The Zebrafish Retina and the Evolution of the Onecut-Mediated Pathway in Cell Type Differentiation
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Maintenance and Sample Collection
2.2. Identification of Zebrafish Orthologs of Ascidian Onecut Target Genes
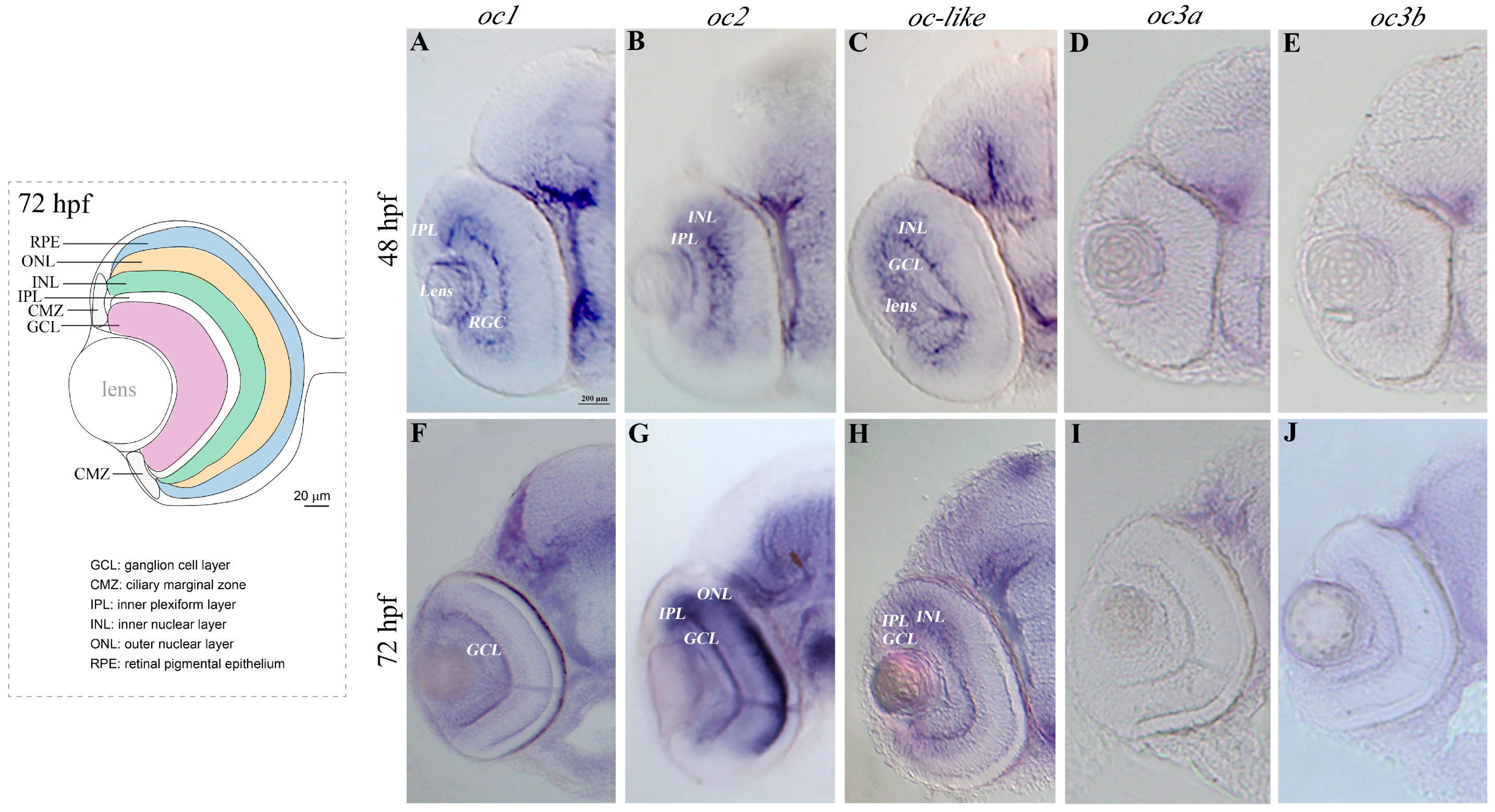
2.3. Whole Mount In Situ Hybridization (WISH) and Vibratome Tissue Sectioning
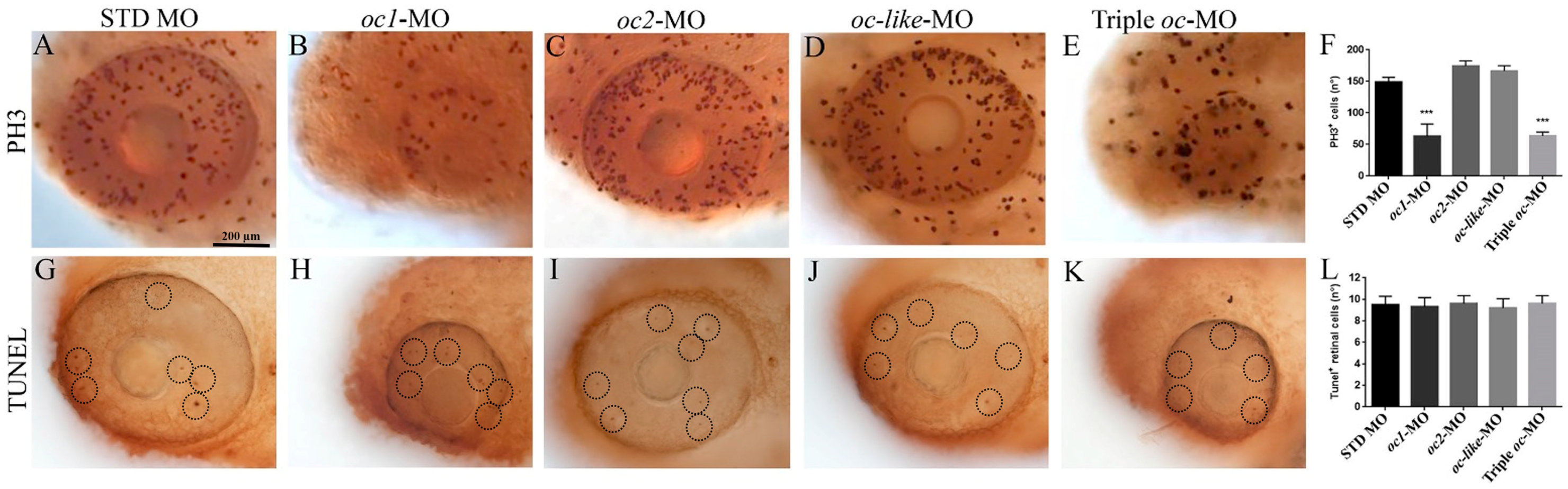
2.4. Morpholino-Mediated Knockdown
2.5. Eye Size Measurement
2.6. TUNEL Staining
2.7. Total RNA Extraction, cDNA Synthesis and Quantitative Real Time PCR (RT-qPCR) Analysis
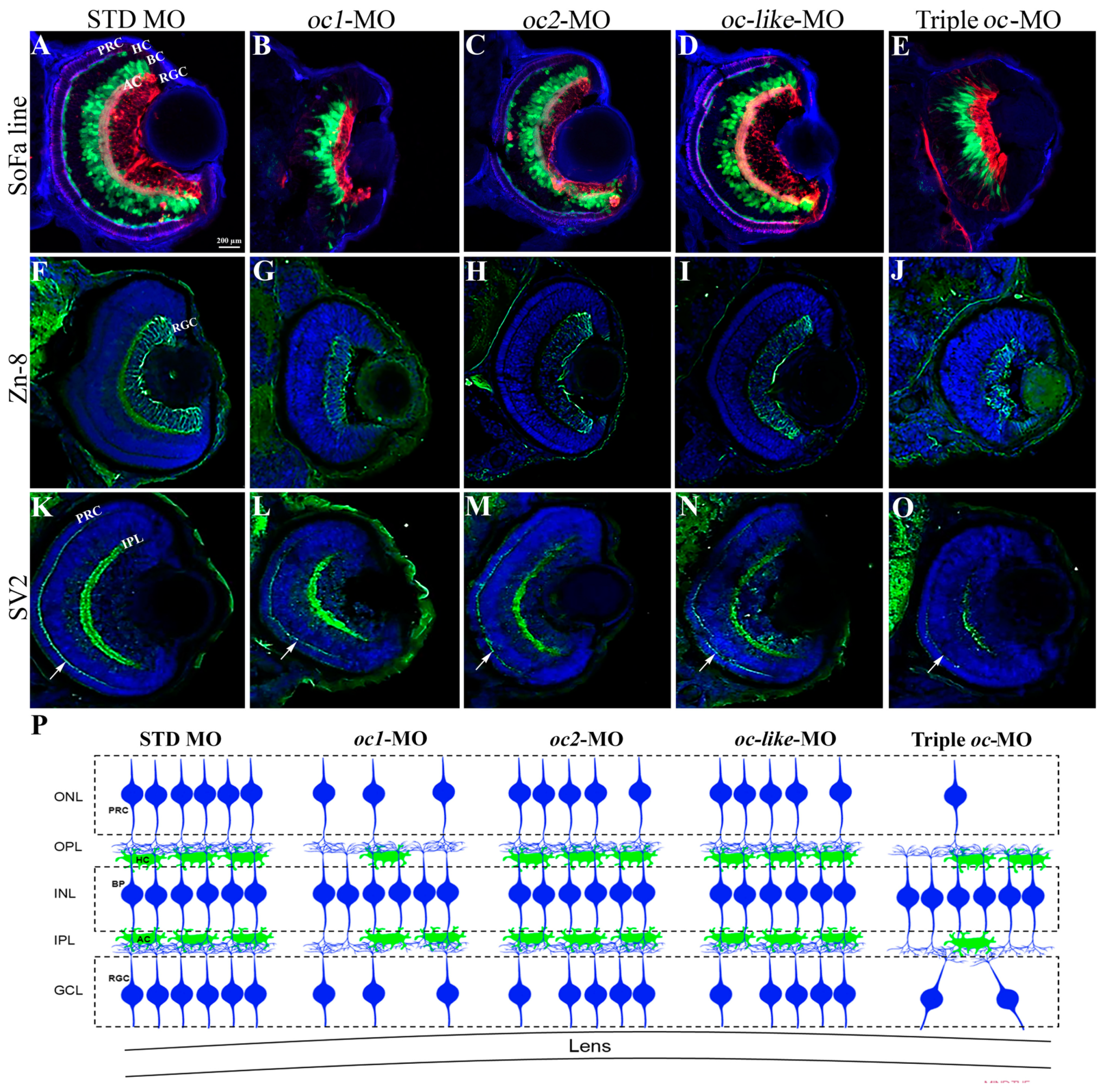
2.8. Cryostat Sectioning and imMUNOFLUORESCENCe
2.9. Whole-Mount Immmunostaining
2.10. Western Blotting
2.11. Tracking of Swimming Behavior
2.12. Image Processing and Statistical Analysis
3. Results
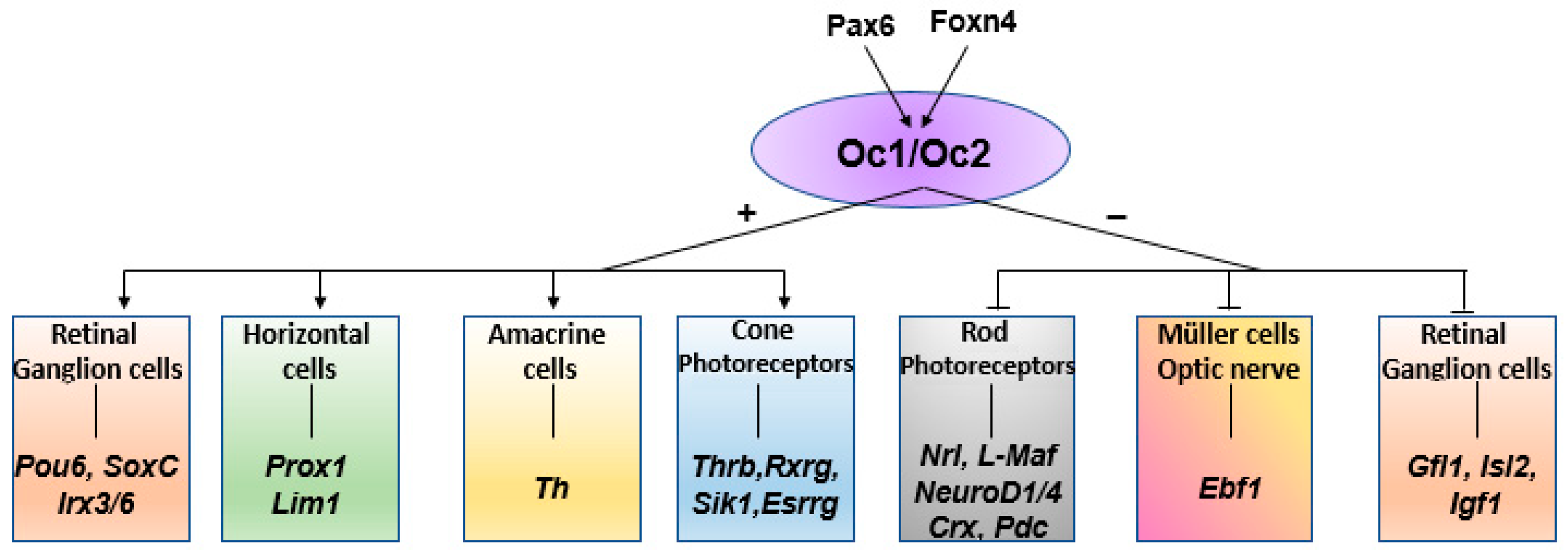
3.1. Zebrafish Onecut Genes in Eye Development
3.2. Functional Role of Onecut Genes in Eye Morphogenesis
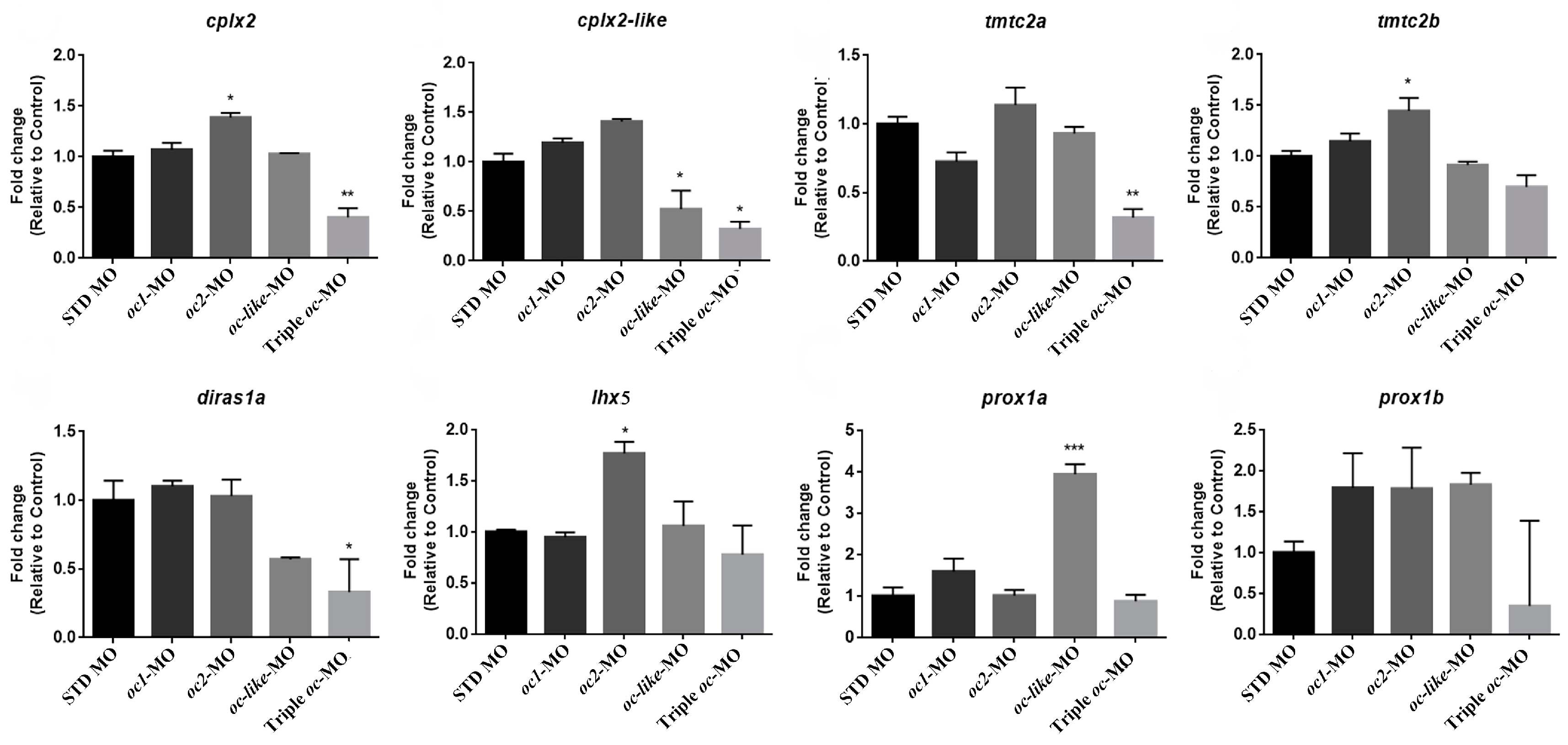
3.3. Onecut Target Genes in Eye Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lamb, T.D. Evolution of Vertebrate Retinal Photoreception. Philos. Trans. R Soc. Lond. B Biol. Sci. 2009, 364, 2911–2924. [Google Scholar] [CrossRef] [PubMed]
- Hennig, A.K.; Peng, G.-H.; Chen, S. Regulation of Photoreceptor Gene Expression by Crx-Associated Transcription Factor Network. Brain Res. 2008, 1192, 114–133. [Google Scholar] [CrossRef]
- Kropp, P.A.; Gannon, M. Onecut Transcription Factors in Development and Disease. Trends Dev. Biol. 2016, 9, 43–57. [Google Scholar] [PubMed]
