Neuronal Progenitors Suffer Genotoxic Stress in the Drosophila Clock Mutant per0
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drosophila Strains and Maintenance
2.2. Antioxidant Feeding
2.3. Immuno-Staining and Confocal Microscopy
2.4. Antibodies
2.5. Measurement of Mitochondrial ROS
2.6. Mitotic Chromosome Preparations
2.7. Immunofluorescence on CNS-Squash Preparations
2.8. Mitochondrial Oxygen Consumption Measurements
2.9. Statistics and Reproducibility
3. Results
3.1. per0 Mutants Are Subject to a High ROS Burden
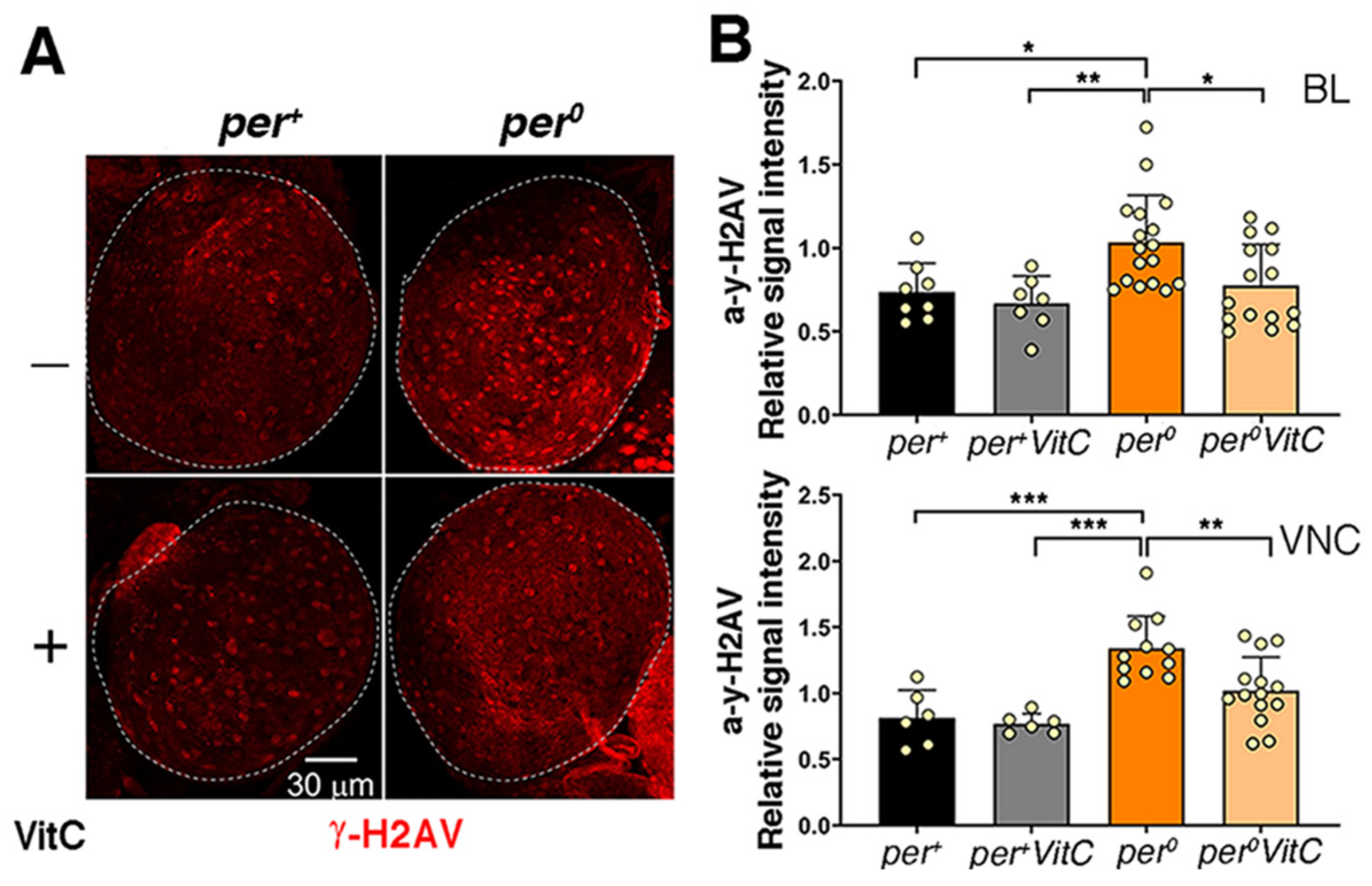
3.2. The CNS of per0 Larvae Shows DNA Damage and Chromosome Aberrations
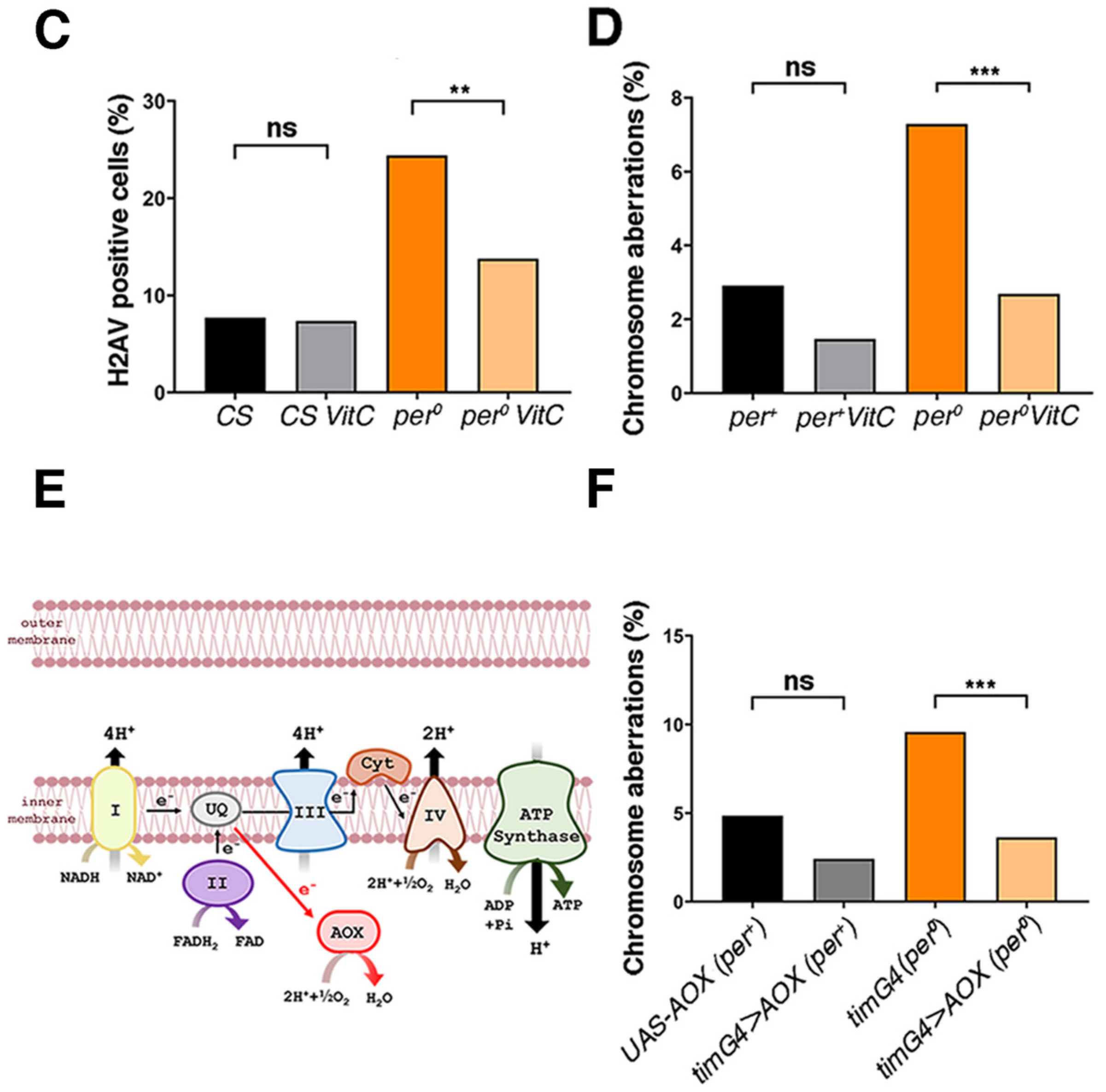
3.3. Buffering of ROS Rescues DNA Damage and Chromosome Aberrations in per0
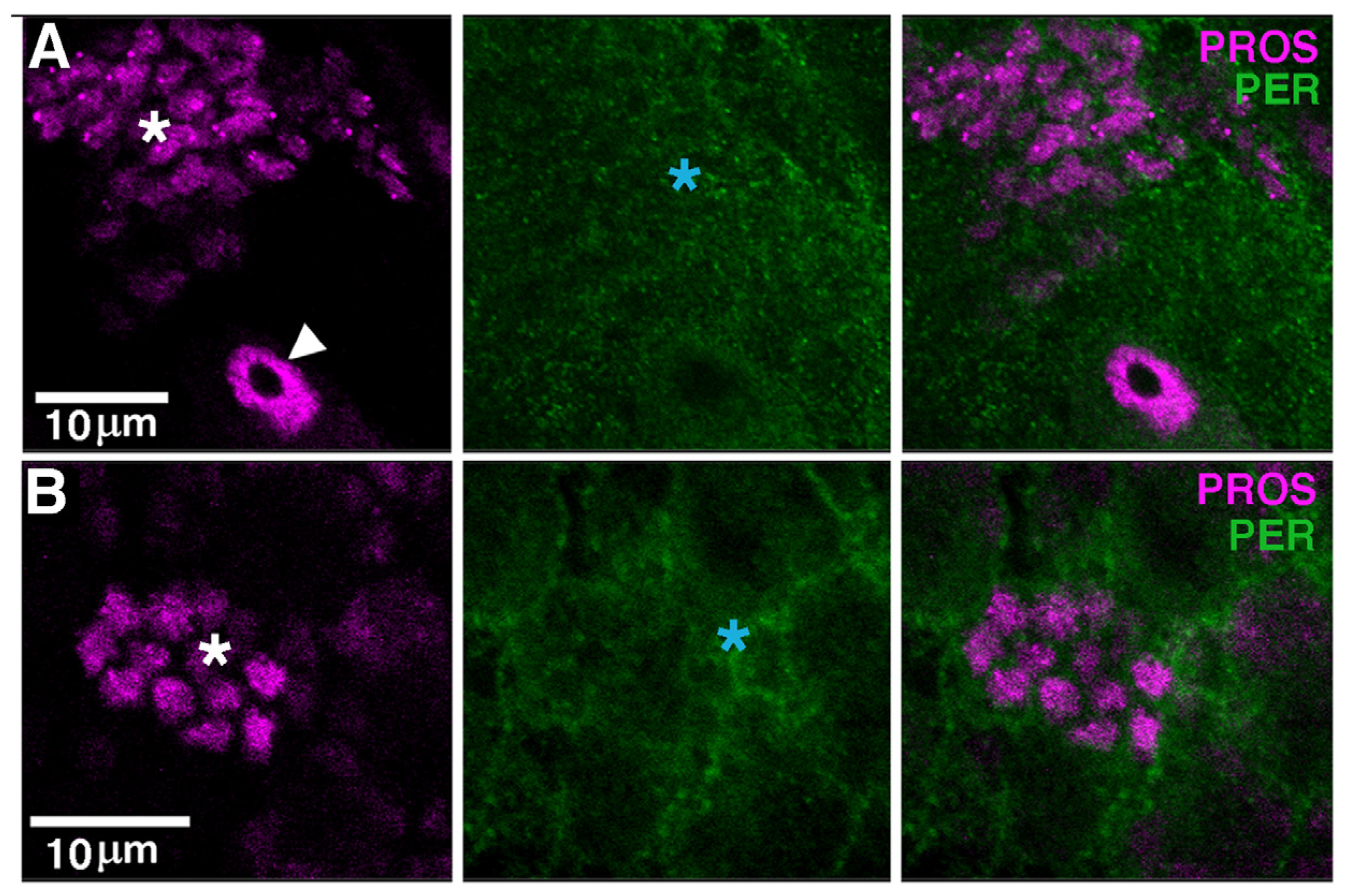
3.4. PER Is Expressed in Glia, Including Cortex
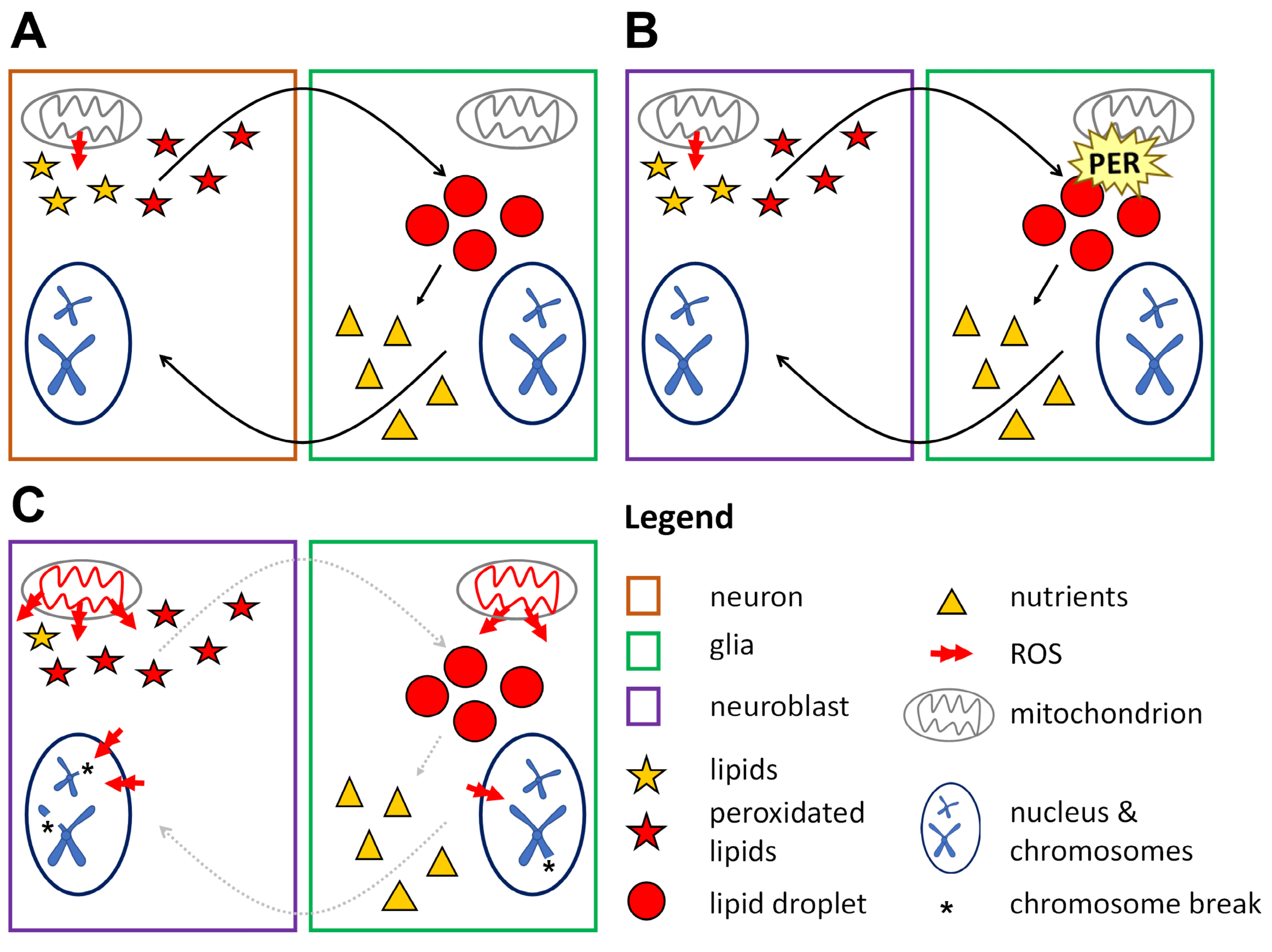
3.5. A Non-Cell Autonomous Genotoxic Effect of per0
3.6. per0 Likely Affects the Chromatin Landscape of Neural Progenitor Cells
4. Discussion
4.1. Lack of PER Results in Higher Levels of ROS
4.2. PER Is Expressed in Larval Glia
4.3. An Altered Epigenetic Landscape in per0 Larvae
4.4. Mechanistic Implications: ROS, ‘Neuroblast Sparing’ and PER
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ozkaya, O.; Rosato, E. The circadian clock of the fly: A neurogenetics journey through time. Adv. Genet. 2012, 77, 79–123. [Google Scholar] [PubMed]
- Patke, A.; Young, M.W.; Axelrod, S. Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 2020, 21, 67–84. [Google Scholar] [CrossRef] [PubMed]
- Claridge-Chang, A.; Wijnen, H.; Naef, F.; Boothroyd, C.; Rajewsky, N.; Young, M.W. Circadian regulation of gene expression systems in the Drosophila head. Neuron 2001, 32, 657–671. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.J.; Rosbash, M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 2001, 107, 567–578. [Google Scholar] [CrossRef]
