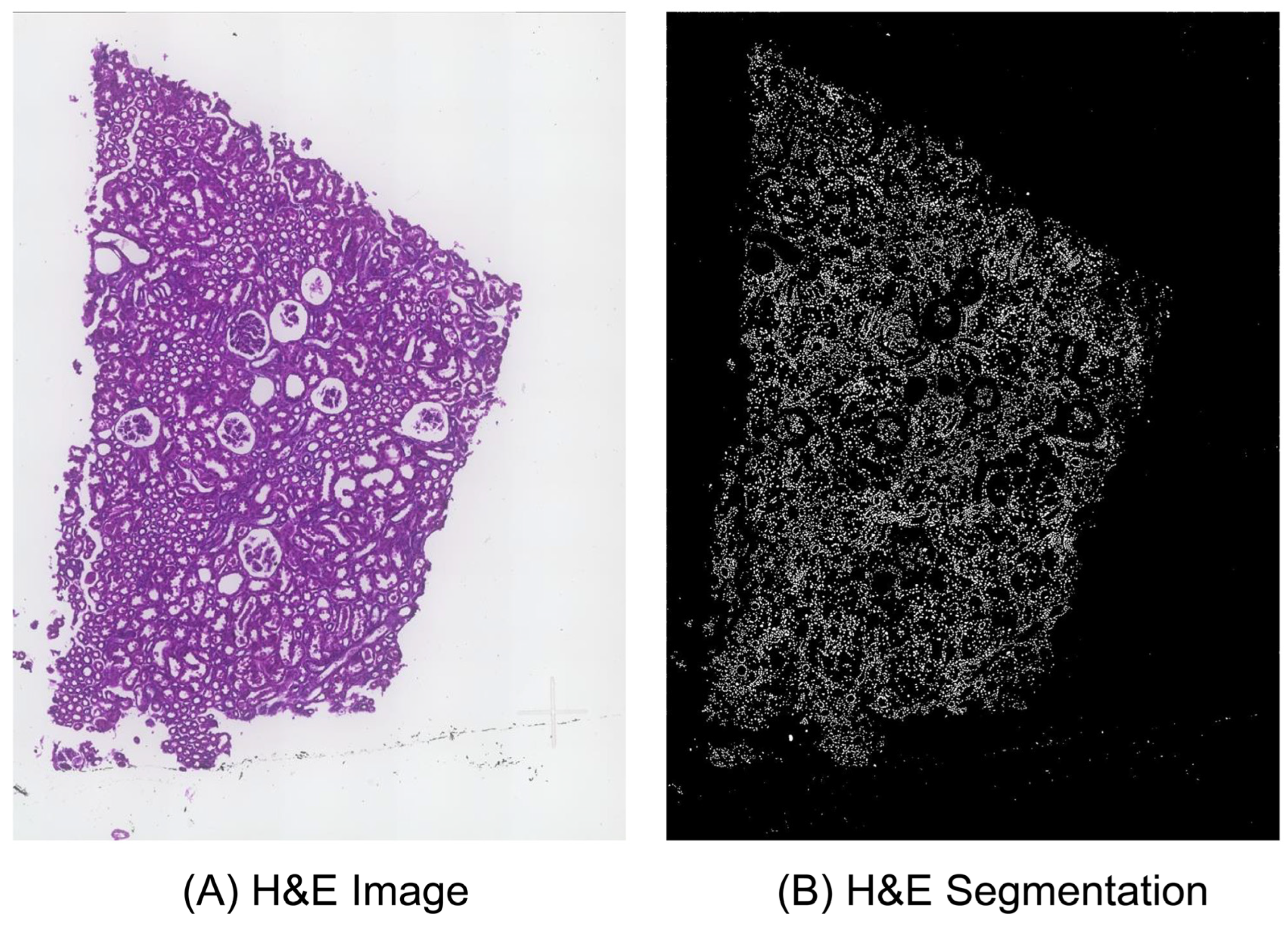
2.1. Image Processing
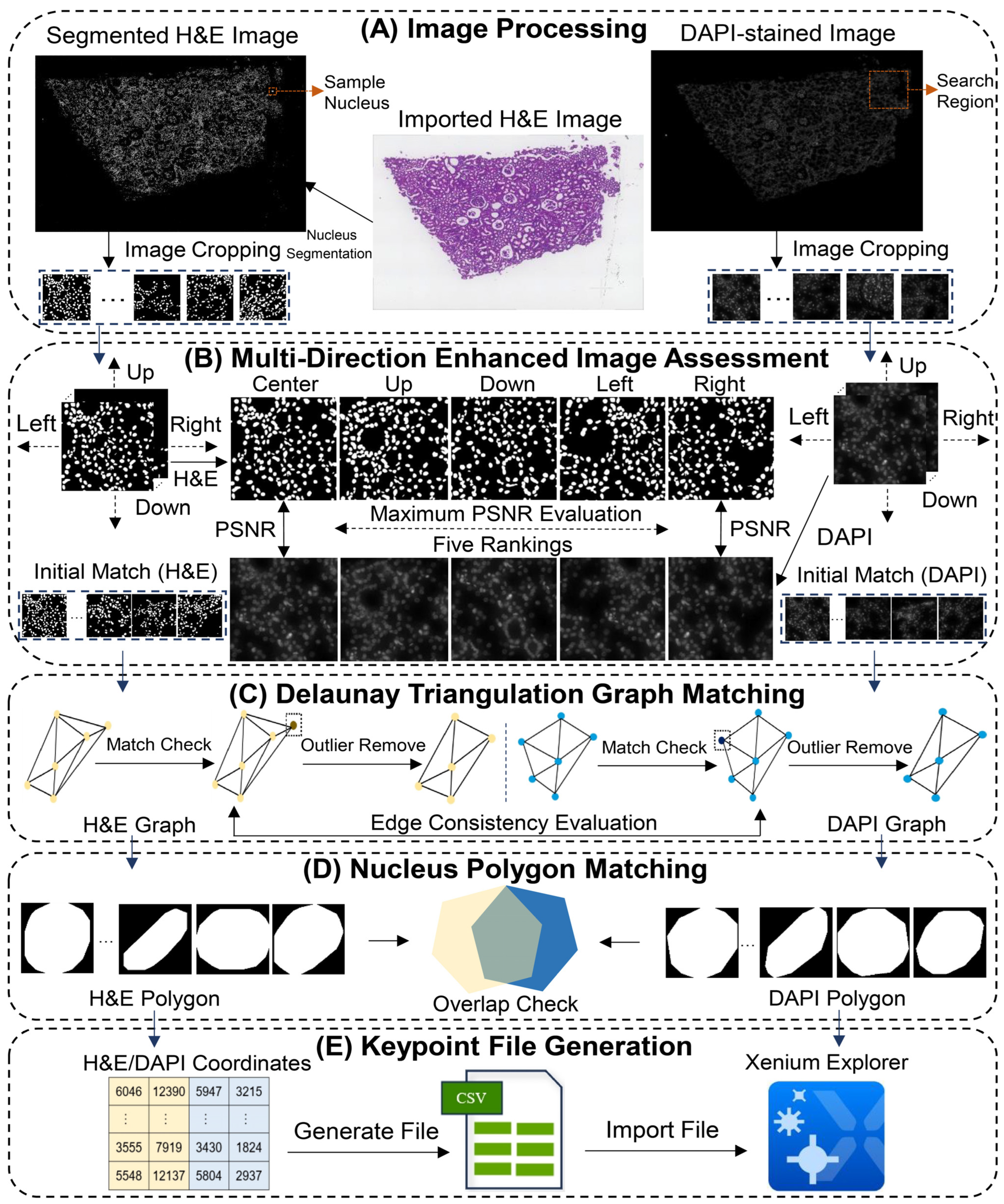
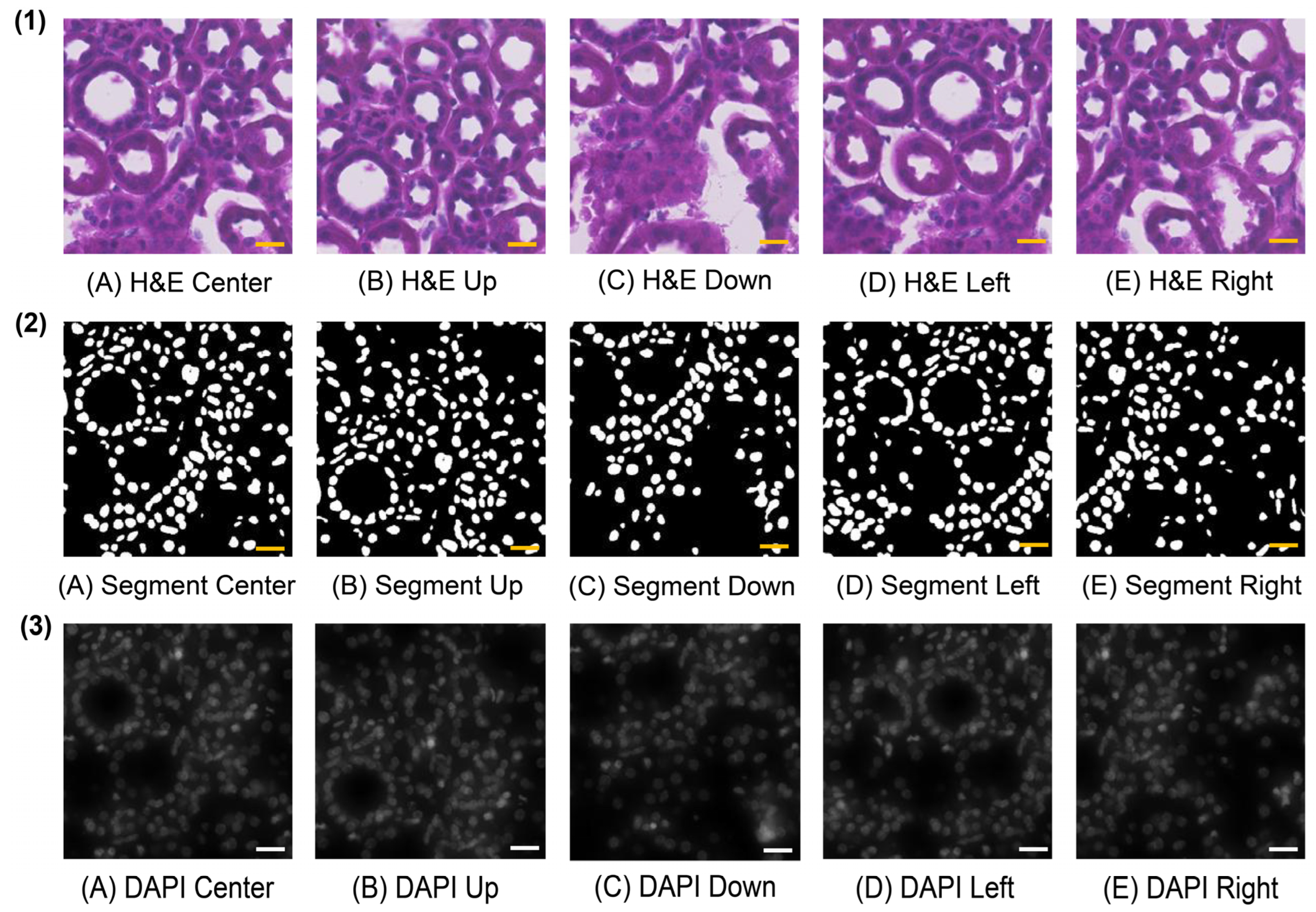
2.2. Multi-Directional Enhanced Image Assessment
2.3. Delaunay Triangulation Graph Matching
2.4. Nucleus Polygon Matching
2.5. Keypoint File Generation
2.6. Datasets and Experimental Settings
3. Results
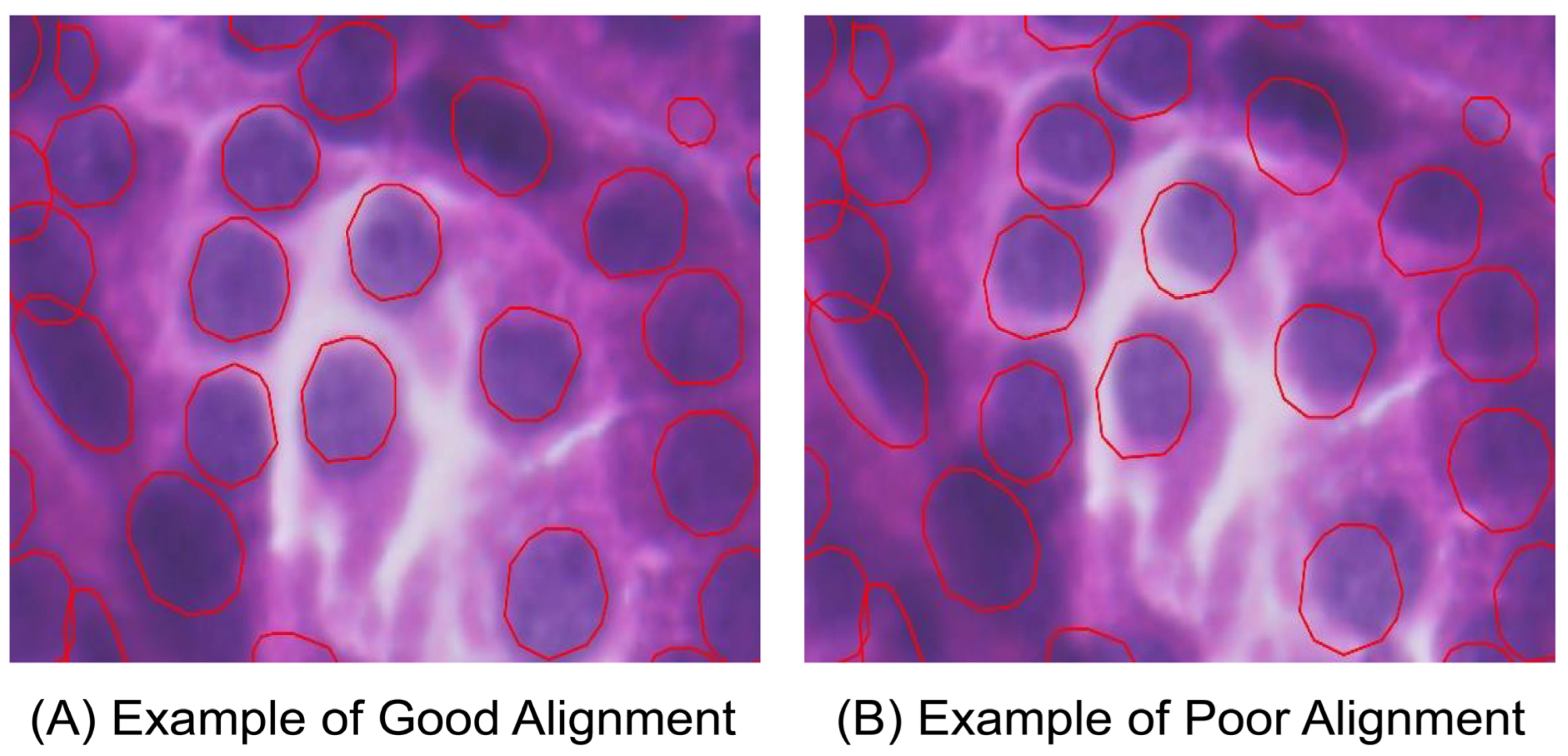
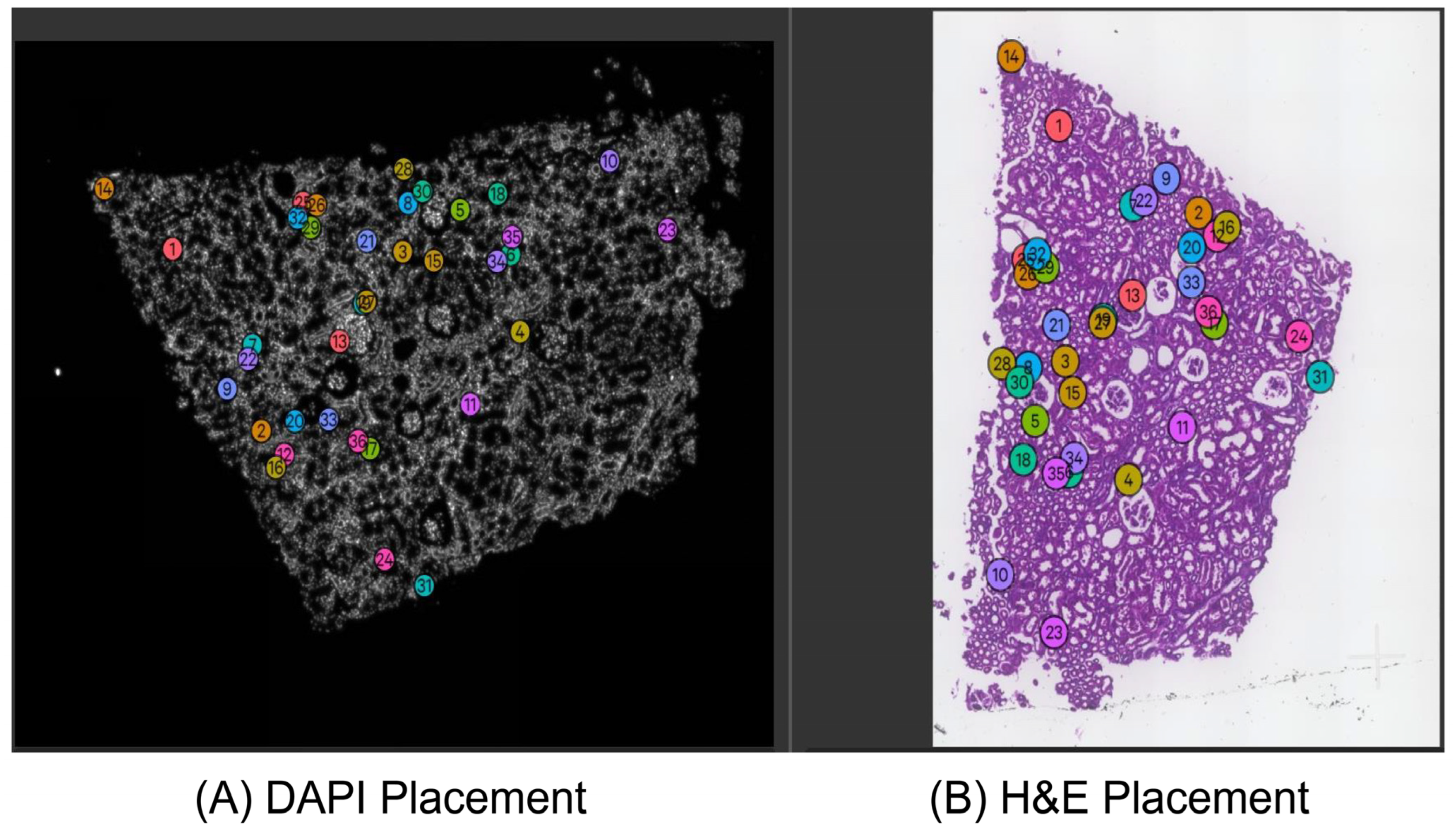
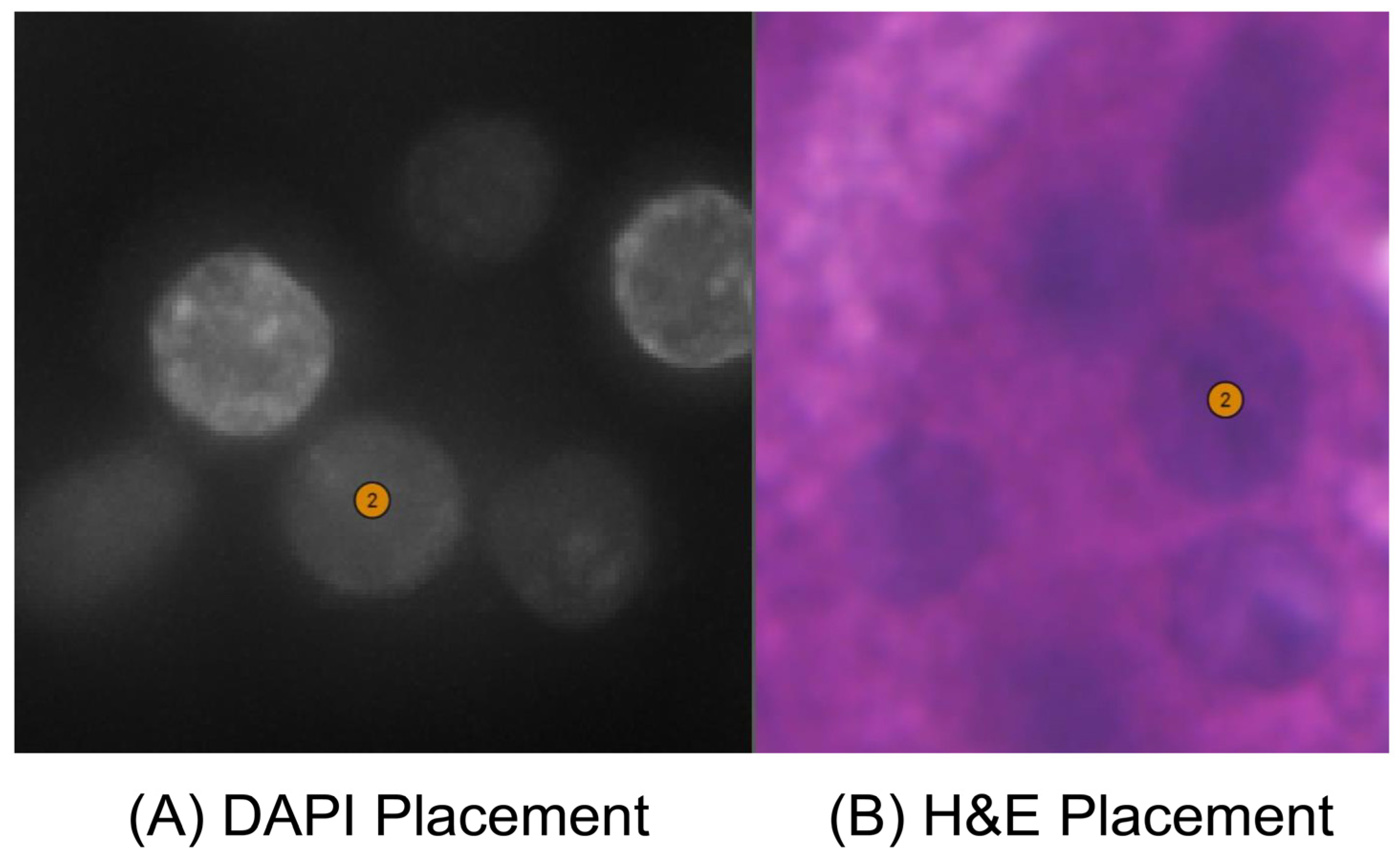
3.1. Image Alignment by Xenium-Align in Xenium Explorer Software
3.2. Description of Image Processing and Multi-Directional Enhanced Image Assessment
3.3. Effectiveness of the Delaunay Triangulation Graph and Nucleus Polygon Matching
3.4. Application of Xenium-Align to the New Test Dataset
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tian, L.; Chen, F.; Macosko, E.Z. The Expanding Vistas of Spatial Transcriptomics. Nat. Biotechnol. 2023, 41, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Moses, L.; Pachter, L. Museum of Spatial Transcriptomics. Nat. Methods 2022, 19, 534–546. [Google Scholar] [CrossRef]
