Clinical Relevance and Interplay between miRNAs in Influencing Glioblastoma Multiforme Prognosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Histological and Molecular Analysis
2.2. MGMT Promoter Methylation
2.3. MGMT Immunohistochemistry
2.4. miRNA Evaluation
2.5. Statistical Analyses
3. Results
3.1. Clinical–Pathological Characteristics of the Cohort and Molecular Data
3.2. Molecular Data: MGMT Promoter Methylation, MGMT IHC, Single-miRNA Expression and Their Association with Survival
3.3. Association between miRNAs
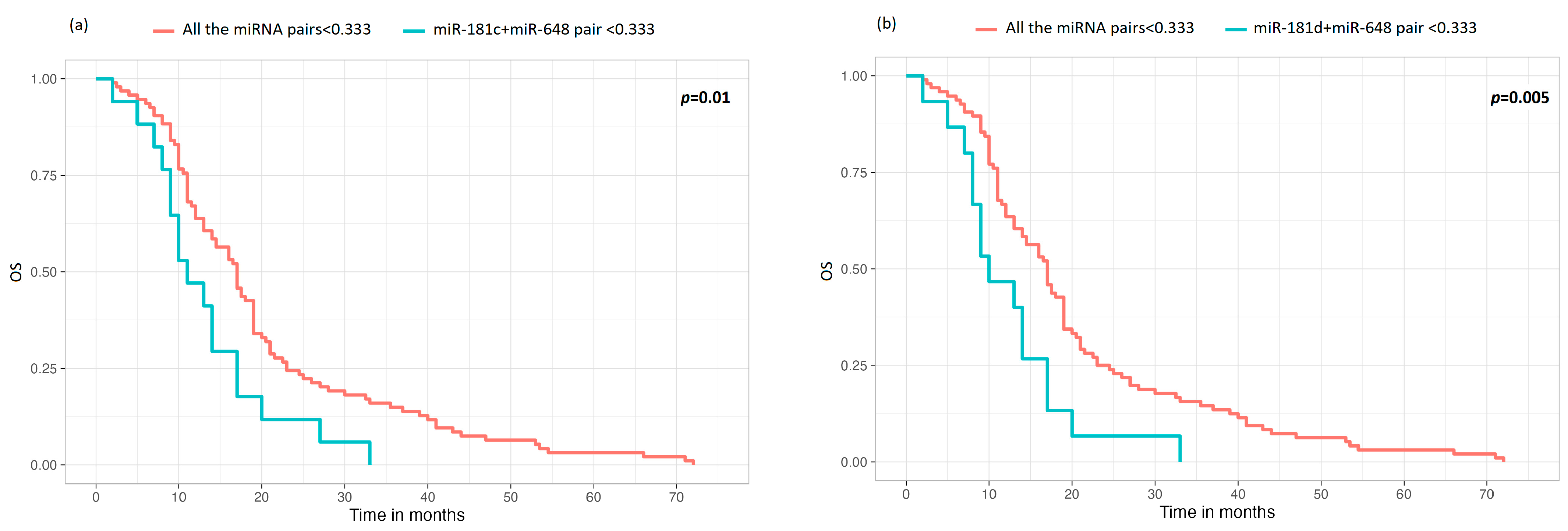
3.4. Comparison of miRNA Pairs with the Group of the Other Four miRNAs in Terms of OS and TTP
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme-Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef]
- Pace, A.; Tanzilli, A.; Benincasa, D. Prognostication in brain tumors. Handb. Clin. Neurol. 2022, 190, 149–161. [Google Scholar]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Stummer, W.; Reulen, H.J.; Meinel, T.; Pichlmeier, U.; Schumacher, W.; Tonn, J.C.; Rohde, V.; Oppel, F.; Turowski, B.; Woiciechowsky, C.; et al. Extent of resection and survival in glioblastoma multiforme: Identification of and adjustment for bias. Neurosurgery 2008, 62, 564–576. [Google Scholar] [CrossRef]
- Wang, L.; Liang, B.; Li, Y.I.; Liu, X.; Huang, J.; Li, Y.M. What is the advance of extent of resection in glioblastoma surgical treatment-a systematic review. Chin. Neurosurg. J. 2019, 5, 2. [Google Scholar] [CrossRef]
- Marchi, F.; Sahnane, N.; Cerutti, R.; Cipriani, D.; Barizzi, J.; Stefanini, F.M.; Epistolio, S.; Cerati, M.; Balbi, S.; Mazzucchelli, L.; et al. The Impact of Surgery in IDH 1 Wild Type Glioblastoma in Relation With the MGMT Deregulation. Front. Oncol. 2020, 9, 1569. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Hamilton, S.R.; Burger, P.C.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA repair geneO6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999, 59, 793–797. [Google Scholar] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Everhard, S.; Tost, J.; El Abdalaoui, H.; Crinière, E.; Busato, F.; Marie, Y.; Gut, I.G.; Sanson, M.; Mokhtari, K.; Laigle-Donadey, F.; et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009, 11, 348–356. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Kushwaha, D.; Koay, D.C.; Reddy, H.; Mao, Y.; Zhou, L.; Ng, K.; Zinn, P.; Carter, B.; Chen, C.C. Post-transcriptional regulation of O(6)- methylguanine-DNA methyltransferase MGMT in glioblastomas. Cancer Biomark. 2011, 10, 185–193. [Google Scholar] [CrossRef]