- Ceriani, M.F.; Hogenesch, J.; Yanovsky, M.; Panda, S.; Straume, M.; Kay, S.A. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 2002, 22, 9305–9319. [Google Scholar] [CrossRef]
- Akhtar, R.A.; Reddy, A.B.; Maywood, E.S.; Clayton, J.D.; King, V.M.; Smith, A.G.; Gant, T.W.; Hastings, M.H.; Kyriacou, C.P. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002, 12, 540–550. [Google Scholar] [CrossRef]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science 2018, 359, eaao0318. [Google Scholar] [CrossRef]
- Doi, M.; Hirayama, J.; Sassone-Corsi, P. Circadian regulator CLOCK is a histone acetyltransferase. Cell 2006, 125, 497–508. [Google Scholar] [CrossRef]
- Duong, H.A.; Weitz, C.J. Temporal orchestration of repressive chromatin modifiers by circadian clock Period complexes. Nat. Struct. Mol. Biol. 2014, 21, 126–132. [Google Scholar] [CrossRef]
- Tartour, K.; Andriani, F.; Folco, E.G.; Letkova, D.; Schneider, R.; Saidi, I.; Sato, T.; Welz, P.S.; Benitah, S.A.; Allier, C.; et al. Mammalian PERIOD2 regulates H2A.Z incorporation in chromatin to orchestrate circadian negative feedback. Nat. Struct. Mol. Biol. 2022, 29, 549–562. [Google Scholar] [CrossRef]
- Michael, A.K.; Stoos, L.; Crosby, P.; Eggers, N.; Nie, X.Y.; Makasheva, K.; Minnich, M.; Healy, K.L.; Weiss, J.; Kempf, G.; et al. Cooperation between bHLH transcription factors and histones for DNA access. Nature 2023, 619, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Chen, Q.; Brovkina, M.; Clowney, E.J.; Yadlapalli, S. Clock-dependent chromatin accessibility rhythms regulate circadian transcription. PLoS Genet. 2024, 20, e1011278. [Google Scholar] [CrossRef]
- Houl, J.H.; Ng, F.; Taylor, P.; Hardin, P.E. CLOCK expression identifies developing circadian oscillator neurons in the brains of Drosophila embryos. BMC Neurosci. 2008, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, S.; Rickert, C.; Berger, C.; Technau, G.M.; Cantera, R. Spatio-temporal pattern of cells expressing the clock genes period and timeless and the lineages of period expressing neurons in the embryonic CNS of Drosophila melanogaster. Gene Expr. Patterns 2010, 10, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Yagita, K.; Horie, K.; Koinuma, S.; Nakamura, W.; Yamanaka, I.; Urasaki, A.; Shigeyoshi, Y.; Kawakami, K.; Shimada, S.; Takeda, J.; et al. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 3846–3851. [Google Scholar] [CrossRef] [PubMed]
- Weger, M.; Diotel, N.; Dorsemans, A.C.; Dickmeis, T.; Weger, B.D. Stem cells and the circadian clock. Dev. Biol. 2017, 431, 111–123. [Google Scholar] [CrossRef]
- Panda, S. Circadian physiology of metabolism. Science 2016, 354, 1008–1015. [Google Scholar] [CrossRef]
- Homem, C.C.F.; Steinmann, V.; Burkard, T.R.; Jais, A.; Esterbauer, H.; Knoblich, J.A. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell 2014, 158, 874–888. [Google Scholar] [CrossRef]
- Fropf, R.; Zhou, H.; Yin, J.C.P. The clock gene period differentially regulates sleep and memory in Drosophila. Neurobiol. Learn. Mem. 2018, 153, 2–12. [Google Scholar] [CrossRef]
- Konopka, R.J.; Wells, S. Drosophila clock mutations affect the morphology of a brain neurosecretory cell group. J. Neurobiol. 1980, 11, 411–415. [Google Scholar] [CrossRef]
- Dorcikova, M.M.; Duret, L.C.; Pottié, E.; Nagoshi, E. Circadian clock disruption promotes the degeneration of dopaminergic neurons in male Drosophila. Nat. Commun. 2023, 14, 5908. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.P.; Berni, J.; Ceriani, M.F. Circadian remodelling of neuronal circuits involved in rhythmic behavior. PLoS Biol. 2008, 6, e69. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Duhart, J.M.; Ceriani, M.F. Neuronal and Glial Clocks Underlying Structural Remodeling of Pacemaker Neurons in Drosophila. Front. Physiol. 2017, 8, 918. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Sehgal, A. AKT and TOR signalling set the pace of the circadian pacemaker. Curr. Biol. 2010, 20, 1203–1208. [Google Scholar] [CrossRef] [PubMed]