- Rao, A.; Barkley, D.; França, G.S.; Yanai, I. Exploring Tissue Architecture Using Spatial Transcriptomics. Nature 2021, 596, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.P.; Jensen, K.B.; Wise, K.; Roach, M.J.; Dezem, F.S.; Ryan, N.K.; Zamojski, M.; Vlachos, I.S.; Knott, S.R.V.; Butler, L.M.; et al. A Comparative Analysis of Imaging-Based Spatial Transcriptomics Platforms. bioRxiv 2023. [Google Scholar] [CrossRef]
- Janesick, A.; Shelansky, R.; Gottscho, A.D.; Wagner, F.; Williams, S.R.; Rouault, M.; Beliakoff, G.; Morrison, C.A.; Oliveira, M.F.; Sicherman, J.T.; et al. 10x Development Teams. High Resolution Mapping of the Tumor Microenvironment Using Integrated Single-Cell, Spatial and In Situ Analysis. Nat. Commun. 2023, 14, 8353. [Google Scholar] [CrossRef]
- Cervilla, S.; Grases, D.; Perez, E.; Real, F.X.; Musulen, E.; Esteller, M.; Porta-Pardo, E. Benchmarking of spatial transcriptomics platforms across six cancer types. bioRxiv 2025. [Google Scholar] [CrossRef]
- Watson, B.R.; Paul, B.; Rahman, R.U.; Amir-Zilberstein, L.; Segerstolpe, Å.; Epstein, E.T.; Murphy, S.; Geistlinger, L.; Lee, T.; Shih, A.; et al. Spatial Transcriptomics of Healthy and Fibrotic Human Liver at Single-Cell Resolution. Nat. Commun. 2025, 16, 319. [Google Scholar] [CrossRef]
