N6-Methyladenosine RNA Modification Regulates the Differential Muscle Development in Large White and Ningxiang Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. RNA Extraction and m6A MeRIP
2.3. Library Preparation
2.4. Sequencing and Analysis
2.5. Differential Expression Analysis
2.6. MeRIP-Seq Analysis
2.7. RNA m6A Dot Blot Assay
2.8. Histology Staining
2.9. Porcine Skeletal Muscle Satellite Cell Isolation and Culture
2.10. Western Blotting
2.11. Cell Immunofluorescence Staining
2.12. RNA Immunoprecipitation qPCR
2.13. Real-Time PCR Analysis
2.14. siRNA Synthesis and Cell Transfection
2.15. Luciferase Assay
3. Results
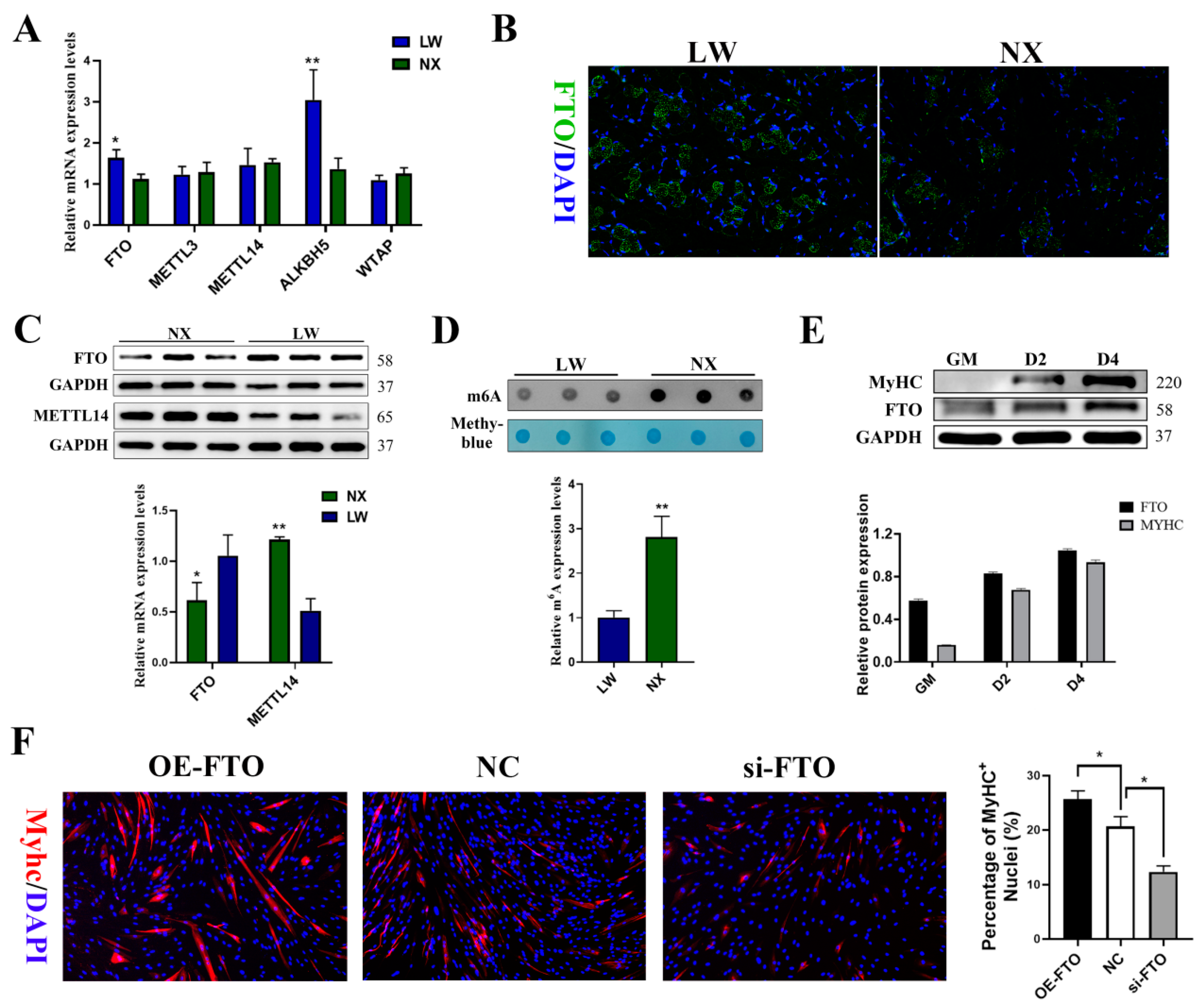
3.1. Differences between LW and NX Pigs in the Neonatal Stages of Muscle Development
3.2. m6A Podifications Regulated Porcine Muscle Development
3.3. Global Features of mRNA m6A Modification in LW and NX Porcine Muscles
3.4. Analysis of Differential m6A Modification Genes in NX and LW Pigs
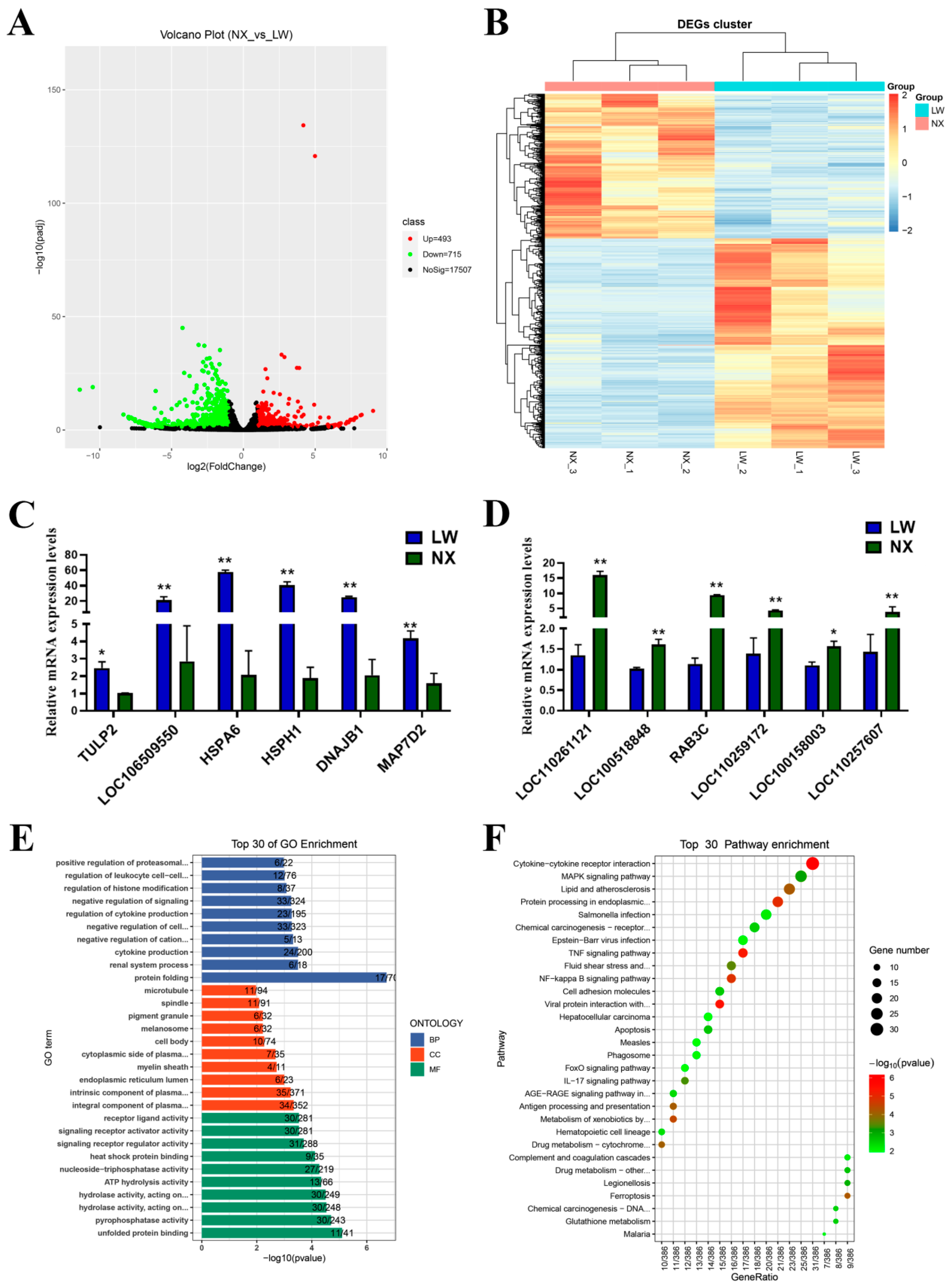
3.5. Analysis of Differentially Expressed Genes in NX and LW Pigs
3.6. Conjoint Analyses of MeRIP-Seq and RNA-Seq Data
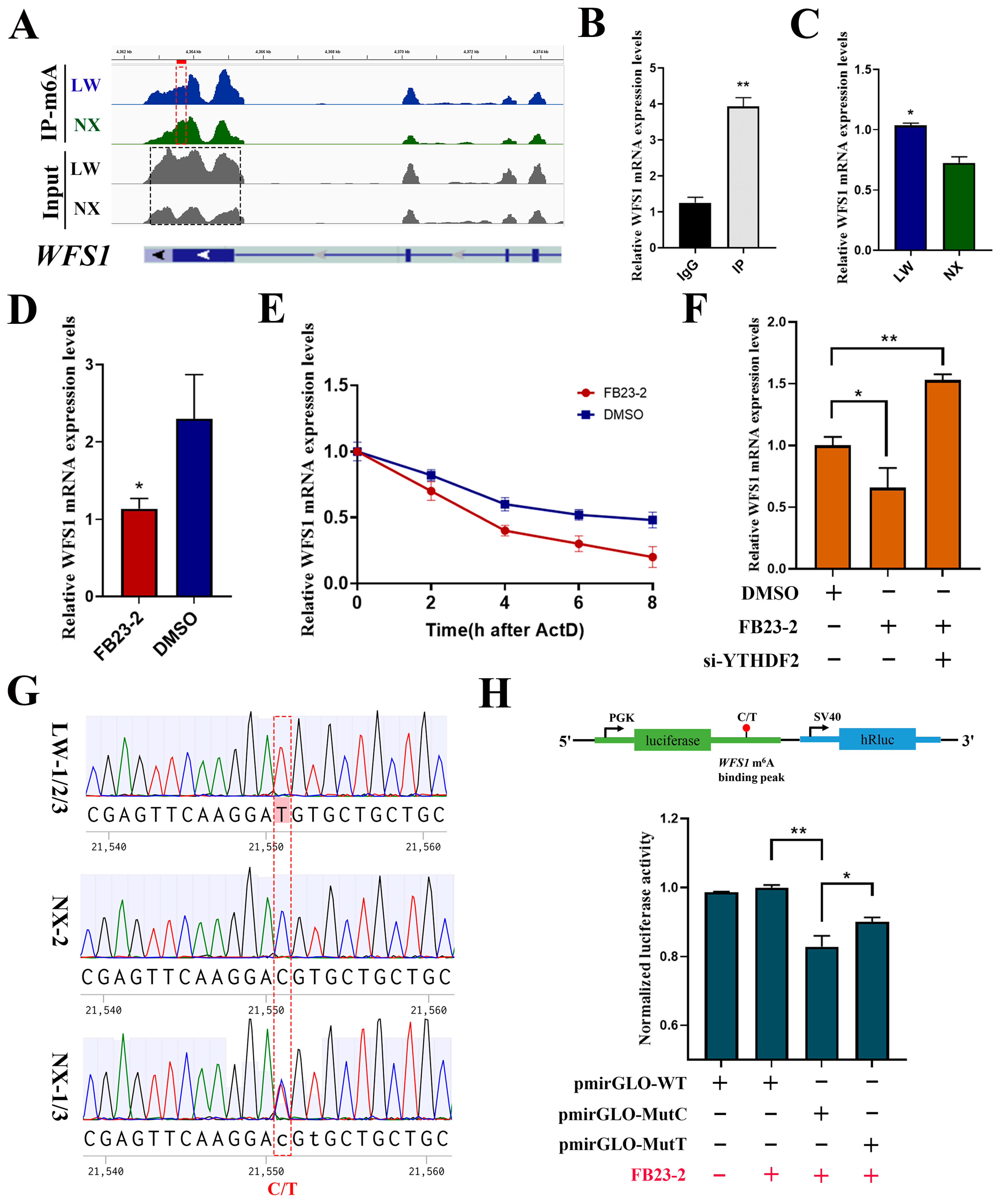
3.7. m6A Modification Regulated Differential Expression of WFS1 in NX and LW Porcine Muscle
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Prather, R.S. Pig genomics for biomedicine. Nat. Biotechnol. 2013, 31, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Mo, D.; Li, A.; Gong, W.; Xiao, S.; Zhang, Y.; Qin, L.; Niu, Y.; Guo, Y.; Liu, X.; et al. Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness. PLoS ONE 2011, 6, e19774. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, J.; Liu, H.; Xi, Y.; Xue, M.; Liu, W.; Zhuang, Z.; Lei, M. Dynamic transcriptome profiles of skeletal muscle tissue across 11 developmental stages for both Tongcheng and Yorkshire pigs. BMC Genom. 