- Katewa, S.D.; Akagi, K.; Bose, N.; Rakshit, K.; Camarella, T.; Zheng, X.; Hall, D.; Davis, S.; Nelson, C.S.; Brem, R.B.; et al. Peripheral Circadian Clocks Mediate Dietary Restriction-Dependent Changes in Lifespan and Fat Metabolism in Drosophila. Cell Metab. 2016, 23, 143–154. [Google Scholar] [CrossRef]
- Giebultowicz, J.M. Circadian regulation of metabolism and healthspan in Drosophila. Free Radic. Biol. Med. 2018, 119, 62–68. [Google Scholar] [CrossRef]
- Krishnan, N.; Davis, A.J.; Giebultowicz, J.M. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2008, 374, 299–303. [Google Scholar] [CrossRef]
- Krishnan, N.; Kretzschmar, D.; Rakshit, K.; Chow, E.; Giebultowicz, J.M. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging 2009, 1, 937–948. [Google Scholar] [CrossRef]
- Schäbler, S.; Amatobi, K.M.; Horn, M.; Rieger, D.; Helfrich-Förster, C.; Mueller, M.J.; Wegener, C.; Fekete, A. Loss of function in the Drosophila clock gene period results in altered intermediary lipid metabolism and increased susceptibility to starvation. Cell Mol. Life Sci. 2020, 77, 4939–4956. [Google Scholar] [CrossRef] [PubMed]
- Amatobi, K.M.; Ozbek-Unal, A.G.; Schäbler, S.; Deppisch, P.; Helfrich-Förster, C.; Mueller, M.J.; Wegener, C.; Fekete, A. The circadian clock is required for rhythmic lipid transport in Drosophila in interaction with diet and photic condition. J. Lipid Res. 2023, 64, 100417. [Google Scholar] [CrossRef]
- Homem, C.C.F.; Knoblich, J.A. Drosophila neuroblasts: A model for stem cell biology. Development 2012, 139, 4297–4310. [Google Scholar] [CrossRef] [PubMed]
- Bayraktar, O.A.; Doe, C.Q. Combinatorial temporal patterning in progenitors expands neural diversity. Nature 2013, 498, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Helfrich-Förster, C.; Hall, J.C. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: Newly identified pacemaker candidates and novel features of clock gene product cycling. J. Neurosci. 1997, 17, 6745–6760. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Hall, J.C. Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 2000, 422, 66–94. [Google Scholar] [CrossRef]