- Isnard, P.; Humphreys, B.D. Spatial Transcriptomics: Integrating Morphology and Molecular Mechanisms of Kidney Diseases. Am. J. Pathol. 2025, 195, 23–39. [Google Scholar] [CrossRef]
- Gulati, G.S.; D’Silva, J.P.; Liu, Y.; Wang, L.; Newman, A.M. Profiling Cell Identity and Tissue Architecture with Single-Cell and Spatial Transcriptomics. Nat. Rev. Mol. Cell Biol. 2025, 26, 11–31. [Google Scholar] [CrossRef]
- Takano, Y.; Suzuki, J.; Nomura, K.; Fujii, G.; Zenkoh, J.; Kawai, H.; Kuze, Y.; Kashima, Y.; Nagasawa, S.; Nakamura, Y.; et al. Spatially Resolved Gene Expression Profiling of Tumor Microenvironment Reveals Key Steps of Lung Adenocarcinoma Development. Nat. Commun. 2024, 15, 10637–10654. [Google Scholar] [CrossRef]
- Nagasawa, S.; Zenkoh, J.; Suzuki, Y.; Suzuki, A. Spatial Omics Technologies for Understanding Molecular Status Associated with Cancer Progression. Cancer Sci. 2024, 115, 3208–3217. [Google Scholar] [CrossRef] [PubMed]
- Mennillo, E.; Lotstein, M.L.; Lee, G.; Johri, V.; Ekstrand, C.; Tsui, J.; Hou, J.; Leet, D.E.; He, J.Y.; Mahadevan, U.; et al. Single-Cell Spatial Transcriptomics of Fixed, Paraffin-Embedded Biopsies Reveals Colitis-Associated Cell Networks. bioRxiv 2024. [Google Scholar] [CrossRef]