2015, 16, 377. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, J.; Fan, X.; Chen, J.; Wang, Z.; Liu, X.; Yi, G.; Liu, Y.; Niu, Y.; Zhang, L.; et al. The genome variation and developmental transcriptome maps reveal genetic differentiation of skeletal muscle in pigs. PLoS Genet. 2021, 17, e1009910. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Oerum, S.; Meynier, V.; Catala, M.; Tisné, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021, 49, 7239–7255. [Google Scholar] [CrossRef]
- Bushkin, G.G.; Pincus, D.; Morgan, J.T.; Richardson, K.; Lewis, C.; Chan, S.H.; Bartel, D.P.; Fink, G.R. m(6)A modification of a 3′ UTR site reduces RME1 mRNA levels to promote meiosis. Nat. Commun. 2019, 10, 3414. [Google Scholar] [CrossRef]
- Gu, C.; Wang, Z.; Zhou, N.; Li, G.; Kou, Y.; Luo, Y.; Wang, Y.; Yang, J.; Tian, F. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol. Cancer 2019, 18, 168. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hsu, P.J.; Chen, Y.S.; Yang, Y.G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Klukovich, R.; Peng, H.; Wang, Z.; Yu, T.; Zhang, Y.; Zheng, H.; Klungland, A.; Yan, W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. USA 2018, 115, E325–E333. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, Y.; Sun, B.F.; Shi, Y.; Yang, X.; Xiao, W.; Hao, Y.J.; Ping, X.L.; Chen, Y.S.; Wang, W.J.; et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014, 24, 1403–1419. [Google Scholar] [CrossRef]
- Fustin, J.M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Deng, K.; Fan, Y.; Liang, Y.; Cai, Y.; Zhang, G.; Deng, M.; Wang, Z.; Lu, J.; Shi, J.; Wang, F.; et al. FTO-mediated demethylation of GADD45B promotes myogenesis through the activation of p38 MAPK pathway. Mol. Ther. Nucleic Acids 2021, 26, 34–48. [Google Scholar] [CrossRef]