- Stanewsky, R.; Frisch, B.; Brandes, C.; Hamblen-Coyle, M.J.; Rosbash, M.; Hall, J.C. Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: Mapping elements of the PER protein involved in circadian cycling. J. Neurosci. 1997, 17, 676–696. [Google Scholar] [CrossRef]
- Grima, B.; Chélot, E.; Xia, R.; Rouyer, F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 2004, 431, 869–873. [Google Scholar] [CrossRef]
- Emery, P.; So, W.V.; Kaneko, M.; Hall, J.C.; Rosbash, M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 1998, 95, 669–679. [Google Scholar] [CrossRef]
- Delventhal, R.; O’Connor, R.M.; Pantalia, M.M.; Ulgherait, M.; Kim, H.X.; Basturk, M.K.; Canman, J.C.; Shirasu-Hiza, M. Dissection of central clock function in Drosophila through cell-specific CRISPR-mediated clock gene disruption. Elife 2019, 8, e48308. [Google Scholar] [CrossRef]
- Yang, C.P.; Fu, C.C.; Sugino, K.; Liu, Z.; Ren, Q.; Liu, L.Y.; Yao, X.; Lee, L.P.; Lee, T. Transcriptomes of lineage-specific Drosophila neuroblasts profiled by genetic targeting and robotic sorting. Development 2016, 143, 411–421. [Google Scholar] [CrossRef]
- Fernandez-Ayala, D.J.; Sanz, A.; Vartiainen, S.; Kemppainen, K.K.; Babusiak, M.; Mustalahti, E.; Costa, R.; Tuomela, T.; Zeviani, M.; Chung, J.; et al. Expression of the Ciona intestinalis alternative oxidase (AOX) in Drosophila complements defects in mitochondrial oxidative phosphorylation. Cell Metab. 2009, 9, 449–460. [Google Scholar] [CrossRef]
- Vaccaro, A.; Kaplan Dor, Y.; Nambara, K.; Pollina, E.A.; Lin, C.; Greenberg, M.E.; Rogulja, D. Sleep Loss Can Cause Death through Accumulation of Reactive Oxygen Species in the Gut. Cell 2020, 181, 1307–1328. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Fanti, L.; Pimpinelli, S. Immunostaining of squash preparations of chromosomes of larval brains. Methods Mol. Biol. 2004, 247, 353–361. [Google Scholar] [PubMed]