- Lee, H.; Kim, S.Y.; Kwon, N.J.; Jo, S.J.; Kwon, O.; Kim, J.I. Single-Cell and Spatial Transcriptome Analysis of Dermal Fibroblast Development in Perinatal Mouse Skin: Dynamic Lineage Differentiation and Key Driver Genes. J. Investig. Dermatol. 2024, 144, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Xiao, J.; Tam, S.S.T.; Cai, M.; Sugimura, R.; Wang, Y.; Wan, X.; Lin, Z.; Wu, A.R.; Yang, C. Integrating Spatial and Single-Cell Transcriptomics Data Using Deep Generative Models with SpatialScope. Nat. Commun. 2023, 14, 7848–7869. [Google Scholar] [CrossRef]
- Gogoberidze, N.; Cimini, B.A. Defining the Boundaries: Challenges and Advances in Identifying Cells in Microscopy Images. Curr. Opin. Biotechnol. 2024, 85, 103055. [Google Scholar] [CrossRef]
- Liu, P.; Li, J.; Chang, J.; Hu, P.; Sun, Y.; Jiang, Y.; Zhang, F.; Shao, H. Software Tools for 2D Cell Segmentation. Cells 2024, 13, 352. [Google Scholar] [CrossRef]
- Stringer, C.; Wang, T.; Michaelos, M.; Pachitariu, M. Cellpose: A Generalist Algorithm for Cellular Segmentation. Nat. Methods 2021, 18, 100–106. [Google Scholar] [CrossRef]
