Association of Genomic Alterations with the Presence of Serum Monoclonal Proteins in Chronic Lymphocytic Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Serum Immunofixation Electrophoresis
2.3. Next-Generation Sequencing
2.4. Statistical Analysis
3. Results
3.1. Clinical and Laboratory Characteristics
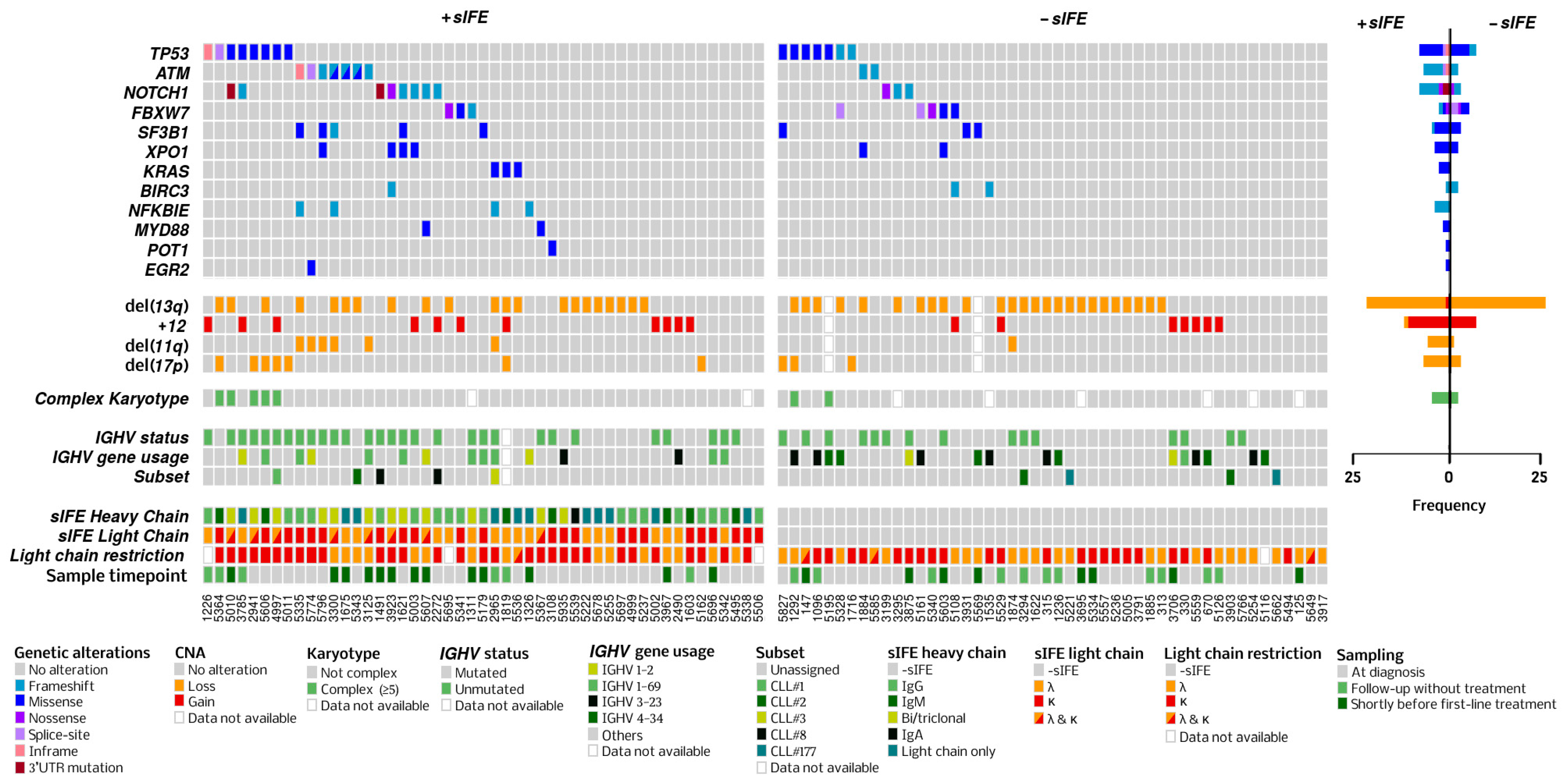
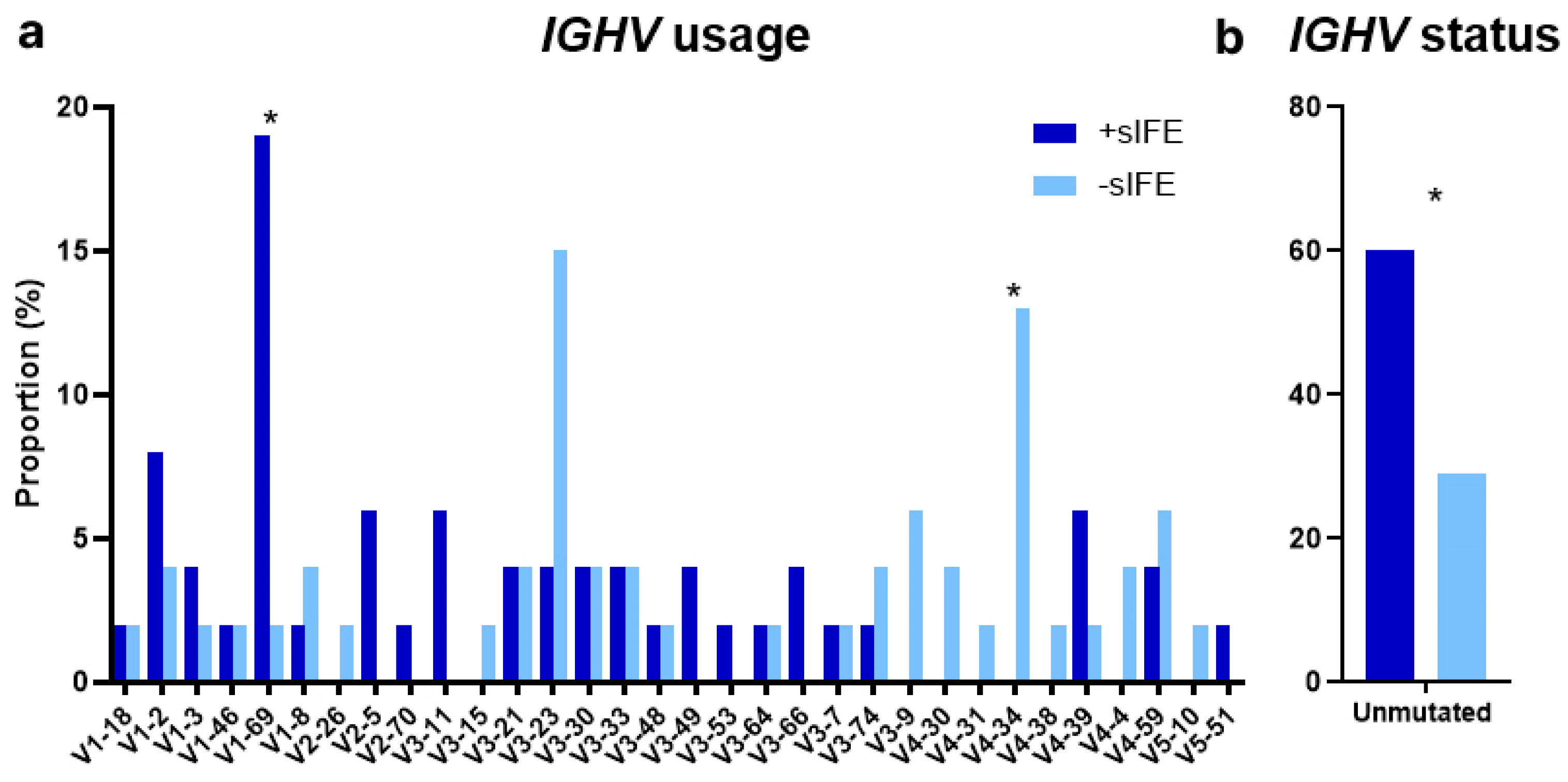
3.2. Genomic Characteristics
3.3. Heavy-Chain Immunoglobulin Isotype
3.4. Light-Chain Immunoglobulin Isotype
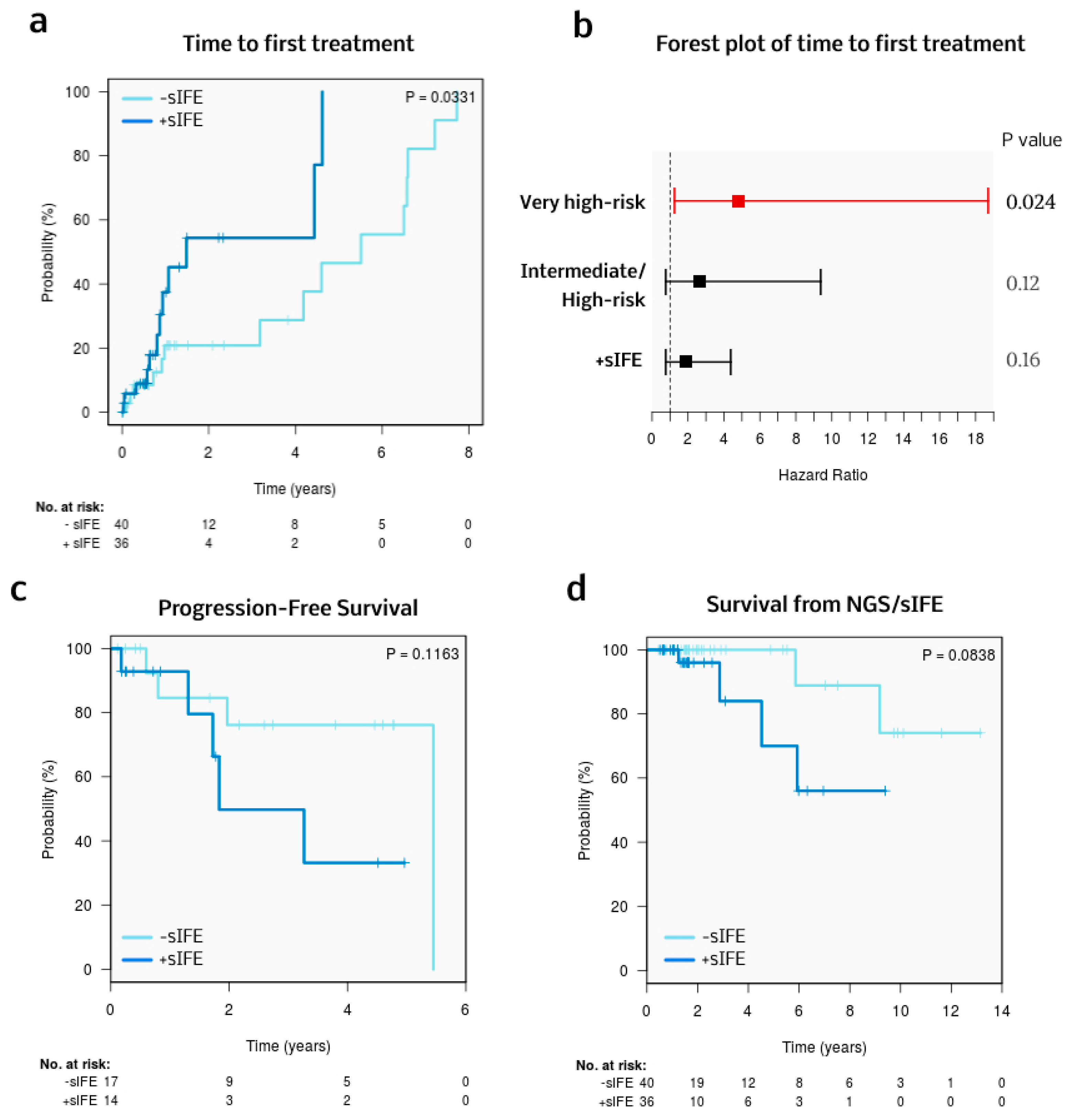
3.5. Treatment and Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. B Cells and Antibodies. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. Available online: https://www.ncbi.nlm.nih.gov/books/NBK21054/ (accessed on 3 March 2024).
