Standardized Protocol for Resazurin-Based Viability Assays on A549 Cell Line for Improving Cytotoxicity Data Reliability
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Conditions (2D)
2.2. Resazurin Working Solution Preparation
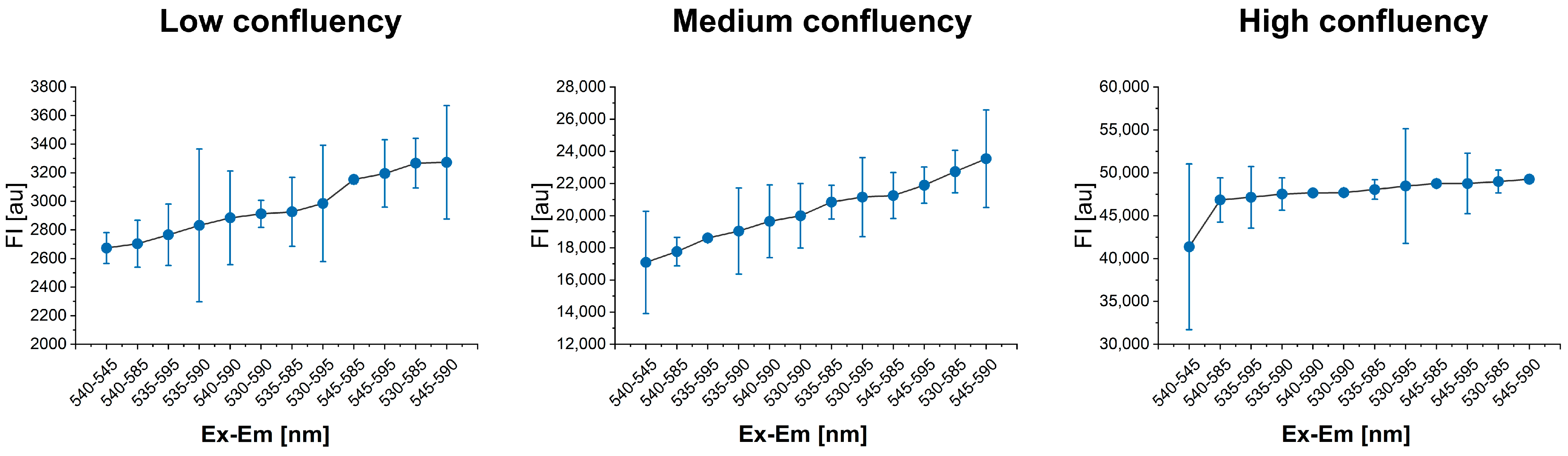
2.3. Identification of Optimal Excitation and Emission Wavelengths
2.4. Evaluation of Optimal Incubation Time
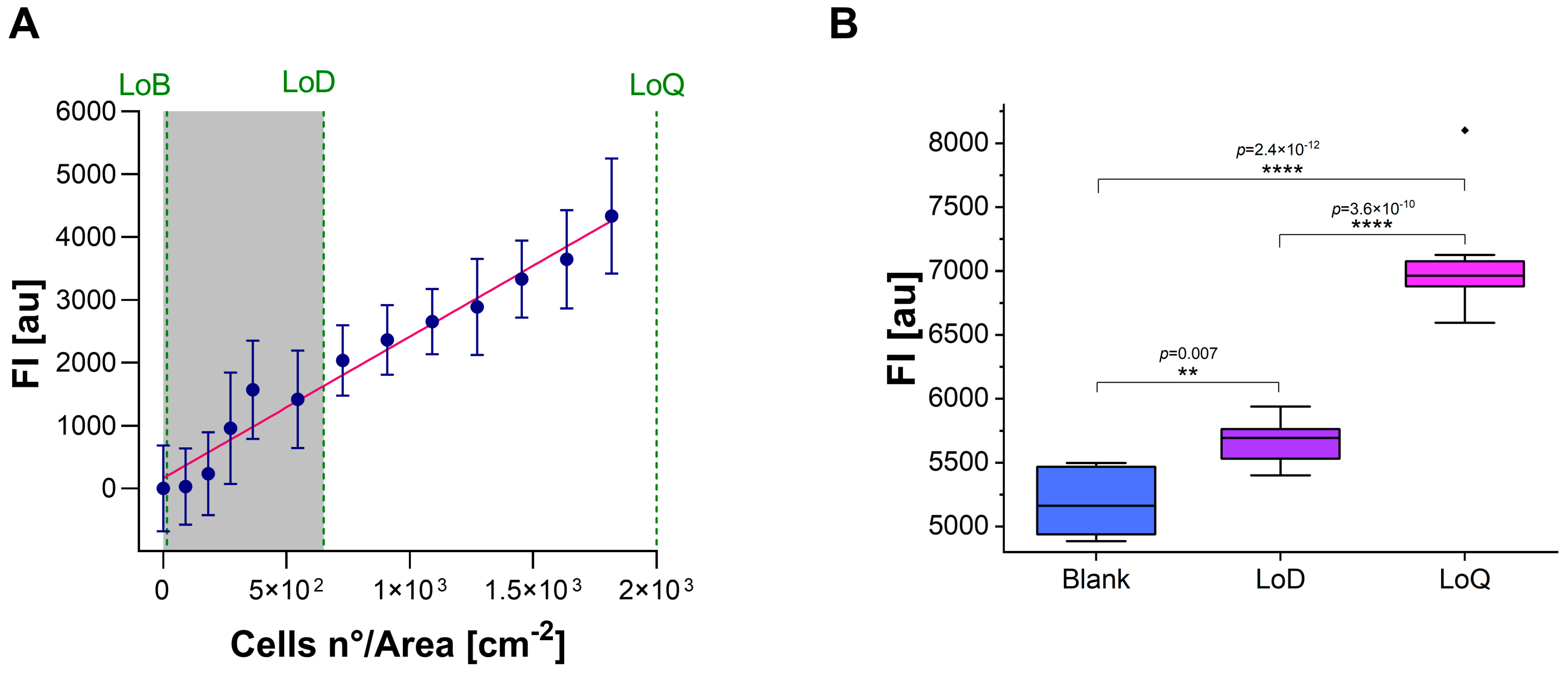
2.5. Limit of Blank, Limit of Detection, and Limit of Quantification
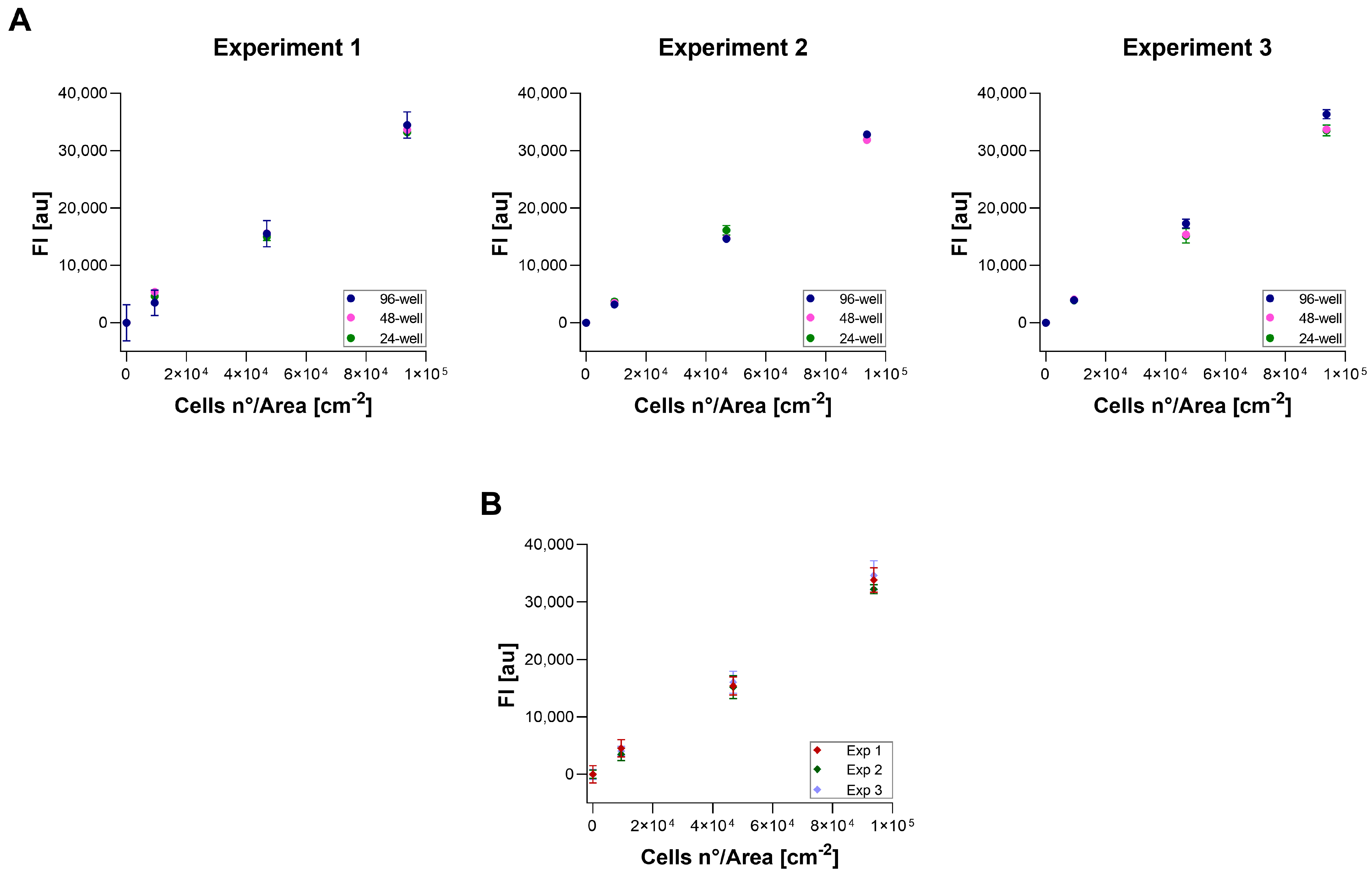
2.6. Repeatability, Reproducibility, and Measurement Uncertainty
2.7. Fibrin Gel Scaffold Preparation (3D Cell Culture)
- Bottom layer: Fibrin gel without cells, which prevents cells from sinking to the bottom of the plate during gelling. In each well, 135 µL of fibrinogen WS were mixed with 15 µL of thrombin WS and incubated at 37 °C for about 30 min.
- Top layer: Fibrin gel with cells. A549 cell line was detached and counted as previously described. Cells were resuspended in fibrinogen WS at different selected cell concentrations. After the complete gelling of the bottom layer, in each well, 135 µL of fibrinogen WS-A549 were mixed with 15 µL of thrombin WS and incubated at 37 °C for 12 h. After the complete gelling of the top layer, 150 µL of complete DMEM (10% FBS) was added over the scaffolds to avoid dehydration and allow cell growth.
2.8. Residual Resazurin on the Cell-Seeded Scaffold
- B1/S1: days 1, 4, 7, and 11 (4 times);
- B2/S2: days 4, 7, and 11 (3 times);
- B3/S3: days 7 and 11 (2 times);
- B4/S4: day 11 (1 time).
