Functional Classification and Interaction Selectivity Landscape of the Human SH3 Domain Superfamily
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics, Databases, and Structural Analysis
2.2. Constructs, Peptides, and Proteins
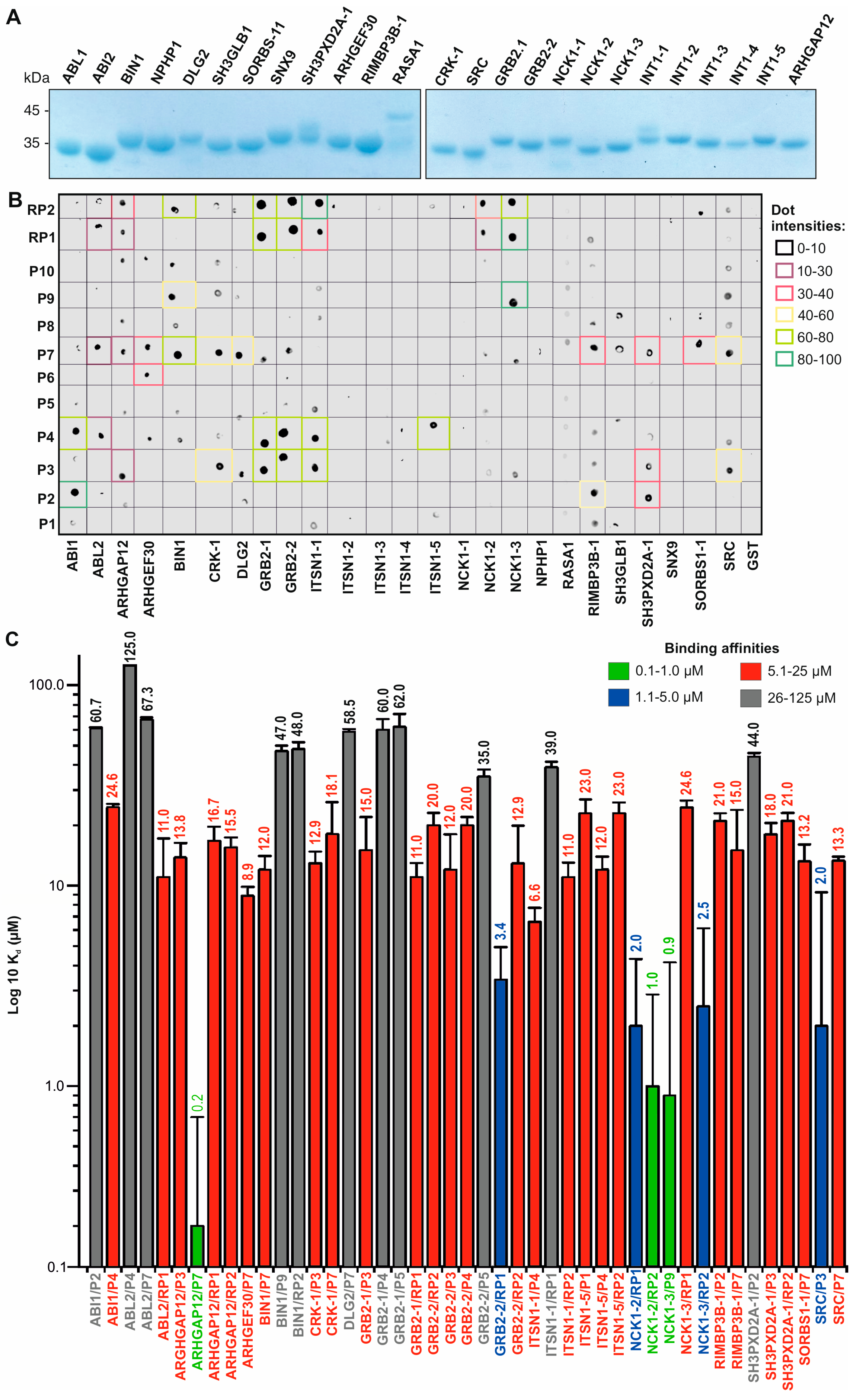
2.3. Pull-Down and Fluorescence Dot Blot Analysis
2.4. Fluorescence Polarization
2.5. Trandfection and Immunoprecipitation Analysis
3. Results
3.1. Sequence–Structure–Function Classification of Human SH3 Domains
3.2. PRM Selectivities of Different SH3 Families
3.3. Affinity and Selectivity of the SH3 Family Proteins for SOS1 PRP
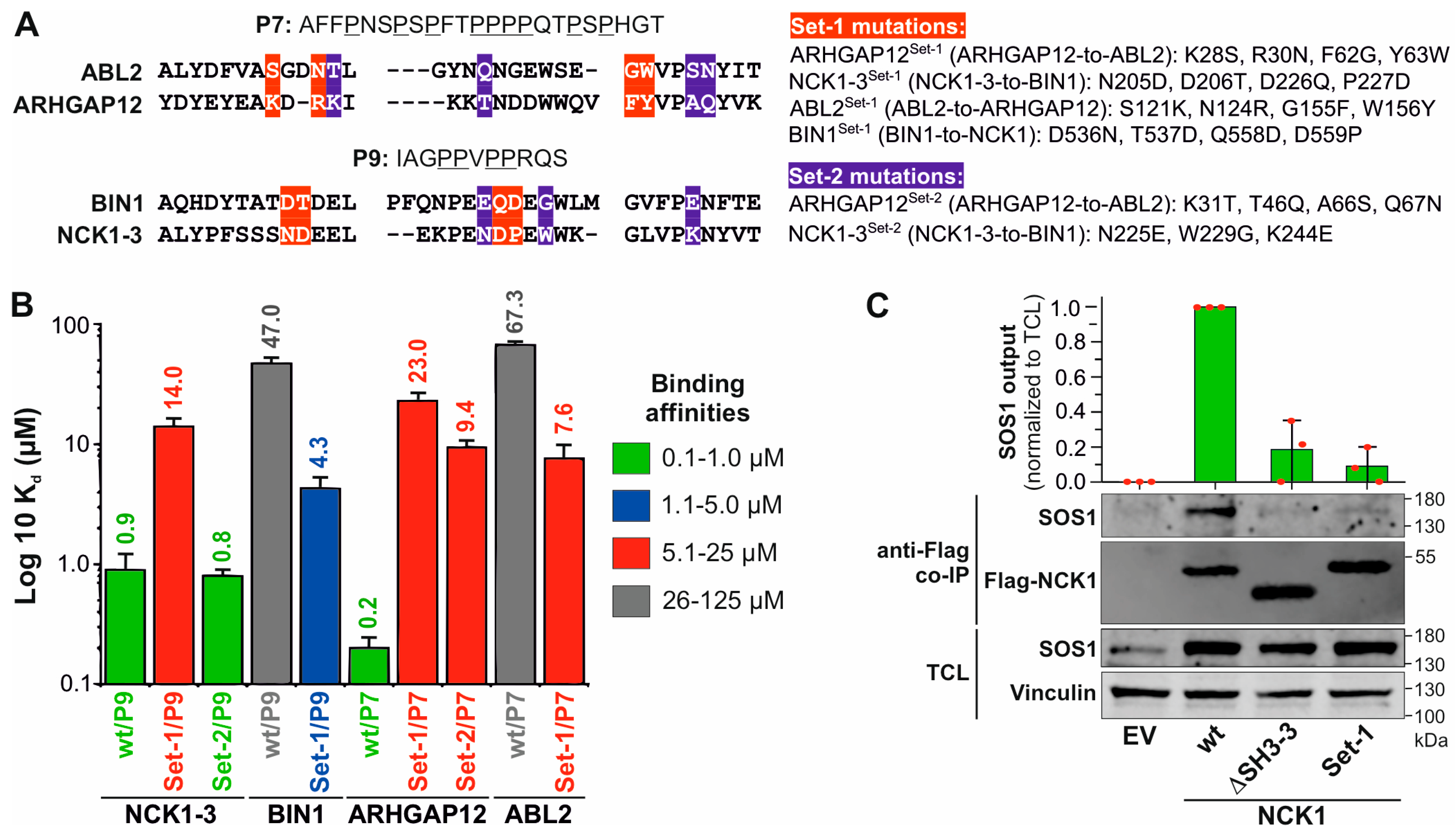
3.4. Non-Conserved Residues Define the Selectivity and Affinity of SH3 Domain–PRM Interactions
3.5. SH3 Domain–PRP Relationships beyond SOS1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mabonga, L.; Kappo, A.P. Protein-protein interaction modulators: Advances, successes and remaining challenges. Biophys. Rev. 2019, 11, 559–581. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A.; Noble, M.; Pauptit, R.; Wierenga, R.; Saraste, M. Crystal structure of a Src-homology 3 (SH3) domain. Nature 1992, 359, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Stahl, M.L.; Ferenz, C.R.; Kelleher, K.L.; Kriz, R.W.; Knopf, J.L. Sequence similarity of phospholipase C with the non-catalytic region of src. Nature 1988, 332, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Mehrabipour, M.; Jasemi, N.S.K.; Dvorsky, R.; Ahmadian, M.R. A Systematic Compilation of Human SH3 Domains: A Versatile Superfamily in Cellular Signaling. Cells 2023, 12, 2054. [Google Scholar] [CrossRef] [PubMed]
- Teyra, J.; Huang, H.; Jain, S.; Guan, X.; Dong, A.; Liu, Y.; Tempel, W.; Min, J.; Tong, Y.; Kim, P.M. Comprehensive analysis of the human SH3 domain family reveals a wide variety of non-canonical specificities. Structure 2017, 25, 1598–1610.e3. [Google Scholar] [CrossRef] [PubMed]
- Berndt, S.; Gurevich, V.V.; Iverson, T. Crystal structure of the SH3 domain of human Lyn non-receptor tyrosine kinase. PLoS ONE 2019, 14, e0215140. [Google Scholar] [CrossRef] [PubMed]
- Bircher, J.E.; Koleske, A.J. Trio family proteins as regulators of cell migration and morphogenesis in development and disease–mechanisms and cellular contexts. J. Cell Sci. 2021, 134, jcs248393. [Google Scholar] [CrossRef]