- Campesan, S.; Del Popolo, I.; Marcou, K.; Straatman-Iwanowska, A.; Repici, M.; Boytcheva, V.K.; Cotton, V.E.; Allcock, N.; Rosato, E.; Kyriacou, C.P.; et al. Bypassing mitochondrial defects rescues Huntington’s phenotypes in Drosophila. Neurobiol. Dis. 2023, 185, 106236. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef]
- Baldi, S.; Becker, P.B. The variant histone H2A.V of Drosophila three roles, two guises. Chromosoma 2013, 122, 245–258. [Google Scholar] [CrossRef]
- Helfrich-Förster, C.; Shafer, O.T.; Wülbeck, C.; Grieshaber, E.; Rieger, D.; Taghert, P. Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J. Comp. Neurol. 2007, 500, 47–70. [Google Scholar] [CrossRef]
- Liu, T.; Mahesh, G.; Houl, J.H.; Hardin, P.E. Circadian Activators Are Expressed Days before They Initiate Clock Function in Late Pacemaker Neurons from Drosophila. J. Neurosci. 2015, 35, 8662–8671. [Google Scholar] [CrossRef]
- Ramon-Cañellas, P.; Peterson, H.P.; Morante, J. From Early to Late Neurogenesis: Neural Progenitors and the Glial Niche from a Fly’s Point of View. Neuroscience 2019, 399, 39–52. [Google Scholar] [CrossRef]
- Canat, A.; Atilla, D.; Torres-Padilla, M.E. Hyperosmotic stress induces 2-cell-like cells through ROS and ATR signaling. EMBO Rep. 2023, 24, e56194. [Google Scholar] [CrossRef]
- Kotova, E.; Lodhi, N.; Jarnik, M.; Pinnola, A.D.; Ji, Y.; Tulin, A.V. Drosophila histone H2A variant (H2Av) controls poly(ADP-ribose) polymerase 1 (PARP1) activation in chromatin. Proc. Natl. Acad. Sci. USA 2011, 108, 6205–6210. [Google Scholar] [CrossRef] [PubMed]
- Schotta, G.; Ebert, A.; Krauss, V.; Fischer, A.; Hoffmann, J.; Rea, S.; Jenuwein, T.; Dorn, R.; Reuter, G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002, 21, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Casale, A.M.; Cappucci, U.; Fanti, L.; Piacentini, L. Heterochromatin protein 1 (HP1) is intrinsically required for post-transcriptional regulation of Drosophila Germline Stem Cell (GSC) maintenance. Sci. Rep. 2019, 9, 4372. [Google Scholar] [CrossRef] [PubMed]