- Wahed, A.; Quesada, A.; Dasgupta, A. Monoclonal Gammopathies and Their Detection. In Hematology and Coagulation; Elsevier: Amsterdam, The Netherlands, 2020; pp. 113–126. ISBN 978-0-12-814964-5. [Google Scholar]
- Cox, M.C.; Esposito, F.; Postorino, M.; Venditti, A.; Di Napoli, A. Serum Paraprotein Is Associated with Adverse Prognostic Factors and Outcome, across Different Subtypes of Mature B-Cell Malignancies—A Systematic Review. Cancers 2023, 15, 4440. [Google Scholar] [CrossRef] [PubMed]
- Deegan, M.; Abraham, J.; Sawdyk, M.; Van Slyck, E. High Incidence of Monoclonal Proteins in the Serum and Urine of Chronic Lymphocytic Leukemia Patients. Blood 1984, 64, 1207–1211. [Google Scholar] [CrossRef] [PubMed]
- Pangalis, G.A.; Moutsopoulos, H.M.; Papadopoulos, N.M.; Costello, R.; Kokkinou, S.; Fessas, P. Monoclonal and Oligoclonal Immunoglobulins in the Serum of Patients with B-Chronic Lymphocytic Leukemia. Acta Haematol. 1988, 80, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.A.; Robbins, B.A.; Bylund, D.J.; Piro, L.D.; Saven, A.; Ellison, D.J. Identification of Monoclonal Immunoglobulins and Quantitative Immunoglobulin Abnormalities in Hairy Cell Leukemia and Chronic Lymphocytic Leukemia. Am. J. Clin. Pathol. 1994, 102, 580–585. [Google Scholar] [CrossRef]
- Tsai, H.-T.; Caporaso, N.E.; Kyle, R.A.; Katzmann, J.A.; Dispenzieri, A.; Hayes, R.B.; Marti, G.E.; Albitar, M.; Ghia, P.; Rajkumar, S.V.; et al. Evidence of Serum Immunoglobulin Abnormalities up to 9.8 Years before Diagnosis of Chronic Lymphocytic Leukemia: A Prospective Study. Blood 2009, 114, 4928–4932. [Google Scholar] [CrossRef]
- Xu, W.; Wang, Y.H.; Fan, L.; Fang, C.; Zhu, D.X.; Wang, D.M.; Qiao, C.; Wu, Y.J.; Li, J.Y. Prognostic Significance of Serum Immunoglobulin Paraprotein in Patients with Chronic Lymphocytic Leukemia. Leuk. Res. 2011, 35, 1060–1065. [Google Scholar] [CrossRef]
- Rizzo, D.; Chauzeix, J.; Trimoreau, F.; Woillard, J.B.; Genevieve, F.; Bouvier, A.; Labrousse, J.; Poli, C.; Guerin, E.; Dmytruk, N.; et al. IgM Peak Independently Predicts Treatment-Free Survival in Chronic Lymphocytic Leukemia and Correlates with Accumulation of Adverse Oncogenetic Events. Leukemia 2015, 29, 337–345. [Google Scholar] [CrossRef]
- Corbingi, A.; Innocenti, I.; Tomasso, A.; Pasquale, R.; Visentin, A.; Varettoni, M.; Flospergher, E.; Autore, F.; Morelli, F.; Trentin, L.; et al. Monoclonal Gammopathy and Serum Immunoglobulin Levels as Prognostic Factors in Chronic Lymphocytic Leukaemia. Br. J. Haematol. 2020, 190, 901–908. [Google Scholar] [CrossRef]
- Mozas, P.; Pineyroa, J.A.; Nadeu, F.; Magnano, L.; Rivero, A.; Rivas-Delgado, A.; Bataller, A.; Fabregat, A.; Gine, E.; Baumann, T.; et al. Serum Monoclonal Component in Chronic Lymphocytic Leukemia: Baseline Correlations and Prognostic Impact. Haematologica 2020, 106, 1754–1757. [Google Scholar] [CrossRef]