- Kreth, S.; Limbeck, E.; Hinske, L.C.; Schütz, S.V.; Thon, N.; Hoefig, K.; Egensperger, R.; Kreth, F.W. In human glioblastomas transcript elongation by alternative polyadenylation and miRNA targeting is a potent mechanism of MGMT silencing. Acta Neuropathol. 2013, 125, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Gareev, I.; Beylerli, O.; Liang, Y.; Xiang, H.; Liu, C.; Xu, X.; Yuan, C.; Ahmad, A.; Yang, G. The Role of MicroRNAs in Therapeutic Resistance of Malignant Primary Brain Tumors. Front. Cell Dev. Biol. 2021, 9, 740303. [Google Scholar] [CrossRef]
- Olivier, C.; Oliver, L.; Lalier, L.; Vallette, F.M. Drug Resistance in Glioblastoma: The Two Faces of Oxidative Stress. Front. Mol. Biosci. 2021, 7, 620677. [Google Scholar] [CrossRef]
- Sati, I.S.E.E.; Parhar, I. MicroRNAs Regulate Cell Cycle and Cell Death Pathways in Glioblastoma. Int. J. Mol. Sci. 2021, 22, 13550. [Google Scholar] [CrossRef]
- Ahmed, S.P.; Castresana, J.S.; Shahi, M.H. Role of Circular RNA in Brain Tumor Development. Cells 2022, 11, 2130. [Google Scholar] [CrossRef] [PubMed]
- Mahinfar, P.; Mansoori, B.; Rostamzadeh, D.; Baradaran, B.; Cho, W.C.; Mansoori, B. The Role of microRNAs in Multidrug Resistance of Glioblastoma. Cancers 2022, 14, 3217. [Google Scholar] [CrossRef]
- Makowska, M.; Smolarz, B.; Romanowicz, H. microRNAs (miRNAs) in Glioblastoma Multiforme (GBM)-Recent Literature Review. Int. J. Mol. Sci. 2023, 24, 3521. [Google Scholar] [CrossRef]
- Cardia, A.; Epistolio, S.; Zaed, I.; Sahnane, N.; Cerutti, R.; Cipriani, D.; Barizzi, J.; Spina, P.; Stefanini, F.M.; Cerati, M.; et al. Identification of MGMT Downregulation Induced by miRNA in Glioblastoma and Possible Effect on Temozolomide Sensitivity. J Clin Med. 2023, 12, 2061. [Google Scholar] [CrossRef]
- de Menezes, M.R.; Acioli, M.E.A.; da Trindade, A.C.L.; da Silva, S.P.; de Lima, R.E.; da Silva Teixeira, V.G.; Vasconcelos, L.R.S. Potential role of microRNAs as biomarkers in human glioblastoma: A mini systematic review from 2015 to 2020. Mol. Biol. Rep. 2021, 48, 4647–4658. [Google Scholar] [CrossRef]
- ISO 14155:2020; Clinical Investigation of Medical Devices for Human Subjects—Good Clinical Practice. ISO: Geneva, Switzerland, 2020.