- Hildebrandt, F.; Otto, E.; Rensing, C.; Nothwang, H.G.; Vollmer, M.; Adolphs, J.; Hanusch, H.; Brandis, M. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat. Genet. 1997, 17, 149–153. [Google Scholar] [CrossRef]
- Saunier, S.; Calado, J.; Benessy, F.; Silbermann, F.; Heilig, R.; Weissenbach, J.; Antignac, C. Characterization of the NPHP1 locus: Mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am. J. Hum. Genet. 2000, 66, 778–789. [Google Scholar] [CrossRef]
- Wu, T.; Shi, Z.; Baumgart, T. Mutations in BIN1 associated with centronuclear myopathy disrupt membrane remodeling by affecting protein density and oligomerization. PLoS ONE 2014, 9, e93060. [Google Scholar] [CrossRef]
- Hohendahl, A.; Roux, A.; Galli, V. Structural insights into the centronuclear myopathy-associated functions of BIN1 and dynamin 2. J. Struct. Biol. 2016, 196, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Manso, J.A.; Marcos, T.; Ruiz-Martín, V.; Casas, J.; Alcón, P.; Sánchez Crespo, M.; Bayón, Y.; de Pereda, J.M.; Alonso, A. PSTPIP1-LYP phosphatase interaction: Structural basis and implications for autoinflammatory disorders. Cell Mol. Life Sci. 2022, 79, 131. [Google Scholar] [CrossRef] [PubMed]
- Starnes, T.W.; Bennin, D.A.; Bing, X.; Eickhoff, J.C.; Grahf, D.C.; Bellak, J.M.; Seroogy, C.M.; Ferguson, P.J.; Huttenlocher, A. The F-BAR protein PSTPIP1 controls extracellular matrix degradation and filopodia formation in macrophages. Blood 2014, 123, 2703–2714. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Du, Y.; Chen, L.; Liu, Y.; Xu, W.; Liu, Y.; Li, Y.; Leng, J.; Wang, Y.; Zhang, X.Y.; et al. A recurrent SHANK1 mutation implicated in autism spectrum disorder causes autistic-like core behaviors in mice via downregulation of mGluR1-IP3R1-calcium signaling. Mol. Psychiatry 2022, 27, 2985–2998. [Google Scholar] [CrossRef] [PubMed]
- Leblond, C.S.; Heinrich, J.; Delorme, R.; Proepper, C.; Betancur, C.; Huguet, G.; Konyukh, M.; Chaste, P.; Ey, E.; Rastam, M. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012, 8, e1002521. [Google Scholar] [CrossRef] [PubMed]
- Ball, L.J.; Kühne, R.; Schneider-Mergener, J.; Oschkinat, H. Recognition of Proline-Rich Motifs by Protein–Protein-Interaction Domains. Angew. Chem. Int. Ed. 2005, 44, 2852–2869. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.-C. Specificity and versatility of SH3 and other proline-recognition domains: Structural basis and implications for cellular signal transduction. Biochem. J. 2005, 390, 641–653. [Google Scholar] [CrossRef]