- Cassata, G.; Kagoshima, H.; Prétôt, R.F.; Aspöck, G.; Niklaus, G.; Bürglin, T.R. Rapid Expression Screening of Caenorhabditis Elegans Homeobox Open Reading Frames Using a Two-Step Polymerase Chain Reaction Promoter-Gfp Reporter Construction Technique. Gene 1998, 212, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Lannoy, V.J.; Bürglin, T.R.; Rousseau, G.G.; Lemaigre, F.P. Isoforms of Hepatocyte Nuclear Factor-6 Differ in DNA-Binding Properties, Contain a Bifunctional Homeodomain, and Define the New ONECUT Class of Homeodomain Proteins. J. Biol. Chem. 1998, 273, 13552–13562. [Google Scholar] [CrossRef]
- Otim, O.; Amore, G.; Minokawa, T.; McClay, D.R.; Davidson, E.H. SpHnf6, a Transcription Factor That Executes Multiple Functions in Sea Urchin Embryogenesis. Dev. Biol. 2004, 273, 226–243. [Google Scholar] [CrossRef]
- Leyva-Díaz, E. CUT Homeobox Genes: Transcriptional Regulation of Neuronal Specification and Beyond. Front. Cell Neurosci. 2023, 17, 1233830. [Google Scholar] [CrossRef]
- Nguyen, D.N.; Rohrbaugh, M.; Lai, Z. The Drosophila Homolog of Onecut Homeodomain Proteins Is a Neural-Specific Transcriptional Activator with a Potential Role in Regulating Neural Differentiation. Mech. Dev. 2000, 97, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Vassalli, Q.A.; Colantuono, C.; Nittoli, V.; Ferraioli, A.; Fasano, G.; Berruto, F.; Chiusano, M.L.; Kelsh, R.N.; Sordino, P.; Locascio, A. Onecut Regulates Core Components of the Molecular Machinery for Neurotransmission in Photoreceptor Differentiation. Front. Cell Dev. Biol. 2021, 9, 602450. [Google Scholar] [CrossRef]
- Sunita Prajapati, K.; Gupta, S.; Chaudhri, S.; Kumar, S. Role of ONECUT Family Transcription Factors in Cancer and Other Diseases. Exp. Cell Res. 2024, 438, 114035. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, D.; Chintala, H.; Wu, F.; Fliesler, S.J.; Hu, Z.; Mu, X. Onecut1 and Onecut2 Redundantly Regulate Early Retinal Cell Fates during Development. Proc. Natl. Acad. Sci. USA 2014, 111, E4086–E4095. [Google Scholar] [CrossRef] [PubMed]