- Brell, M.; Tortosa, A.; Verger, E.; Gil, J.M.; Viñolas, N.; Villá, S.; Acebes, J.J.; Caral, L.; Pujol, T.; Ferrer, I.; et al. Prognostic significance of O6-methylguanine-DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin. Cancer Res. 2005, 11, 5167–5174. [Google Scholar] [CrossRef] [PubMed]
- Cao, V.T.; Jung, T.-Y.; Jung, S.; Jin, S.-G.; Moon, K.-S.; Kim, I.-Y.; Kang, S.-S.; Park, C.-S.; Lee, K.-H.; Chae, H.-J. The correlation and prognostic significance of MGMT promoter methylation and MGMT protein in glioblastomas. Neurosurgery 2009, 65, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Forcella, M.; Mozzi, A.; Stefanini, F.M.; Riva, A.; Epistolio, S.; Molinari, F.; Merlo, E.; Monti, E.; Fusi, P.; Frattini, M. Deregulation of sialidases in human normal and tumor tissues. Cancer Biomark. 2018, 21, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Abdallah, M.O.E.; Breuer, P.; Stahl, F.; Bakhit, Y.; Potthoff, A.L.; Pregler, B.E.F.; Schneider, M.; Waha, A.; Wüllner, U.; et al. Downregulation of MGMT expression by targeted editing of DNA methylation enhances temozolomide sensitivity in glioblastoma. Neoplasia 2023, 44, 100929. [Google Scholar] [CrossRef] [PubMed]
- Siegel, S.R.; Mackenzie, J.; Chaplin, G.; Jablonski, N.G.; Griffiths, L. Circulating microRNAs involved in multiple sclerosis. Mol. Biol. Rep. 2012, 39, 6219–6225. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Zheng, G.; Shao, C.; Liu, J.; Lv, H.; Zhang, G. Downregulation of miR-196b Promotes Glioma Cell Sensitivity to Temozolomide Chemotherapy and Radiotherapy. Ann. Clin. Lab. Sci. 2018, 48, 719–725. [Google Scholar]
- Jia, Y.; Tian, Y.; An, S.; Yang, D. Effects of microRNA-195 on the Prognosis of Glioma Patients and the Proliferation and Apoptosis of Human Glioma Cells. Pathol. Oncol. Res. 2020, 26, 753–763. [Google Scholar] [CrossRef]
- Jiang, G.; Mu, J.; Liu, X.; Peng, X.; Zhong, F.; Yuan, W.; Deng, F.; Peng, X.; Peng, S.; Zeng, X. Prognostic value of miR-21 in gliomas: Comprehensive study based on meta-analysis and TCGA dataset validation. Sci. Rep. 2020, 10, 4220. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, H.Y.; He, Z.Y. MicroRNA-181c provides neuroprotection in an intracerebral hemorrhage model. Neural Regen. Res. 2020, 15, 1274–1282. [Google Scholar]
- Tang, D.; Gao, W.; Yang, J.; Liu, J.; Zhao, J.; Ge, J.; Chen, Q.; Liu, B. miR-181d promotes cell proliferation via the IGF1/PI3K/AKT axis in glioma. Mol. Med. Rep. 2020, 22, 3804–3812. [Google Scholar] [CrossRef]
- Kos, A.; Olde Loohuis, N.; Meinhardt, J.; van Bokhoven, H.; Kaplan, B.B.; Martens, G.J.; Aschrafi, A. MicroRNA-181 promotes synaptogenesis and attenuates axonal outgrowth in cortical neurons. Cell Mol. Life Sci. 2016, 73, 3555–3567. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Ho, H.L.; Lin, S.C.; Ho, T.D.; Hsu, C.Y. Upregulation of miR-125b, miR-181d, and miR-221 Predicts Poor Prognosis in MGMT Promoter-Unmethylated Glioblastoma Patients. Am. J. Clin. Pathol. 2018, 149, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Song, L.D.; Weng, J.C.; Li, C.B.; Wang, L.; Wu, Z.; Zhang, J.T. MicroRNA-195 Functions as a Tumor Suppressor by Directly Targeting Fatty Acid Synthase in Malignant Meningioma. World Neurosurg. 2020, 136, e355–e364. [Google Scholar] [CrossRef] [PubMed]