- Cesareni, G.; Gimona, M. Modular Protein Domains; Wiley: Hoboken, NJ, USA, 2005. [Google Scholar]
- Cámara-Artigas, A.; Martínez-Rodríguez, S.; Ortiz-Salmerón, E.; Martín-García, J.M. 3D domain swapping in a chimeric c-Src SH3 domain takes place through two hinge loops. J. Struct. Biol. 2014, 186, 195–203. [Google Scholar] [CrossRef]
- Carducci, M.; Perfetto, L.; Briganti, L.; Paoluzi, S.; Costa, S.; Zerweck, J.; Schutkowski, M.; Castagnoli, L.; Cesareni, G. The protein interaction network mediated by human SH3 domains. Biotechnol. Adv. 2012, 30, 4–15. [Google Scholar] [CrossRef]
- Panni, S.; Dente, L.; Cesareni, G. In vitro evolution of recognition specificity mediated by SH3 domains reveals target recognition rules. J. Biol. Chem. 2002, 277, 21666–21674. [Google Scholar] [CrossRef]
- Kazemein Jasemi, N.S.; Herrmann, C.; Magdalena Estirado, E.; Gremer, L.; Willbold, D.; Brunsveld, L.; Dvorsky, R.; Ahmadian, M.R. The intramolecular allostery of GRB2 governing its interaction with SOS1 is modulated by phosphotyrosine ligands. Biochem. J. 2021, 478, 2793–2809. [Google Scholar] [CrossRef] [PubMed]
- Mayer, B.J. SH3 domains: Complexity in moderation. J. Cell Sci. 2001, 114, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Kay, B.K. SH3 domains come of age. FEBS Lett. 2012, 586, 2606–2608. [Google Scholar] [CrossRef]
- Landgraf, C.; Panni, S.; Montecchi-Palazzi, L.; Castagnoli, L.; Schneider-Mergener, J.; Volkmer-Engert, R.; Cesareni, G. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2004, 2, e14. [Google Scholar] [CrossRef] [PubMed]
- Bae, D.J.; Seo, J.; Kim, S.Y.; Park, S.Y.; Do Yoo, J.; Pyo, J.H.; Cho, W.; Cho, J.Y.; Kim, S.; Kim, I.S. ArhGAP12 plays dual roles in Stabilin-2 mediated efferocytosis: Regulates Rac1 basal activity and spatiotemporally turns off the Rac1 to orchestrate phagosome maturation. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1595–1607. [Google Scholar] [CrossRef] [PubMed]
- Hibino, H.; Pironkova, R.; Onwumere, O.; Vologodskaia, M.; Hudspeth, A.J.; Lesage, F. RIM binding proteins (RBPs) couple Rab3-interacting molecules (RIMs) to voltage-gated Ca(2+) channels. Neuron 2002, 34, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, T.; Yamagata, T.; Sakai, R.; Ogawa, S.; Honda, H.; Ueno, H.; Hirano, N.; Yazaki, Y.; Hirai, H. CIZ, a zinc finger protein that interacts with p130(cas) and activates the expression of matrix metalloproteinases. Mol. Cell Biol. 2000, 20, 1649–1658. [Google Scholar] [CrossRef]