- Bailey, A.P.; Koster, G.; Guillermier, C.; Hirst, E.M.; MacRae, J.I.; Lechene, C.P.; Postle, A.D.; Gould, A.P. Antioxidant Role for Lipid Droplets in a Stem Cell Niche of Drosophila. Cell 2015, 163, 340–353. [Google Scholar] [CrossRef]
- Van den Ameele, J.; Brand, A.H. Neural stem cell temporal patterning and brain tumour growth rely on oxidative phosphorylation. Elife 2019, 8, e47887. [Google Scholar] [CrossRef]
- Da-Ré, C.; De Pittà, C.; Zordan, M.A.; Teza, G.; Nestola, F.; Zeviani, M.; Costa, R.; Bernardi, P. UCP4C mediates uncoupled respiration in larvae of Drosophila melanogaster. EMBO Rep. 2014, 15, 586–591. [Google Scholar] [CrossRef]
- Ulgherait, M.; Chen, A.; McAllister, S.F.; Kim, H.X.; Delventhal, R.; Wayne, C.R.; Garcia, C.J.; Recinos, Y.; Oliva, M.; Canman, J.C.; et al. Circadian regulation of mitochondrial uncoupling and lifespan. Nat. Commun. 2020, 11, 1927. [Google Scholar] [CrossRef]
- Zhao, J.; Warman, G.R.; Stanewsky, R.; Cheeseman, J.F. Development of the Molecular Circadian Clock and Its Light Sensitivity in Drosophila melanogaster. J. Biol. Rhythm. 2019, 34, 272–282. [Google Scholar] [CrossRef]
- Swaminathan, J.; Baxter, E.M.; Corces, V.G. The role of histone H2Av variant replacement and histone H4 acetylation in the establishment of Drosophila heterochromatin. Genes Dev. 2005, 19, 65–76. [Google Scholar] [CrossRef]
- Hardy, S.; Robert, F. Random deposition of histone variants: A cellular mistake or a novel regulatory mechanism? Epigenetics 2010, 5, 368–372. [Google Scholar] [CrossRef]
- Fu, Y.; Sinha, M.; Peterson, C.L.; Weng, Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008, 4, e1000138. [Google Scholar] [CrossRef] [PubMed]
- Simmons, J.R.; An, R.; Amankwaa, B.; Zayac, S.; Kemp, J.; Labrador, M. Phosphorylated histone variant γH2Av is associated with chromatin insulators in Drosophila. PLoS Genet. 2022, 18, e1010396. [Google Scholar] [CrossRef] [PubMed]