- Jesionek-Kupnicka, D.; Braun, M.; Trąbska-Kluch, B.; Czech, J.; Szybka, M.; Szymańska, B.; Kulczycka-Wojdala, D.; Bieńkowski, M.; Kordek, R.; Zawlik, I. MiR-21, miR-34a, miR-125b, miR-181d and miR-648 levels inversely correlate with MGMT and TP53 expression in primary glioblastoma patients. Arch. Med. Sci. 2019, 15, 504–512. [Google Scholar] [CrossRef]

| miR-181c | p | ||||
|---|---|---|---|---|---|
| <0.333 | 0.333–3 | >3 | |||
| miR-181d | <0.333 | 30/112 | 8/112 | 0/112 | 0.0005 |
| (26.8%) | (7.1%) | (0%) | |||
| 0.333–3 | 9/112 | 53/112 | 9/112 | ||
| (8.0%) | (47.3%) | (8.0%) | |||
| >3 | 0/112 | 1/112 | 2/112 | ||
| (0%) | (0.9%) | (1.8%) | |||
| miR-21 | <0.333 | 1/112 | 0/112 | 0/112 | 0.0009 |
| (0.9%) | (0%) | (0%) | |||
| 0.333–3 | 15/112 | 5/112 | 0/112 | ||
| (13.4%) | (4.5%) | (0%) | |||
| >3 | 23/112 | 57/112 | 11/112 | ||
| (20.5%) | (50.9%) | (9.8%) | |||
| miR-195 | <0.333 | 18/112 | 1/112 | 0/112 | 0.0005 |
| (16.1%) | (0.9%) | (0%) | |||
| 0.333–3 | 21/112 | 48/112 | 0/112 | ||
| (18.8%) | (42.9%) | (0%) | |||
| >3 | 0/112 | 13/112 | 11/112 | ||
| (0%) | (11.6%) | (9.8%) | |||
| miR-196b | <0.333 | 7/112 | 0/112 | 0/112 | 0.0005 |
| (6.2%) | (0%) | (0%) | |||
| 0.333–3 | 9/112 | 1/112 | 0/112 | ||
| (8.0%) | (0.9%) | (0%) | |||
| >3 | 23/112 | 61/112 | 11/112 | ||
| (20.5%) | (54.5%) | (9.8%) | |||
| miR-648 | <0.333 | 17/112 | 11/112 | 0/112 | 0.0145 |
| (15.2%) | (9.8%) | (0%) | |||
| 0.333–3 | 20/112 | 47/112 | 10/112 | ||
| (17.9%) | (42.0%) | (8.9%) | |||
| >3 | 2/112 | 4/112 | 1/112 | ||
| (1.8%) | (3.6%) | (0.9%) | |||
| miR-181d | p | ||||
| <0.333 | 0.333–3 | >3 | |||
| miR-21 | <0.333 | 1/112 | 0/112 | 0/112 | 0.0345 |
| (0.9%) | (0%) | (0%) | |||
| 0.333–3 | 13/112 | 7/112 | 0/112 | ||
| (11.6%) | (6.2%) | (0%) | |||
| >3 | 24/112 | 64/112 | 3/112 | ||
| (21.4%) | (57.1%) | (2.7%) | |||
| miR-195 | <0.333 | 16/112 | 3/112 | 0/112 | 0.0005 |
| (14.3%) | (2.7%) | (0%) | |||
| 0.333–3 | 21/112 | 47/112 | 1/112 | ||
| (18.7%) | (42.0%) | (0.9%) | |||
| >3 | 1/112 | 21/112 | 2/112 | ||
| (0.9%) | (18.7%) | (1.8%) | |||
| miR-196b | <0.333 | 6/112 | 1/112 | 0/112 | 0.0025 |
| (5.4%) | (0.9%) | (0%) | |||
| 0.333–3 | 9/112 | 1/112 | 0/112 | ||
| (8.0%) | (0.9%) | (0%) | |||
| >3 | 23/112 | 69/112 | 3/112 | ||
| (20.54%) | (61.6%) | (2.7%) | |||
| miR-648 | <0.333 | 15/112 | 13/112 | 0/112 | 0.0390 |
| (13.4%) | (11.6%) | (0%) | |||
| 0.333–3 | 21/112 | 54/112 | 2/112 | ||
| (18.7%) | (48.2%) | (1.8%) | |||
| >3 | 2/112 | 4/112 | 1/112 | ||
| (1.8%) | (3.6%) | (0.9%) | |||