- Salem, D.; Fecek, R.J. Role of microtubule actin crosslinking factor 1 (MACF1) in bipolar disorder pathophysiology and potential in lithium therapeutic mechanism. Transl. Psychiatry 2023, 13, 221. [Google Scholar] [CrossRef]
- Rasmussen, A.H.; Rasmussen, H.B.; Silahtaroglu, A. The DLGAP family: Neuronal expression, function and role in brain disorders. Mol. Brain 2017, 10, 43. [Google Scholar] [CrossRef]
- Bustelo, X.R.; Sauzeau, V.; Berenjeno, I.M. GTP-binding proteins of the Rho/Rac family: Regulation, effectors and functions in vivo. Bioessays 2007, 29, 356–370. [Google Scholar] [CrossRef]
- Risse, S.L.; Vaz, B.; Burton, M.F.; Aspenström, P.; Piekorz, R.P.; Brunsveld, L.; Ahmadian, M.R. SH3-mediated targeting of Wrch1/RhoU by multiple adaptor proteins. Biol. Chem. 2013, 394, 421–432. [Google Scholar] [CrossRef] [PubMed]
- López-Colomé, A.M.; Lee-Rivera, I.; Benavides-Hidalgo, R.; López, E. Paxillin: A crossroad in pathological cell migration. J. Hematol. Oncol. 2017, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Webb, D.J.; Schroeder, M.J.; Brame, C.J.; Whitmore, L.; Shabanowitz, J.; Hunt, D.F.; Horwitz, A.R. Paxillin phosphorylation sites mapped by mass spectrometry. J. Cell Sci. 2005, 118, 4925–4929. [Google Scholar] [CrossRef] [PubMed]
- Kreienkamp, H.-J.; Liew, C.W.; Bächner, D.; Mameza, M.-G.; Soltau, M.; Quitsch, A.; Christenn, M.; Wente, W.; Richter, D. Physiology of somatostatin receptors: From genetics to molecular analysis. In Somatostatin; Springer: Berlin/Heidelberg, Germany, 2004; pp. 185–202. [Google Scholar]
- Verschueren, E.; Spiess, M.; Gkourtsa, A.; Avula, T.; Landgraf, C.; Mancilla, V.T.; Huber, A.; Volkmer, R.; Winsor, B.; Serrano, L. Evolution of the SH3 domain specificity Landscape in Yeasts. PLoS ONE 2015, 10, e0129229. [Google Scholar] [CrossRef] [PubMed]
- Cesareni, G.; Panni, S.; Nardelli, G.; Castagnoli, L. Can we infer peptide recognition specificity mediated by SH3 domains? FEBS Lett. 2002, 513, 38–44. [Google Scholar] [CrossRef]
- Chook, Y.M.; Gish, G.D.; Kay, C.M.; Pai, E.F.; Pawson, T. The Grb2-mSos1 complex binds phosphopeptides with higher affinity than Grb2. J. Biol. Chem. 1996, 271, 30472–30478. [Google Scholar] [CrossRef]