- Puente, X.S.; Beà, S.; Valdés-Mas, R.; Villamor, N.; Gutiérrez-Abril, J.; Martín-Subero, J.I.; Munar, M.; Rubio-Pérez, C.; Jares, P.; Aymerich, M.; et al. Non-Coding Recurrent Mutations in Chronic Lymphocytic Leukaemia. Nature 2015, 526, 519–524. [Google Scholar] [CrossRef]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Böttcher, S.; et al. Mutations Driving CLL and Their Evolution in Progression and Relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Knisbacher, B.A.; Lin, Z.; Hahn, C.K.; Nadeu, F.; Duran-Ferrer, M.; Stevenson, K.E.; Tausch, E.; Delgado, J.; Barbera-Mourelle, A.; Taylor-Weiner, A.; et al. Molecular Map of Chronic Lymphocytic Leukemia and Its Impact on Outcome. Nat. Genet. 2022, 54, 1664–1674. [Google Scholar] [CrossRef] [PubMed]
- Sutton, L.-A.; Ljungstrom, V.; Mansouri, L.; Young, E.; Cortese, D.; Navrkalova, V.; Malcikova, J.; Muggen, A.F.; Trbusek, M.; Panagiotidis, P.; et al. Targeted Next-Generation Sequencing in Chronic Lymphocytic Leukemia: A High-Throughput yet Tailored Approach Will Facilitate Implementation in a Clinical Setting. Haematologica 2015, 100, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Sutton, L.-A.; Ljungström, V.; Enjuanes, A.; Cortese, D.; Skaftason, A.; Tausch, E.; Kozubik, K.S.; Nadeu, F.; Armand, M.; Malcikova, J.; et al. Comparative Analysis of Targeted Next-Generation Sequencing Panels for the Detection of Gene Mutations in Chronic Lymphocytic Leukemia: An ERIC Multi-Center Study. Haematologica 2020, 106, 682–691. [Google Scholar] [CrossRef] [PubMed]
- López-Oreja, I.; López-Guerra, M.; Correa, J.; Mozas, P.; Muntañola, A.; Muñoz, L.; Salgado, A.-C.; Ruiz-Gaspà, S.; Costa, D.; Beà, S.; et al. All-CLL: A Capture-Based Next-Generation Sequencing Panel for the Molecular Characterization of Chronic Lymphocytic Leukemia. HemaSphere 2023, 7, e962. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.D.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th Edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef]
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; De Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A Report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef]
- Eichhorst, B.; Robak, T.; Montserrat, E.; Ghia, P.; Niemann, C.U.; Kater, A.P.; Gregor, M.; Cymbalista, F.; Buske, C.; Hillmen, P.; et al. Chronic Lymphocytic Leukaemia: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2021, 32, 23–33. [Google Scholar] [CrossRef]
- Crespo, M.; Medina-Pérez, A.; Muntañola, A.; Abrisqueta, P.; Alcoceba, M.; Bosch, F.; Delgado, J.; de la Serna, J.; Espinet, B.; González-Díaz, M.; et al. Guía de Práctica Clínica para el Diagnóstico y Tratamiento de la Leucemia Linfocítica Crónica y el Linfoma Linfocítico de Células Pequeñas; Grupo Español de Leucemia Linfocítica Crónica: Madrid, Spain, 2024; ISBN 978-84-09-58591-5. [Google Scholar]
- Nadeu, F.; Mas-de-les-Valls, R.; Navarro, A.; Royo, R.; Martín, S.; Villamor, N.; Suárez-Cisneros, H.; Mares, R.; Lu, J.; Enjuanes, A.; et al. IgCaller for Reconstructing Immunoglobulin Gene Rearrangements and Oncogenic Translocations from Whole-Genome Sequencing in Lymphoid Neoplasms. Nat. Commun. 2020, 11, 3390. [Google Scholar] [CrossRef]
- Brochet, X.; Lefranc, M.-P.; Giudicelli, V. IMGT/V-QUEST: The Highly Customized and Integrated System for IG and TR Standardized V-J and V-D-J Sequence Analysis. Nucleic Acids Res. 2008, 36, W503–W508. [Google Scholar] [CrossRef]