| miR-21 | p | ||||
| <0.333 | 0.333–3 | >3 | |||
| miR-195 | <0.333 | 1/112 | 9/112 | 9/112 | 0.0005 |
| (0.9%) | (8.0%) | (8.0%) | |||
| 0.333–3 | 0/112 | 11/112 | 58/112 | ||
| (0%) | (9.8%) | (51.8%) | |||
| >3 | 0/112 | 0/112 | 24/112 | ||
| (0%) | (0%) | (21.4%) | |||
| miR-196b | <0.333 | 1/112 | 5/112 | 1/112 | 0.0005 |
| (0.9%) | (4.5%) | (0.9%) | |||
| 0.333–3 | 0/112 | 3/112 | 7/112 | ||
| (0%) | (2.7%) | (6.2%) | |||
| >3 | 0/112 | 12/112 | 83/112 | ||
| (0%) | (10.7%) | (74.1%) | |||
| miR-648 | <0.333 | 1/112 | 6/112 | 21/112 | 0.2269 |
| (0.9%) | (5.4%) | (18.7%) | |||
| 0.333–3 | 0/112 | 14/112 | 63/112 | ||
| (0%) | (12.5%) | (56.2%) | |||
| >3 | 0/112 | 0/112 | 7/112 | ||
| (0%) | (0%) | (6.2%) | |||
| miR-195 | p | ||||
| <0.333 | 0.333–3 | >3 | |||
| miR-196b | <0.333 | 6/112 | 1/112 | 0/112 | 0.0005 |
| (5.4%) | (0.9%) | (0%) | |||
| 0.333–3 | 5/112 | 5/112 | 0/112 | ||
| (4.5%) | (4.5%) | (0%) | |||
| >3 | 8/112 | 63/112 | 24/112 | ||
| (7.1%) | (56.2%) | (21.4%) | |||
| miR-648 | <0.333 | 11/112 | 17/112 | 0/112 | 0.0020 |
| (9.8%) | (15.2%) | (0%) | |||
| 0.333–3 | 7/112 | 48/112 | 22/112 | ||
| (6.2%) | (42.9%) | (19.6%) | |||
| >3 | 1/112 | 4/112 | 2/112 | ||
| (0.9%) | (3.6%) | (1.8%) | |||
| miR-196b | p | ||||
| <0.333 | 0.333–3 | >3 | |||
| miR-648 | <0.333 | 4/112 | 3/112 | 21/112 | 0.2859 |
| (3.6%) | (2.7%) | (18.7%) | |||
| 0.333–3 | 3/112 | 6/112 | 68/112 | ||
| (2.7%) | (5.4%) | (60.7%) | |||
| >3 | 0/112 | 1/112 | 6/112 | ||
| (0%) | (0.9%) | (5.36%) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Epistolio, S.; Dazio, G.; Zaed, I.; Sahnane, N.; Cipriani, D.; Polinelli, F.; Barizzi, J.; Spina, P.; Stefanini, F.M.; Cerati, M.; et al. Clinical Relevance and Interplay between miRNAs in Influencing Glioblastoma Multiforme Prognosis. Cells 2024, 13, 276. https://doi.org/10.3390/cells13030276
Epistolio S, Dazio G, Zaed I, Sahnane N, Cipriani D, Polinelli F, Barizzi J, Spina P, Stefanini FM, Cerati M, et al. Clinical Relevance and Interplay between miRNAs in Influencing Glioblastoma Multiforme Prognosis. Cells. 2024; 13(3):276. https://doi.org/10.3390/cells13030276
Chicago/Turabian StyleEpistolio, Samantha, Giulia Dazio, Ismail Zaed, Nora Sahnane, Debora Cipriani, Francesco Polinelli, Jessica Barizzi, Paolo Spina, Federico Mattia Stefanini, Michele Cerati, and et al. 2024. "Clinical Relevance and Interplay between miRNAs in Influencing Glioblastoma Multiforme Prognosis" Cells 13, no. 3: 276. https://doi.org/10.3390/cells13030276
APA StyleEpistolio, S., Dazio, G., Zaed, I., Sahnane, N., Cipriani, D., Polinelli, F., Barizzi, J., Spina, P., Stefanini, F. M., Cerati, M., Balbi, S., Mazzucchelli, L., Sessa, F., Pesce, G. A., Reinert, M., Cardia, A., Marchi, F., & Frattini, M. (2024). Clinical Relevance and Interplay between miRNAs in Influencing Glioblastoma Multiforme Prognosis. Cells, 13(3), 276. https://doi.org/10.3390/cells13030276