- Wang, X.; Huang, N.; Yang, M.; Wei, D.; Tai, H.; Han, X.; Gong, H.; Zhou, J.; Qin, J.; Wei, X.; et al. FTO is required for myogenesis by positively regulating mTOR-PGC-1α pathway-mediated mitochondria biogenesis. Cell Death Dis. 2017, 8, e2702. [Google Scholar] [CrossRef] [PubMed]
- Petrosino, J.M.; Hinger, S.A.; Golubeva, V.A.; Barajas, J.M.; Dorn, L.E.; Iyer, C.C.; Sun, H.L.; Arnold, W.D.; He, C.; Accornero, F. The m(6)A methyltransferase METTL3 regulates muscle maintenance and growth in mice. Nat. Commun. 2022, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zuo, H.; Wang, Z.; Wang, W.; Qian, X.; Xie, Y.; Peng, D.; Xie, Y.; Hong, L.; You, W.; et al. The m6A reader YTHDC1 regulates muscle stem cell proliferation via PI4K-Akt-mTOR signalling. Cell Prolif. 2023, 56, e13410. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tan, B.; Xiao, L.; Zhao, X.; Zeng, J.; Hong, L.; Yang, J.; Cai, G.; Zheng, E.; Wu, Z.; et al. Comprehensive Analysis of Long Noncoding RNA Modified by m(6)A Methylation in Oxidative and Glycolytic Skeletal Muscles. Int. J. Mol. Sci. 2022, 23, 4600. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, S.; Zhang, X.; Ren, S.; Tang, Z.; Gao, F. Coordinated transcriptional and post-transcriptional epigenetic regulation during skeletal muscle development and growth in pigs. J. Anim. Sci. Biotechnol. 2022, 13, 146. [Google Scholar] [CrossRef]
- Gu, H.; Zhou, Y.; Yang, J.; Li, J.; Peng, Y.; Zhang, X.; Miao, Y.; Jiang, W.; Bu, G.; Hou, L.; et al. Targeted overexpression of PPARγ in skeletal muscle by random insertion and CRISPR/Cas9 transgenic pig cloning enhances oxidative fiber formation and intramuscular fat deposition. FASEB J. 2021, 35, e21308. [Google Scholar] [CrossRef]