- Bystry, V.; Agathangelidis, A.; Bikos, V.; Sutton, L.A.; Baliakas, P.; Hadzidimitriou, A.; Stamatopoulos, K.; Darzentas, N. ARResT/AssignSubsets: A Novel Application for Robust Subclassification of Chronic Lymphocytic Leukemia Based on B Cell Receptor IG Stereotypy. Bioinformatics 2015, 31, 3844–3846. [Google Scholar] [CrossRef] [PubMed]
- Agathangelidis, A.; Chatzidimitriou, A.; Chatzikonstantinou, T.; Tresoldi, C.; Davis, Z.; Giudicelli, V.; Kossida, S.; Belessi, C.; Rosenquist, R.; Ghia, P.; et al. Immunoglobulin Gene Sequence Analysis in Chronic Lymphocytic Leukemia: The 2022 Update of the Recommendations by ERIC, the European Research Initiative on CLL. Leukemia 2022, 36, 1961–1968. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Kröber, A.; Bullinger, L.; Döhner, K.; Bentz, M.; Lichter, P. Genomic Aberrations and Survival in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef] [PubMed]
- Kleinstern, G.; O’Brien, D.R.; Li, X.; Tian, S.; Kabat, B.F.; Rabe, K.G.; Norman, A.D.; Yan, H.; Vachon, C.M.; Boddicker, N.J.; et al. Tumor Mutational Load Predicts Time to First Treatment in Chronic Lymphocytic Leukemia (CLL) and Monoclonal B-cell Lymphocytosis beyond the CLL International Prognostic Index. Am. J. Hematol. 2020, 95, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Mikhaleva, M.; Tyekucheva, S.; Mashima, K.; Fernandes, S.M.; Davids, M.S.; Brown, J.R. Higher Mutational Burden Is an Independent Predictor of Shorter Time to First Treatment in Untreated Chronic Lymphocytic Leukemia Patients. Blood 2023, 142, 3270. [Google Scholar] [CrossRef]
- Kleinstern, G.; Boddicker, N.J.; O’Brien, D.R.; Allmer, C.; Rabe, K.G.; Norman, A.D.; Griffin, R.; Yan, H.; Ma, T.; Call, T.G.; et al. Tumor Mutational Load Is Prognostic for Progression to Therapy among High-Count Monoclonal B-Cell Lymphocytosis. Blood Adv. 2024, 8, 2118–2129. [Google Scholar] [CrossRef]
- Nadeu, F.; Clot, G.; Delgado, J.; Martín-García, D.; Baumann, T.; Salaverria, I.; Beà, S.; Pinyol, M.; Jares, P.; Navarro, A.; et al. Clinical Impact of the Subclonal Architecture and Mutational Complexity in Chronic Lymphocytic Leukemia. Leukemia 2018, 32, 645–653. [Google Scholar] [CrossRef]
- Griffin, R.; Wiedmeier-Nutor, J.E.; Parikh, S.A.; McCabe, C.E.; O’Brien, D.R.; Boddicker, N.J.; Kleinstern, G.; Rabe, K.G.; Bruins, L.; Brown, S.; et al. Differential Prognosis of Single and Multiple TP53 Abnormalities in High-Count MBL and Untreated CLL. Blood Adv. 2023, 7, 3169–3179. [Google Scholar] [CrossRef]
- Malcikova, J.; Tausch, E.; Rossi, D.; Sutton, L.A.; Soussi, T.; Zenz, T.; Kater, A.P.; Niemann, C.U.; Gonzalez, D.; Davi, F.; et al. ERIC Recommendations for TP53 Mutation Analysis in Chronic Lymphocytic Leukemia—Update on Methodological Approaches and Results Interpretation. Leukemia 2018, 32, 1070–1080. [Google Scholar] [CrossRef]
- Roco, J.A.; Mesin, L.; Binder, S.C.; Nefzger, C.; Gonzalez-Figueroa, P.; Canete, P.F.; Ellyard, J.; Shen, Q.; Robert, P.A.; Cappello, J.; et al. Class-Switch Recombination Occurs Infrequently in Germinal Centers. Immunity 2019, 51, 337–350.e7. [Google Scholar] [CrossRef] [PubMed]
| sIFE at the Time of NGS | ||||
|---|---|---|---|---|
| Characteristic | All Patients (n = 97) | Negative (n = 48, 49%) | Positive (n = 49, 51%) | p Value |
| Age in years, median (range) | 69 (41–96) | 68 (44–96) | 70 (41–94) | NS |