- Schmidt, U.; Weigert, M.; Broaddus, C.; Myers, G. Cell Detection with Star-Convex Polygons. Med. Image Comput. Comput. Assist. Interv. (MICCAI) 2018, 11071, 265–273. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Med. Image Comput. Comput.-Assist. Interv. (MICCAI) Pt. III 2015, 9351, 234–241. [Google Scholar]
- Bannon, D.; Moen, E.; Schwartz, M.; Borba, E.; Kudo, T.; Greenwald, N.; Vijayakumar, V.; Chang, B.; Pao, E.; Osterman, E.; et al. DeepCell Kiosk: Scaling Deep Learning–Enabled Cellular Image Analysis with Kubernetes. Nat. Methods 2021, 18, 43–45. [Google Scholar] [CrossRef]
- Palla, G.; Spitzer, H.; Klein, M.; Fischer, D.; Schaar, A.C.; Kuemmerle, L.B.; Rybakov, S.; Ibarra, I.L.; Holmberg, O.; Virshup, I.; et al. Squidpy: A Scalable Framework for Spatial Omics Analysis. Nat. Methods 2022, 19, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Thung, K.; Raveendran, P. A Survey of Image Quality Measures. In Proceedings of the 2009 International Conference for Technical Postgraduates (TECHPOS), Kuala Lumpur, Malaysia, 14–15 December 2009; pp. 1–4. [Google Scholar]