- Goetz, J.J.; Martin, G.M.; Chowdhury, R.; Trimarchi, J.M. Onecut1 and Onecut2 Play Critical Roles in the Development of the Mouse Retina. PLoS ONE 2014, 9, e110194. [Google Scholar] [CrossRef]
- Wu, F.; Sapkota, D.; Li, R.; Mu, X. Onecut 1 and Onecut 2 Are Potential Regulators of Mouse Retinal Development. J. Comp. Neurol. 2012, 520, 952–969. [Google Scholar] [CrossRef]
- Wu, F.; Li, R.; Umino, Y.; Kaczynski, T.J.; Sapkota, D.; Li, S.; Xiang, M.; Fliesler, S.J.; Sherry, D.M.; Gannon, M.; et al. Onecut1 Is Essential for Horizontal Cell Genesis and Retinal Integrity. J. Neurosci. 2013, 33, 13053–13065, 13065a. [Google Scholar] [CrossRef] [PubMed]
- Klimova, L.; Antosova, B.; Kuzelova, A.; Strnad, H.; Kozmik, Z. Onecut1 and Onecut2 Transcription Factors Operate Downstream of Pax6 to Regulate Horizontal Cell Development. Dev. Biol. 2015, 402, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Kreplova, M.; Kuzelova, A.; Antosova, B.; Zilova, L.; Jägle, H.; Kozmik, Z. Dose-Dependent Regulation of Horizontal Cell Fate by Onecut Family of Transcription Factors. PLoS ONE 2020, 15, e0237403. [Google Scholar] [CrossRef]
- Lonfat, N.; Wang, S.; Lee, C.; Garcia, M.; Choi, J.; Park, P.J.; Cepko, C. Cis-Regulatory Dissection of Cone Development Reveals a Broad Role for Otx2 and Oc Transcription Factors. Development 2021, 148, dev198549. [Google Scholar] [CrossRef] [PubMed]