- Herrero-Garcia, E.; O’Bryan, J.P. Intersectin scaffold proteins and their role in cell signaling and endocytosis. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2017, 1864, 23–30. [Google Scholar] [CrossRef]
- Alfaidi, M.; Scott, M.L.; Orr, A.W. Sinner or Saint?: Nck adaptor proteins in vascular biology. Front. Cell Dev. Biol. 2021, 9, 688388. [Google Scholar] [CrossRef]
- Yu, X.; Liang, C.; Zhang, Y.; Zhang, W.; Chen, H. Inhibitory short peptides targeting EPS8/ABI1/SOS1 tri-complex suppress invasion and metastasis of ovarian cancer cells. BMC Cancer 2019, 19, 878. [Google Scholar] [CrossRef]
- Hahn, S.; Kim, D. Transient protein-protein interaction of the SH3-peptide complex via closely located multiple binding sites. PLoS ONE 2012, 7, e32804. [Google Scholar] [CrossRef]
- Mayer, B.J.; Saksela, K. SH3 domains. In Modular Protein Domains; Wiley: Hoboken, NJ, USA, 2005; pp. 37–58. [Google Scholar]
- Dionne, U.; Percival, L.J.; Chartier, F.J.; Landry, C.R.; Bisson, N. SRC homology 3 domains: Multifaceted binding modules. Trends Biochem. Sci. 2022, 47, 772–784. [Google Scholar] [CrossRef] [PubMed]
- Dionne, U.; Bourgault, É.; Dubé, A.K.; Bradley, D.; Chartier, F.J.M.; Dandage, R.; Dibyachintan, S.; Després, P.C.; Gish, G.D.; Pham, N.T.H.; et al. Protein context shapes the specificity of SH3 domain-mediated interactions in vivo. Nat. Commun. 2021, 12, 1597. [Google Scholar] [CrossRef] [PubMed]
- Zarrinpar, A.; Park, S.-H.; Lim, W.A. Optimization of specificity in a cellular protein interaction network by negative selection. Nature 2003, 426, 676–680. [Google Scholar] [CrossRef]
- Humphries, A.C.; Donnelly, S.K.; Way, M. Cdc42 and the Rho GEF intersectin-1 collaborate with Nck to promote N-WASP-dependent actin polymerisation. J. Cell Sci. 2014, 127 Pt 3, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Shutes, A.; Berzat, A.C.; Cox, A.D.; Der, C.J. Atypical mechanism of regulation of the Wrch-1 Rho family small GTPase. Curr. Biol. 2004, 14, 2052–2056. [Google Scholar] [CrossRef] [PubMed]
- Lettau, M.; Pieper, J.; Gerneth, A.; Lengl-Janssen, B.; Voss, M.; Linkermann, A.; Schmidt, H.; Gelhaus, C.; Leippe, M.; Kabelitz, D.; et al. The adapter protein Nck: Role of individual SH3 and SH2 binding modules for protein interactions in T lymphocytes. Protein Sci. 2010, 19, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Gu, W.; Helms, V. Mechanism of fast peptide recognition by SH3 domains. Angew. Chem. Int. Ed. 2008, 47, 7626–7630. [Google Scholar] [CrossRef]