- Horeé, A.; Ziou, D. Image Quality Metrics: PSNR vs SSIM. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 2366–2369. [Google Scholar]
- Palubinskas, G. Mystery Behind Similarity Measures MSE and SSIM. In Proceedings of the 2014 IEEE International Conference on Image Processing (ICIP), Paris, France, 27–30 October 2014; pp. 575–579. [Google Scholar]
- Huynh-Thu, Q.; Ghanbari, M. Scope of Validity of PSNR in Image/Video Quality Assessment. Electron. Lett. 2008, 44, 800–801. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image Quality Assessment: From Error Visibility to Structural Similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.L.; Li, Z.; Tan, Y.H.; Rahardja, S.; Yeo, C. A Perceptually Relevant MSE-Based Image Quality Metric. IEEE Trans. Image Process. 2013, 22, 4447–4459. [Google Scholar]
- Horn, B.K.P. Closed-Form Solution of Absolute Orientation Using Unit Quaternions. J. Opt. Soc. Am. A 1987, 4, 629–642. [Google Scholar] [CrossRef]
- 10X Genomics Company. Image Alignment in Xenium Explorer. Available online: https://www.10xgenomics.com/cn/support/software/xenium-explorer/latest/tutorials/xe-image-alignment (accessed on 24 June 2025).