| Male sex, n (%) | 57 (59) | 27 (56) | 30 (61) | NS |
| Diagnosis | ||||
| CLL, n (%) | 89 (92) | 43 (90) | 46 (94) | NS |
| MBL, n (%) | 5 (5) | 4 (8) | 1 (2) | |
| SLL, n (%) | 3 (3) | 1 (2) | 2 (4) | |
| ECOG PS ≥ 1, n (%) (n = 89) | 8 (9) | 2 (4) | 6 (14) | NS |
| B symptoms, n (%) | 2 (2) | 0 | 2 (4) | NS |
| Binet stage C, n (%) | 13 (13) | 5 (10) | 8 (16) | NS |
| Rai stage III-IV, n (%) | 14 (14) | 5 (10) | 9 (18) | NS |
| Lymphadenopathy (CT), n (%) | 35 (36) | 14 (30) | 21 (43) | NS |
| ALC > 15 × 109/L, n (%) | 56 (58) | 27 (56) | 29 (59) | NS |
| LDH above ULN, n (%) (n = 92) | 13 (14) | 7 (15) | 6 (13) | NS |
| β2-microglobulin above ULN, n (%) (n = 91) | 52 (57) | 21 (47) | 31 (67) | NS |
| Complex karyotype (n = 88) | 7 (8) | 2 (5) | 5 (11) | NS |
| FISH, n (%) | ||||
| Normal | 15 (16) | 8 (18) | 7 (15) | NS |
| del(13)(q14.3) | 37 (40) | 22 (49) | 15 (32) | |
| Trisomy 12 | 13 (14) | 5 (11) | 8 (17) | |
| del(11)(q22.3) | 8 (9) | 2 (4) | 6 (13) | |
| del(17)(p13.1) | 19 (21) | 8 (18) | 11 (23) | |
| CLL-IPI, n (%) (n = 87) | ||||
| Low | 20 (23) | 13 (32) | 7 (15) | |
| Intermediate | 16 (18) | 10 (24) | 6 (13) | 0.055 |
| High | 33 (38) | 13 (32) | 20 (44) | |
| Very high | 18 (21) | 5 (12) | 13 (28) | |
| Diagnosis period | ||||
| 2003–2007 | 3 (3) | 1 (2) | 2 (4) | |
| 2008–2012 | 13 (13) | 9 (19) | 4 (8) | NS |
| 2013–2017 | 19 (20) | 10 (21) | 9 (19) | |
| 2018–2023 | 62 (64) | 28 (58) | 34 (69) | |
| NGS and sIFE date | ||||
| 2003–2007 | 0 | 0 | 0 | |
| 2008–2012 | 3 (3) | 3 (6) | 0 | NS |
| 2013–2017 | 14 (14) | 7 (15) | 7 (14) | |
| 2018–2023 | 80 (83) | 38 (79) | 42 (86) | |
| sIFE at the Time of NGS | ||||
|---|---|---|---|---|
| Pathogenic/Likely Pathogenic Mutation | All Patients (n = 97) | Negative (n = 48, 49%) | Positive (n = 49, 51%) | p Value |
| Presence of any mutation, n (%) | 51 (53) | 19 (40) | 32 (65) | 0.0196 |
| Number of mutations, median (range) | 1 (0–4) | 0 (0–3) | 1 (0–4) | 0.0069 |
| Number of mutated genes, median (range) | 1 (0–3) | 0 (0–2) | 1 (0–3) | 0.0061 |
| ATM, n (%) | 9 (9) | 2 (4) | 7 (14) | NS |
| BCL2, n (%) | 0 | 0 | 0 | - |
| BIRC3, n (%) | 3 (5) | 2 (4) | 1 (2) | NS |
| BTK, n (%) | 0 | 0 | 0 | - |
| CXCR4, n (%) | 0 | 0 | 0 | - |
| EGR2, n (%) | 1 (1) | 0 | 1 (2) | - |
| FBXW7, n (%) | 8 (8) | 5 (10) | 3 (6) | NS |
| KRAS, n (%) | 3 (3) | 0 | 3 (6) | NS |
| MYD88, n (%) | 2 (2) | 0 | 2 (4) | NS |
| NFKBIE, n (%) | 4 (4) | 0 | 4 (8) | NS |
| NOTCH1, n (%) | 11 (11) | 3 (6) | 8 (16) | NS |
| PLCG2, n (%) | 0 | 0 | 0 | - |
| POT1, n (%) | 1 (1) | 0 | 1 (2) | - |
| SF3B1, n (%) | 8 (8) | 3 (6) | 5 (10) | NS |
| TP53, n (%) | 15 (15) | 7 (15) | 8 (16) | NS |
| XPO1, n (%) | 6 (6) | 2 (4) | 4 (8) | NS |
| Mutated Gene, n (%) | -sIFE/Non-IgG +sIFE (n = 69, 71%) | IgG +sIFE (n = 28, 29%) | p | -sIFE/Non-IgM +sIFE (n = 83, 86%) | IgM +sIFE (n = 14, 14%) | p | -sIFE/Monoclonal +sIFE (n = 85, 88%) | Bi/Triclonal (n = 12, 12%) | p |
|---|---|---|---|---|---|---|---|---|---|
| ATM | 4 (6) | 5 (18) | NS | 7 (8) | 2 (14) | NS | 6 (7) | 3 (25) | NS |
| FBXW7 | 5 (7) | 3 (11) | NS | 8 (10) | 0 | NS | 7 (8) | 1 (8) | NS |
| NFKBIE | 2 (3) | 2 (7) | NS | 4 (5) | 0 | NS | 3 (4) | 1 (8) | NS |
| NOTCH1 | 6 (7) | 6 (21) | NS | 10 (12) | 2 (14) | NS | 8 (9) | 4 (33) | 0.04 |