- Fujitani, Y.; Fujitani, S.; Luo, H.; Qiu, F.; Burlison, J.; Long, Q.; Kawaguchi, Y.; Edlund, H.; MacDonald, R.J.; Furukawa, T.; et al. Ptf1a Determines Horizontal and Amacrine Cell Fates during Mouse Retinal Development. Development 2006, 133, 4439–4450. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Mo, Z.; Yang, X.; Price, S.M.; Shen, M.M.; Xiang, M. Foxn4 Controls the Genesis of Amacrine and Horizontal Cells by Retinal Progenitors. Neuron 2004, 43, 795–807. [Google Scholar] [CrossRef]
- Emerson, M.M.; Surzenko, N.; Goetz, J.J.; Trimarchi, J.; Cepko, C.L. Otx2 and Onecut1 Promote the Fates of Cone Photoreceptors and Horizontal Cells and Repress Rod Photoreceptors. Dev. Cell 2013, 26, 59–72. [Google Scholar] [CrossRef]
- Jean-Charles, N.; Buenaventura, D.F.; Emerson, M.M. Identification and Characterization of Early Photoreceptor Cis-Regulatory Elements and Their Relation to Onecut1. Neural Dev. 2018, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, E.; Pezzotti, M.R.; Locascio, A.; Branno, M. Onecut Is a Direct Neural-Specific Transcriptional Activator of Rx in Ciona Intestinalis. Dev. Biol. 2011, 355, 358–371. [Google Scholar] [CrossRef]
- Aleström, P.; D’Angelo, L.; Midtlyng, P.J.; Schorderet, D.F.; Schulte-Merker, S.; Sohm, F.; Warner, S. Zebrafish: Housing and Husbandry Recommendations. Lab. Anim. 2020, 54, 213–224. [Google Scholar] [CrossRef]
- Ambrosino, L.; Ruggieri, V.; Bostan, H.; Miralto, M.; Vitulo, N.; Zouine, M.; Barone, A.; Bouzayen, M.; Frusciante, L.; Pezzotti, M.; et al. Multilevel Comparative Bioinformatics to Investigate Evolutionary Relationships and Specificities in Gene Annotations: An Example for Tomato and Grapevine. BMC Bioinform. 2018, 19, 435. [Google Scholar] [CrossRef] [PubMed]