2.9. Optical Microscopy
2.10. Statistical Analysis
3. Results and Discussion
3.1. Optimal λEx-λEm Wavelengths Identification
3.2. Optimal Incubation Time Evaluation
3.3. Calculation of LoB, LoD, and LoQ
3.4. Repeatability, Reproducibility, and Measurement Uncertainty
3.5. Resazurin Assay on 3D Cell Culture
3.6. Residual Resazurin on the Fibrin Gel Scaffold
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lieber, M.; Todaro, G.; Smith, B.; Szakal, A.; Nelson-Rees, W. A Continuous Tumor-cell Line from a Human Lung Carcinoma with Properties of Type II Alveolar Epithelial Cells. Int. J. Cancer 1976, 17, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, M.; Barnes, P.F. Chemokine Production by a Human Alveolar Epithelial Cell Line in Response to Mycobacterium Tuberculosis. Infect. Immun. 1998, 66, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Thomas, L.H.; Friedland, J.S.; Sharland, M.; Becker, S. Respiratory Syncytial Virus-Induced RANTES Production from Human Bronchial Epithelial Cells Is Dependent on Nuclear Factor-Kappa B Nuclear Binding and Is Inhibited by Adenovirus-Mediated Expression of Inhibitor of Kappa B Alpha. J. Immunol. 1998, 161, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In Vitro Cultivation of Human Tumors: Establishment of Cell Lines Derived From a Series of Solid Tumors2. JNCI J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Garcia-de-Alba, C. Repurposing A549 Adenocarcinoma Cells: New Options for Drug Discovery. Am. J. Respir. Cell Mol. Biol. 2021, 64, 405–406. [Google Scholar] [CrossRef]
- Vieira-da-Silva, B.; Castanho, M.A.R.B. Resazurin Reduction-Based Assays Revisited: Guidelines for Accurate Reporting of Relative Differences on Metabolic Status. Molecules 2023, 28, 2283. [Google Scholar] [CrossRef]
- O’Brien, J.; Wilson, I.; Orton, T.; Pognan, F. Investigation of the Alamar Blue (Resazurin) Fluorescent Dye for the Assessment of Mammalian Cell Cytotoxicity: Resazurin as a Cytotoxicity Assay. Eur. J. Biochem. 2000, 267, 5421–5426. [Google Scholar] [CrossRef]
- Lavogina, D.; Lust, H.; Tahk, M.-J.; Laasfeld, T.; Vellama, H.; Nasirova, N.; Vardja, M.; Eskla, K.-L.; Salumets, A.; Rinken, A.; et al. Revisiting the Resazurin-Based Sensing of Cellular Viability: Widening the Application Horizon. Biosensors 2022, 12, 196. [Google Scholar] [CrossRef]
- Rampersad, S.N. Multiple Applications of Alamar Blue as an Indicator of Metabolic Function and Cellular Health in Cell Viability Bioassays. Sensors 2012, 12, 12347–12360. [Google Scholar] [CrossRef]
- Hirsch, C.; Schildknecht, S. In Vitro Research Reproducibility: Keeping Up High Standards. Front. Pharmacol. 2019, 10, 1484. [Google Scholar] [CrossRef]
- Reynolds, P.S. Between Two Stools: Preclinical Research, Reproducibility, and Statistical Design of Experiments. BMC Res. Notes 2022, 15, 73. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. 1,500 Scientists Lift the Lid on Reproducibility. Nature 2016, 533, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Haibe-Kains, B.; El-Hachem, N.; Birkbak, N.J.; Jin, A.C.; Beck, A.H.; Aerts, H.J.W.L.; Quackenbush, J. Inconsistency in Large Pharmacogenomic Studies. Nature 2013, 504, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Freedman, L.P.; Cockburn, I.M.; Simcoe, T.S. The Economics of Reproducibility in Preclinical Research. PLoS Biol. 2015, 13, e1002165. [Google Scholar] [CrossRef] [PubMed]
- Drude, N.I.; Martinez Gamboa, L.; Danziger, M.; Dirnagl, U.; Toelch, U. Improving Preclinical Studies through Replications. eLife 2021, 10, e62101. [Google Scholar] [CrossRef]
- Fosse, V.; Oldoni, E.; Bietrix, F.; Budillon, A.; Daskalopoulos, E.P.; Fratelli, M.; Gerlach, B.; Groenen, P.M.A.; Hölter, S.M.; Menon, J.M.L.; et al. Recommendations for Robust and Reproducible Preclinical Research in Personalised Medicine. BMC Med. 2023, 21, 14. [Google Scholar] [CrossRef]
- Petiti, J.; Revel, L.; Divieto, C. Standard Operating Procedure to Optimize Resazurin-Based Viability Assays. Biosensors 2024, 14, 156. [Google Scholar] [CrossRef]
- CLSI. Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures, 2nd ed.; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2012; Available online: https://clsi.org/standards/products/method-evaluation/documents/ep17/ (accessed on 16 October 2024).
- Chandran, S.; Singh, R.S.P. Comparison of Various International Guidelines for Analytical Method Validation. Pharmazie 2007, 62, 4–14. [Google Scholar]
- Committee for Medicinal Products for Human Use. ICH Guideline Q2(R2) on Validation of Analytical Procedures 2022; European Medicines Agency (EMA): Amsterdam, The Netherlands; Available online: https://www.ema.europa.eu/en/ich-guideline-q2r2-validation-analytical-procedures (accessed on 16 October 2024).