| SF3B1 | 3 (4) | 5 (18) | 0.04 | 7 (8) | 1 (7) | NS | 5 (6) | 3 (25) | NS |
| TP53 | 11 (16) | 4 (14) | NS | 10 (12) | 5 (36) | 0.04 | 12 (14) | 3 (25) | NS |
| XPO1 | 2 (3) | 4 (14) | NS | 5 (6) | 1 (7) | NS | 3 (4) | 3 (25) | 0.03 |
| sIFE at the Time of NGS | ||||
|---|---|---|---|---|
| First Line of Treatment * | Patients (n = 76) | Negative (n = 40, 53%) | Positive (n = 36, 47%) | p Value |
| Untreated, n (%) | 45 (60) | 24 (60) | 21 (58) | NS |
| BTKi, n (%) | 29 (38) | 15 (37) | 14 (39) | |
| Ibrutinib | 20 (70) | 11 (74) | 9 (65) | |
| Acalabrutinib | 5 (17) | 2 (13) | 3 (21) | |
| Zanubrutinib | 4 (13) | 2 (13) | 2 (14) | |
| BCL2i, n (%) | 1 (1) | 0 | 1 (3) | |
| Immunochemotherapy n (%) | 1 (1) | 1 (3) | 0 | |
| Univariable Analysis | Multivariable Analysis (75 Cases, 27 Events) | ||||
|---|---|---|---|---|---|
| Parameter | Risk Category | HR (95% CI) | p | HR (95% CI) | p Value |
| Sex | Male | 0.72 (0.34–1.53) | NS | NI a | |
| Age | >65 years | 1.44 (0.63–3.32) | NS | NI b | |
| Stage | Binet B-C and/or Rai I-IV | 3.47 (1.39–8.68) | 0.008 | NI b | |
| ECOG PS | ≥1 | 1.85 (0.47–7.27) | NS | NI a | |
| ALC | >15 × 109/L | 1.02 (0.49–2.13) | NS | NI a | |
| B2M levels | >3.5 mg/L | 3.13 (1.33–7.4) | 0.01 | NI b | |
| IGHV status | Unmutated | 4.16 (1.69–10.25) | 0.001 | NI b | |
| del(17p) and/or TP53 mutation | Present | 1.28 (0.56–2.9) | NS | NI b | |
| CLL-IPI risk group | Intermediate/ High-risk | 2.98 (0.85–10.39) | NS | 2.68 (0.76–9.4) | NS |
| Very high-risk | 6.28 (1.68–23.52) | 0.006 | 4.8 (1.32–18.71) | 0.02 | |
| sIFE | Positive | 2.6 (1.14–6.08) | 0.03 | 1.86 (0.79–4.41) | NS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piñeyroa, J.A.; López-Oreja, I.; Nadeu, F.; Martínez-Farran, A.; Aróstegui, J.I.; López-Guerra, M.; Correa, J.G.; Fabregat, A.; Villamor, N.; Monge-Escatín, I.; et al. Association of Genomic Alterations with the Presence of Serum Monoclonal Proteins in Chronic Lymphocytic Leukemia. Cells 2024, 13, 1839. https://doi.org/10.3390/cells13221839
Piñeyroa JA, López-Oreja I, Nadeu F, Martínez-Farran A, Aróstegui JI, López-Guerra M, Correa JG, Fabregat A, Villamor N, Monge-Escatín I, et al. Association of Genomic Alterations with the Presence of Serum Monoclonal Proteins in Chronic Lymphocytic Leukemia. Cells. 2024; 13(22):1839. https://doi.org/10.3390/cells13221839
Chicago/Turabian StylePiñeyroa, Juan A., Irene López-Oreja, Ferran Nadeu, Ares Martínez-Farran, Juan Ignacio Aróstegui, Mónica López-Guerra, Juan Gonzalo Correa, Aleix Fabregat, Neus Villamor, Ines Monge-Escatín, and et al. 2024. "Association of Genomic Alterations with the Presence of Serum Monoclonal Proteins in Chronic Lymphocytic Leukemia" Cells 13, no. 22: 1839. https://doi.org/10.3390/cells13221839
APA StylePiñeyroa, J. A., López-Oreja, I., Nadeu, F., Martínez-Farran, A., Aróstegui, J. I., López-Guerra, M., Correa, J. G., Fabregat, A., Villamor, N., Monge-Escatín, I., Albiol, N., Costa, D., Aymerich, M., Beà, S., Campo, E., Delgado, J., Colomer, D., & Mozas, P. (2024). Association of Genomic Alterations with the Presence of Serum Monoclonal Proteins in Chronic Lymphocytic Leukemia. Cells, 13(22), 1839. https://doi.org/10.3390/cells13221839