- De Felice, B.; Copia, L.; Guida, M. Gene Expression Profiling in Zebrafish Embryos Exposed to Diclofenac, an Environmental Toxicant. Mol. Biol. Rep. 2012, 39, 2119–2128. [Google Scholar] [CrossRef]
- Nittoli, V.; Sepe, R.M.; Coppola, U.; D’Agostino, Y.; De Felice, E.; Palladino, A.; Vassalli, Q.A.; Locascio, A.; Ristoratore, F.; Spagnuolo, A.; et al. A Comprehensive Analysis of Neurotrophins and Neurotrophin Tyrosine Kinase Receptors Expression during Development of Zebrafish. J. Comp. Neurol. 2018, 526, 1057–1072. [Google Scholar] [CrossRef] [PubMed]
- van Huet, R.A.C.; Siemiatkowska, A.M.; Özgül, R.K.; Yücel, D.; Hoyng, C.B.; Banin, E.; Blumenfeld, A.; Rotenstreich, Y.; Riemslag, F.C.C.; den Hollander, A.I.; et al. Retinitis Pigmentosa Caused by Mutations in the Ciliary MAK Gene Is Relatively Mild and Is Not Associated with Apparent Extra-Ocular Features. Acta Ophthalmol. 2015, 93, 83–94. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Pujic, Z.; Omori, Y.; Tsujikawa, M.; Thisse, B.; Thisse, C.; Malicki, J. Reverse Genetic Analysis of Neurogenesis in the Zebrafish Retina. Dev. Biol. 2006, 293, 330–347. [Google Scholar] [CrossRef] [PubMed]
- Verduzco, D.; Amatruda, J.F. Analysis of Cell Proliferation, Senescence and Cell Death in Zebrafish Embryos. Methods Cell Biol. 2011, 101, 19–38. [Google Scholar] [CrossRef]
- Thisse, B.; Heyer, V.; Lux, A.; Alunni, V.; Degrave, A.; Seiliez, I.; Kirchner, J.; Parkhill, J.-P.; Thisse, C. Spatial and Temporal Expression of the Zebrafish Genome by Large-Scale in Situ Hybridization Screening. Methods Cell Biol. 2004, 77, 505–519. [Google Scholar] [CrossRef]