- Ing, G.; Stewart, A.; Battaglia, G.; Ruiz-Perez, L. SimpliPyTEM: An Open-Source Python Library and App to Simplify Transmission Electron Microscopy and In Situ-TEM Image Analysis. PLoS ONE 2023, 18, e0285691. [Google Scholar] [CrossRef]
- Marconato, L.; Palla, G.; Yamauchi, K.A.; Virshup, I.; Heidari, E.; Treis, T.; Vierdag, W.; Toth, M.; Stockhaus, S.; Shrestha, R.B.; et al. SpatialData: An Open and Universal Data Framework for Spatial Omics. Nat. Methods 2025, 22, 58–62. [Google Scholar] [CrossRef]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Lin, Y.; Liang, Y.; Wang, D.; Chang, Y.; Ma, Q.; Wang, Y.; He, F.; Xu, D. A Contrastive Learning Approach to Integrate Spatial Transcriptomics and Histological Images. Comput. Struct. Biotechnol. J. 2024, 23, 1786–1795. [Google Scholar] [CrossRef]
- Elshakhs, Y.S.; Deliparaschos, K.M.; Charalambous, T.; Oliva, G.; Zolotas, A. A Comprehensive Survey on Delaunay Triangulation: Applications, Algorithms, and Implementations over CPUs, GPUs, and FPGAs. IEEE Access 2024, 12, 12562–12585. [Google Scholar] [CrossRef]
- Li, X.; Calinescu, G.; Wan, P.; Wang, Y. Localized Delaunay Triangulation with Application in Ad Hoc Wireless Networks. IEEE Trans. Parallel Distrib. Syst. 2003, 14, 1035–1047. [Google Scholar]
- Lee, D.T.; Schachter, B.J. Two Algorithms for Constructing a Delaunay Triangulation. Int. J. Comput. Inf. Sci. 1980, 9, 219–242. [Google Scholar] [CrossRef]
- Wu, Z.; Trevino, A.E.; Wu, E.; Swanson, K.; Kim, H.J.; D’Angio, H.B.; Zou, J. Graph Deep Learning for the Characterization of Tumour Microenvironments from Spatial Protein Profiles in Tissue Specimens. Nat. Biomed. Eng. 2022, 6, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Lertrattanapanich, S.; Bose, N.K. High Resolution Image Formation from Low Resolution Frames Using Delaunay Triangulation. IEEE Trans. Image Process. 2002, 11, 1427–1441. [Google Scholar] [CrossRef]
- De Floriani, L.; Magillo, P.; Puppo, E. Applications of Computational Geometry to Geographic Information Systems. In Handbook of Computational Geometry; Sack, J.-R., Urrutia, J., Eds.; Elsevier Science & Technology: Amsterdam, The Netherlands, 1999; pp. 333–388. [Google Scholar]
- Zhao, M.; An, B.; Wu, Y.; Chen, B.; Sun, S. A Robust Delaunay Triangulation Matching for Multispectral/Multidate Remote Sensing Image Registration. IEEE Geosci. Remote Sens. Lett. 2015, 12, 711–715. [Google Scholar] [CrossRef]
- Jiang, S.; Jiang, W. Reliable Image Matching via Photometric and Geometric Constraints Structured by Delaunay Triangulation. ISPRS J. Photogramm. Remote Sens. 2019, 153, 1–20. [Google Scholar] [CrossRef]
- Wu, Z.; Trevino, A.E.; Wu, E.; Swanson, K.; Kim, H.J.; D’Angio, H.B.; Preska, R.; Charville, G.W.; Dalerba, P.D.; Egloff, A.M. SPACE-GM: Geometric Deep Learning of Disease-Associated Microenvironments from Multiplex Spatial Protein Profiles. bioRxiv 2022. [Google Scholar] [CrossRef]
- Chen, S.; Ding, C.; Liu, M.; Cheng, J.; Tao, D. CPP-Net: Context-Aware Polygon Proposal Network for Nucleus Segmentation. IEEE Trans. Image Process. 2023, 32, 980–994. [Google Scholar] [CrossRef]
- Weigert, M.; Schmidt, U.; Haase, R.; Sugawara, K.; Myers, G. Star-Convex Polyhedra for 3D Object Detection and Segmentation in Microscopy. In Proceedings of the 2020 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Snowmass Village, CO, USA, 1–5 March 2020; IEEE: New York, NY, USA, 2020; pp. 3666–3673. [Google Scholar]
- Barber, C.B.; Dobkin, D.P.; Huhdanpaa, H. The Quickhull Algorithm for Convex Hulls. ACM Trans. Math. Softw. 1996, 22, 469–483. [Google Scholar] [CrossRef]
- Mirzazadeh, R.; Andrusivova, Z.; Larsson, L.; Newton, P.T.; Galicia, L.A.; Abalo, X.M.; Avijgan, M.; Kvastad, L.; Denadai-Souza, A.; Stakenborg, N.; et al. Spatially Resolved Transcriptomic Profiling of Degraded and Challenging Fresh Frozen Samples. Nat. Commun. 2023, 14, 509. [Google Scholar] [CrossRef]