- Divieto, C.; Sassi, M.P. A First Approach to Evaluate the Cell Dose in Highly Porous Scaffolds by Using a Nondestructive Metabolic Method. Future Sci. OA 2015, 1, FSO58. [Google Scholar] [CrossRef]
- Divieto, C.; Revel, L.; Sassi, G.; Sassi, M.P. Uncertainty Analysis of Cell Counting by Metabolic Assays. J. Phys. Conf. Ser. 2013, 459, 012051. [Google Scholar] [CrossRef]
- Revel, L.; Santiano, M. RT 15/2022 Manual Micropipettes: Internal Procedure for Periodic Verification, Data Acquisition and Processing Software; INRIM: Torino, Italy, 2022. [Google Scholar]
- Caria, S.; Petiti, J.; Pegoraro, M.; Revel, L.; Mortati, L.; Divieto, C. 17/2023: Fibrin Gel as Tissue-Mimicking Material: Synthesis, Optimization and Characterization; INRIM: Torino, Italy, 2023. [Google Scholar]
- Czylkowska, A.; Rogalewicz, B.; Szczesio, M.; Raducka, A.; Gobis, K.; Szymański, P.; Czarnecka, K.; Camargo, B.C.; Szczytko, J.; Babich, A.; et al. Antitumor Activity against A549 Cancer Cells of Three Novel Complexes Supported by Coating with Silver Nanoparticles. Int. J. Mol. Sci. 2022, 23, 2980. [Google Scholar] [CrossRef] [PubMed]
- Graves, O.K.; Kim, W.; Özcan, M.; Ashraf, S.; Turkez, H.; Yuan, M.; Zhang, C.; Mardinoglu, A.; Li, X. Discovery of Drug Targets and Therapeutic Agents Based on Drug Repositioning to Treat Lung Adenocarcinoma. Biomed. Pharmacother. 2023, 161, 114486. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P.A. Why Most Published Research Findings Are False. PLoS Med 2005, 2, e124. [Google Scholar] [CrossRef] [PubMed]
- Wichman, C.; Smith, L.M.; Yu, F. A Framework for Clinical and Translational Research in the Era of Rigor and Reproducibility. J. Clin. Trans. Sci. 2021, 5, e31. [Google Scholar] [CrossRef]
- Prasanna, B.; Kothapalli, P.; Vasanthan, M. The Role of Quality Assurance in Clinical Trials: Safeguarding Data Integrity and Compliance. Cureus 2024, 16, e67573. [Google Scholar] [CrossRef]
- Borowski, E.J.; Borwein, J.M. Collins dictionary of Mathematics. In Collins Dictionary of Mathematics; Harper Collins: New York, NY, USA, 2012. [Google Scholar]
- Barwick, V. Preparation of Calibration Curves—A Guide to Best Practice [Internet]. 2003. Available online: https://www.lgcgroup.com/media/2997/preparation-of-calibration-curves.pdf?ipignore=true (accessed on 16 October 2024).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petiti, J.; Caria, S.; Revel, L.; Pegoraro, M.; Divieto, C. Standardized Protocol for Resazurin-Based Viability Assays on A549 Cell Line for Improving Cytotoxicity Data Reliability. Cells 2024, 13, 1959. https://doi.org/10.3390/cells13231959
Petiti J, Caria S, Revel L, Pegoraro M, Divieto C. Standardized Protocol for Resazurin-Based Viability Assays on A549 Cell Line for Improving Cytotoxicity Data Reliability. Cells. 2024; 13(23):1959. https://doi.org/10.3390/cells13231959
Chicago/Turabian StylePetiti, Jessica, Sabrina Caria, Laura Revel, Mattia Pegoraro, and Carla Divieto. 2024. "Standardized Protocol for Resazurin-Based Viability Assays on A549 Cell Line for Improving Cytotoxicity Data Reliability" Cells 13, no. 23: 1959. https://doi.org/10.3390/cells13231959
APA StylePetiti, J., Caria, S., Revel, L., Pegoraro, M., & Divieto, C. (2024). Standardized Protocol for Resazurin-Based Viability Assays on A549 Cell Line for Improving Cytotoxicity Data Reliability. Cells, 13(23), 1959. https://doi.org/10.3390/cells13231959