- Matthews, R.P.; Lorent, K.; Russo, P.; Pack, M. The Zebrafish Onecut Gene Hnf-6 Functions in an Evolutionarily Conserved Genetic Pathway That Regulates Vertebrate Biliary Development. Dev. Biol. 2004, 274, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Hendzel, M.J.; Wei, Y.; Mancini, M.A.; Van Hooser, A.; Ranalli, T.; Brinkley, B.R.; Bazett-Jones, D.P.; Allis, C.D. Mitosis-Specific Phosphorylation of Histone H3 Initiates Primarily within Pericentromeric Heterochromatin during G2 and Spreads in an Ordered Fashion Coincident with Mitotic Chromosome Condensation. Chromosoma 1997, 106, 348–360. [Google Scholar] [CrossRef]
- Saka, Y.; Smith, J.C. Spatial and Temporal Patterns of Cell Division during Early Xenopus Embryogenesis. Dev. Biol. 2001, 229, 307–318. [Google Scholar] [CrossRef]
- Almeida, A.D.; Boije, H.; Chow, R.W.; He, J.; Tham, J.; Suzuki, S.C.; Harris, W.A. Spectrum of Fates: A New Approach to the Study of the Developing Zebrafish Retina. Development 2014, 141, 1971–1980. [Google Scholar] [CrossRef]
- Yeh, C.-W.; Hsu, L.-S. Zebrafish Diras1 Promoted Neurite Outgrowth in Neuro-2a Cells and Maintained Trigeminal Ganglion Neurons In Vivo via Rac1-Dependent Pathway. Mol. Neurobiol. 2016, 53, 6594–6607. [Google Scholar] [CrossRef]
- Bradford, Y.M.; Van Slyke, C.E.; Ruzicka, L.; Singer, A.; Eagle, A.; Fashena, D.; Howe, D.G.; Frazer, K.; Martin, R.; Paddock, H.; et al. Zebrafish Information Network, the Knowledgebase for Danio Rerio Research. Genetics 2022, 220, iyac016. [Google Scholar] [CrossRef]
- Nowak, M.A.; Boerlijst, M.C.; Cooke, J.; Smith, J.M. Evolution of Genetic Redundancy. Nature 1997, 388, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Kvon, E.Z.; Waymack, R.; Elabd, M.G.; Wunderlich, Z. Enhancer Redundancy in Development and Disease. Nat. Rev. Genet. 2021, 22, 324. [Google Scholar] [CrossRef]