- Dannhorn, A.; Kazanc, E.; Flint, L.; Guo, F.; Carter, A.; Hall, A.R.; Jones, S.A.; Poulogiannis, G.; Barry, S.T.; Sansom, O.J.; et al. Morphological and Molecular Preservation through Universal Preparation of Fresh-Frozen Tissue Samples for Multimodal Imaging Workflows. Nat. Protoc. 2024, 19, 2685–2711. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Zhang, D.; Gao, Y.; Tao, B.; Zhang, D.; Bao, S.; Enninful, A.; Wang, Y.; Li, H.; Su, G.; et al. Spatially Exploring RNA Biology in Archival Formalin-Fixed Paraffin-Embedded Tissues. Cell 2024, 187, 6760–6779.e24. [Google Scholar] [CrossRef] [PubMed]
- Wehrle, E.; Günther, D.; Mathavan, N.; Singh, A.; Müller, R. Protocol for Preparing Formalin-Fixed Paraffin-Embedded Musculoskeletal Tissue Samples from Mice for Spatial Transcriptomics. Star Protoc. 2024, 5, 102986. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, H.; Arikawa, K.; Yamazaki, M.; Wagatsuma, R.; Ide, K.; Samuel, A.Z.; Takamochi, K.; Suzuki, K.; Hayashi, T.; Hosokawa, M.; et al. Reproducible and Sensitive Micro-Tissue RNA Sequencing from Formalin-Fixed Paraffin-Embedded Tissues for Spatial Gene Expression Analysis. Sci. Rep. 2022, 12, 19511. [Google Scholar] [CrossRef]
- Lou, W.; Wan, X.; Li, G.; Lou, X.; Li, C.; Gao, F.; Li, H. Structure Embedded Nucleus Classification for Histopathology Images. IEEE Trans. Med. Imaging 2024, 43, 3149–3160. [Google Scholar] [CrossRef]
| Sample | Nucleus Segmentation | DAPI Search | Evaluation and Matching | Run Time | Keypoint Number |
|---|---|---|---|---|---|
| F59 | Cellpose Model | crop_radius_ratio = 0.125, extracted_region_min = 50 | crop_radius_pixel = 400, center_move_pixel = 300, cell_num_each_epoch = 100, overlap_ave_threshold = 0.9, keypoints_min = 15 | 4.915 | 36 |
| 20012 | 3.397 | 24 | |||
| 26429 | 22.949 | 18 | |||
| 36816 | 3.543 | 17 | |||
| 3723 | 20.949 | 18 | |||
| 3775 | StarDist Model | crop_radius_ratio = 0.06, extracted_region_min = 50 | 26.197 | 19 | |
| 3781 | Cellpose Model | crop_radius_ratio = 0.125, extracted_region_min = 50 | 22.854 | 16 | |
| 38111 | StarDist Model | crop_radius_ratio = 0.06, extracted_region_min = 50 | 36.425 | 16 | |
| 40440 | Cellpose Model | crop_radius_ratio = 0.5, extracted_region_min = 50 | 15.253 | 21 | |
| 40610 | Cellpose Model | crop_radius_ratio = 0.06, extracted_region_min = 50 | 8.749 | 34 | |
| 40775 | crop_radius_ratio = 0.5, extracted_region_min = 50 | 21.559 | 22 | ||
| 5582 | 8.151 | 16 |
| Sample | F59 | 20012 | 26429 | 36816 | 3723 | 3775 |
| N_Keypoints | 36 | 24 | 18 | 17 | 18 | 19 |
| N_Accuate | 35 | 24 | 18 | 17 | 18 | 19 |
| N_Threshold | 11 (0.3) | 2 (0.1) | 5 (0.3) | 5 (0.3) | 5 (0.3) | 6 (0.3) |
| Sample | 3781 | 38111 | 40440 | 40610 | 40775 | 5582 |
| N_Keypoints | 16 | 16 | 21 | 34 | 22 | 16 |
| N_Accuate | 15 | 16 | 21 | 34 | 22 | 16 |
| N_Threshold | 3 (0.2) | 3 (0.2) | 4 (0.2) | 10 (0.3) | 4 (0.2) | 5 (0.3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.; Wang, Y.; Wang, J.; Raina, M.; Ferreira, R.M.; Eadon, M.T.; Liang, Y.; Xu, D. Defining Keypoints to Align H&E Images and Xenium DAPI-Stained Images Automatically. Cells 2025, 14, 1000. https://doi.org/10.3390/cells14131000
Lin Y, Wang Y, Wang J, Raina M, Ferreira RM, Eadon MT, Liang Y, Xu D. Defining Keypoints to Align H&E Images and Xenium DAPI-Stained Images Automatically. Cells. 2025; 14(13):1000. https://doi.org/10.3390/cells14131000
Chicago/Turabian StyleLin, Yu, Yan Wang, Juexin Wang, Mauminah Raina, Ricardo Melo Ferreira, Michael T. Eadon, Yanchun Liang, and Dong Xu. 2025. "Defining Keypoints to Align H&E Images and Xenium DAPI-Stained Images Automatically" Cells 14, no. 13: 1000. https://doi.org/10.3390/cells14131000
APA StyleLin, Y., Wang, Y., Wang, J., Raina, M., Ferreira, R. M., Eadon, M. T., Liang, Y., & Xu, D. (2025). Defining Keypoints to Align H&E Images and Xenium DAPI-Stained Images Automatically. Cells, 14(13), 1000. https://doi.org/10.3390/cells14131000