- Yin, J.; Li, Y.; Tian, Y.; Zhou, F.; Ma, J.; Xia, S.; Yang, T.; Ma, L.; Zeng, Q.; Liu, G.; et al. Obese Ningxiang pig-derived microbiota rewires carnitine metabolism to promote muscle fatty acid deposition in lean DLY pigs. Innovation 2023, 4, 100486. [Google Scholar] [CrossRef]
- Yin, S.; Song, G.; Gao, N.; Gao, H.; Zeng, Q.; Lu, P.; Zhang, Q.; Xu, K.; He, J. Identifying Genetic Architecture of Carcass and Meat Quality Traits in a Ningxiang Indigenous Pig Population. Genes 2023, 14, 1308. [Google Scholar] [CrossRef]
- Rudar, M.; Fiorotto, M.L.; Davis, T.A. Regulation of Muscle Growth in Early Postnatal Life in a Swine Model. Annu. Rev. Anim. Biosci. 2019, 7, 309–335. [Google Scholar] [CrossRef]
- Tian, S.; Lai, J.; Yu, T.; Li, Q.; Chen, Q. Regulation of Gene Expression Associated with the N6-Methyladenosine (m6A) Enzyme System and Its Significance in Cancer. Front. Oncol. 2020, 10, 623634. [Google Scholar] [CrossRef]
- Robson, M.I.; de Las Heras, J.I.; Czapiewski, R.; Thành, P.L.; Booth, D.G.; Kelly, D.A.; Webb, S.; Kerr, A.R.W.; Schirmer, E.C. Tissue-Specific Gene Repositioning by Muscle Nuclear Membrane Proteins Enhances Repression of Critical Developmental Genes during Myogenesis. Mol. Cell 2016, 62, 834–847. [Google Scholar] [CrossRef] [PubMed]
- Eimre, M.; Paju, K.; Peet, N.; Kadaja, L.; Tarrend, M.; Kasvandik, S.; Seppet, J.; Ivask, M.; Orlova, E.; Kõks, S. Increased Mitochondrial Protein Levels and Bioenergetics in the Musculus Rectus Femoris of Wfs1-Deficient Mice. Oxid. Med. Cell Longev. 2018, 2018, 3175313. [Google Scholar] [CrossRef] [PubMed]