- Mueller, K.P.; Neuhauss, S.C.F. Light Perception: More Than Meets the Eyes. Curr. Biol. 2012, 22, R912–R914. [Google Scholar] [CrossRef][Green Version]
- Vaithianathan, T.; Henry, D.; Akmentin, W.; Matthews, G. Functional Roles of Complexin in Neurotransmitter Release at Ribbon Synapses of Mouse Retinal Bipolar Neurons. J. Neurosci. 2015, 35, 4065–4070. [Google Scholar] [CrossRef]
- Reim, K.; Regus-Leidig, H.; Ammermüller, J.; El-Kordi, A.; Radyushkin, K.; Ehrenreich, H.; Brandstätter, J.H.; Brose, N. Aberrant Function and Structure of Retinal Ribbon Synapses in the Absence of Complexin 3 and Complexin 4. J. Cell Sci. 2009, 122, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, I.; Mühlhans, J.; Dedek, K.; Reim, K.; Brandstätter, J.H.; Ammermüller, J. The Absence of Complexin 3 and Complexin 4 Differentially Impacts the ON and OFF Pathways in Mouse Retina. Eur. J. Neurosci. 2012, 36, 2470–2481. [Google Scholar] [CrossRef] [PubMed]
- Tada, M.; Gengyo-Ando, K.; Kobayashi, T.; Fukuyama, M.; Mitani, S.; Kontani, K.; Katada, T. Neuronally Expressed Ras-Family GTPase Di-Ras Modulates Synaptic Activity in Caenorhabditis Elegans. Genes Cells 2012, 17, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Sunryd, J.C.; Cheon, B.; Graham, J.B.; Giorda, K.M.; Fissore, R.A.; Hebert, D.N. TMTC1 and TMTC2 Are Novel Endoplasmic Reticulum Tetratricopeptide Repeat-Containing Adapter Proteins Involved in Calcium Homeostasis. J. Biol. Chem. 2014, 289, 16085–16099. [Google Scholar] [CrossRef] [PubMed]