- Spielmann, M.; Lupiáñez, D.G.; Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018, 19, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Batut, P.J.; Bing, X.Y.; Sisco, Z.; Raimundo, J.; Levo, M.; Levine, M.S. Genome organization controls transcriptional dynamics during development. Science 2022, 375, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Lanet, E.; Maurange, C. Building a brain under nutritional restriction: Insights on sparing and plasticity from Drosophila studies. Front Physiol. 2014, 5, 117. [Google Scholar] [CrossRef]
- Uchida, K. 4-Hydroxy-2-nonenal: A product and mediator of oxidative stress. Prog. Lipid Res. 2003, 42, 318–343. [Google Scholar] [CrossRef]
- Kis, V.; Barti, B.; Lippai, M.; Sass, M. Specialized cortex glial cells accumulate lipid droplets in Drosophila melanogaster. PLoS ONE 2015, 10, e0131250. [Google Scholar] [CrossRef]
- Haynes, P.R.; Pyfrom, E.S.; Li, Y.; Stein, C.; Cuddapah, V.A.; Jacobs, J.A.; Yue, Z.; Sehgal, A. A neuron-glia lipid metabolic cycle couples daily sleep to mitochondrial homeostasis. Nat. Neurosci. 2024, 27, 666–678. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colonna Romano, N.; Marchetti, M.; Marangoni, A.; Leo, L.; Retrosi, D.; Rosato, E.; Fanti, L. Neuronal Progenitors Suffer Genotoxic Stress in the Drosophila Clock Mutant per0. Cells 2024, 13, 1944. https://doi.org/10.3390/cells13231944
Colonna Romano N, Marchetti M, Marangoni A, Leo L, Retrosi D, Rosato E, Fanti L. Neuronal Progenitors Suffer Genotoxic Stress in the Drosophila Clock Mutant per0. Cells. 2024; 13(23):1944. https://doi.org/10.3390/cells13231944
Chicago/Turabian StyleColonna Romano, Nunzia, Marcella Marchetti, Anna Marangoni, Laura Leo, Diletta Retrosi, Ezio Rosato, and Laura Fanti. 2024. "Neuronal Progenitors Suffer Genotoxic Stress in the Drosophila Clock Mutant per0" Cells 13, no. 23: 1944. https://doi.org/10.3390/cells13231944
APA StyleColonna Romano, N., Marchetti, M., Marangoni, A., Leo, L., Retrosi, D., Rosato, E., & Fanti, L. (2024). Neuronal Progenitors Suffer Genotoxic Stress in the Drosophila Clock Mutant per0. Cells, 13(23), 1944. https://doi.org/10.3390/cells13231944