- Cagalinec, M.; Zahradníková, A.; Zahradníková, A., Jr.; Kováčová, D.; Paulis, L.; Kureková, S.; Hot’ka, M.; Pavelková, J.; Plaas, M.; Novotová, M.; et al. Calcium Signaling and Contractility in Cardiac Myocyte of Wolframin Deficient Rats. Front. Physiol. 2019, 10, 172. [Google Scholar] [CrossRef] [PubMed]
- Kõks, S. Genomics of Wolfram Syndrome 1 (WFS1). Biomolecules 2023, 13, 1346. [Google Scholar] [CrossRef]
- Richard, E.M.; Brun, E.; Korchagina, J.; Crouzier, L.; Affortit, C.; Alves, S.; Cazevieille, C.; Mausset-Bonnefont, A.L.; Lenoir, M.; Puel, J.L.; et al. Wfs1(E864K) knock-in mice illuminate the fundamental role of Wfs1 in endocochlear potential production. Cell Death Dis. 2023, 14, 387. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, H.; Xu, K.; Yu, Z.; Ren, Z.; Chen, F.; Zhou, C.; Zeng, W.; Ren, H.; Yin, Y.; Bi, Y. N6-Methyladenosine RNA Modification Regulates the Differential Muscle Development in Large White and Ningxiang Pigs. Cells 2024, 13, 1744. https://doi.org/10.3390/cells13201744
Gu H, Xu K, Yu Z, Ren Z, Chen F, Zhou C, Zeng W, Ren H, Yin Y, Bi Y. N6-Methyladenosine RNA Modification Regulates the Differential Muscle Development in Large White and Ningxiang Pigs. Cells. 2024; 13(20):1744. https://doi.org/10.3390/cells13201744
Chicago/Turabian StyleGu, Hao, Kang Xu, Zhao Yu, Zufeng Ren, Fan Chen, Changfan Zhou, Wei Zeng, Hongyan Ren, Yulong Yin, and Yanzhen Bi. 2024. "N6-Methyladenosine RNA Modification Regulates the Differential Muscle Development in Large White and Ningxiang Pigs" Cells 13, no. 20: 1744. https://doi.org/10.3390/cells13201744
APA StyleGu, H., Xu, K., Yu, Z., Ren, Z., Chen, F., Zhou, C., Zeng, W., Ren, H., Yin, Y., & Bi, Y. (2024). N6-Methyladenosine RNA Modification Regulates the Differential Muscle Development in Large White and Ningxiang Pigs. Cells, 13(20), 1744. https://doi.org/10.3390/cells13201744