- Choquet, H.; Paylakhi, S.; Kneeland, S.C.; Thai, K.K.; Hoffmann, T.J.; Yin, J.; Kvale, M.N.; Banda, Y.; Tolman, N.G.; Williams, P.A.; et al. A Multiethnic Genome-Wide Association Study of Primary Open-Angle Glaucoma Identifies Novel Risk Loci. Nat. Commun. 2018, 9, 2278. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.B.; Sunryd, J.C.; Mathavan, K.; Weir, E.; Larsen, I.S.B.; Halim, A.; Clausen, H.; Cousin, H.; Alfandari, D.; Hebert, D.N. Endoplasmic Reticulum Transmembrane Protein TMTC3 Contributes to O-Mannosylation of E-Cadherin, Cellular Adherence, and Embryonic Gastrulation. Mol. Biol. Cell 2020, 31, 167–183. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vassalli, Q.A.; Fasano, G.; Nittoli, V.; Gagliardi, E.; Sepe, R.M.; Donizetti, A.; Aniello, F.; Sordino, P.; Kelsh, R.; Locascio, A. The Zebrafish Retina and the Evolution of the Onecut-Mediated Pathway in Cell Type Differentiation. Cells 2024, 13, 2071. https://doi.org/10.3390/cells13242071
Vassalli QA, Fasano G, Nittoli V, Gagliardi E, Sepe RM, Donizetti A, Aniello F, Sordino P, Kelsh R, Locascio A. The Zebrafish Retina and the Evolution of the Onecut-Mediated Pathway in Cell Type Differentiation. Cells. 2024; 13(24):2071. https://doi.org/10.3390/cells13242071
Chicago/Turabian StyleVassalli, Quirino Attilio, Giulia Fasano, Valeria Nittoli, Eleonora Gagliardi, Rosa Maria Sepe, Aldo Donizetti, Francesco Aniello, Paolo Sordino, Robert Kelsh, and Annamaria Locascio. 2024. "The Zebrafish Retina and the Evolution of the Onecut-Mediated Pathway in Cell Type Differentiation" Cells 13, no. 24: 2071. https://doi.org/10.3390/cells13242071
APA StyleVassalli, Q. A., Fasano, G., Nittoli, V., Gagliardi, E., Sepe, R. M., Donizetti, A., Aniello, F., Sordino, P., Kelsh, R., & Locascio, A. (2024). The Zebrafish Retina and the Evolution of the Onecut-Mediated Pathway in Cell Type Differentiation. Cells, 13(24), 2071. https://doi.org/10.3390/cells13242071