- Yu, H.; Chen, J.K.; Feng, S.; Dalgarno, D.C.; Brauer, A.W.; Schrelber, S.L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 1994, 76, 933–945. [Google Scholar] [CrossRef]
- McDonald, C.B.; Seldeen, K.L.; Deegan, B.J.; Farooq, A. SH3 domains of Grb2 adaptor bind to PXψPXR motifs within the Sos1 nucleotide exchange factor in a discriminate manner. Biochemistry 2009, 48, 4074–4085. [Google Scholar] [CrossRef]
- Jaiswal, M.; Dvorsky, R.; Amin, E.; Risse, S.L.; Fansa, E.K.; Zhang, S.-C.; Taha, M.S.; Gauhar, A.R.; Nakhaei-Rad, S.; Kordes, C. Functional cross-talk between ras and rho pathways: A Ras-specific GTPase-activating protein (p120RasGAP) competitively inhibits the RhoGAP activity of deleted in liver cancer (DLC) tumor suppressor by masking the catalytic arginine finger. J. Biol. Chem. 2014, 289, 6839–6849. [Google Scholar] [CrossRef]
- Fan, P.-D.; Goff, S.P. Abl interactor 1 binds to sos and inhibits epidermal growth factor-and v-Abl-induced activation of extracellular signal-regulated kinases. Mol. Cell. Biol. 2000, 20, 7591–7601. [Google Scholar] [CrossRef] [PubMed]
- Tong, X.K.; Hussain, N.K.; de Heuvel, E.; Kurakin, A.; Abi-Jaoude, E.; Quinn, C.C.; Olson, M.F.; Marais, R.; Baranes, D.; Kay, B.K.; et al. The endocytic protein intersectin is a major binding partner for the Ras exchange factor mSos1 in rat brain. EMBO J. 2000, 19, 1263–1271. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Esteban, L.; Vass, W.C.; Upadhyaya, C.; Papageorge, A.G.; Yienger, K.; Ward, J.M.; Lowy, D.R.; Santos, E. The Sos1 and Sos2 Ras-specific exchange factors: Differences in placental expression and signaling properties. Embo J. 2000, 19, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Milfay, D.; Williams, L.T. Binding of NCK to SOS and activation of ras-dependent gene expression. Mol. Cell Biol. 1995, 15, 1169–1174. [Google Scholar] [CrossRef] [PubMed]
- Chardin, P.; Camonis, J.H.; Gale, N.W.; Van Aelst, L.; Schlessinger, J.; Wigler, M.H.; Bar-Sagi, D. Human Sos1: A guanine nucleotide exchange factor for Ras that binds to GRB2. Science 1993, 260, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Birge, R.B.; Kalodimos, C.; Inagaki, F.; Tanaka, S. Crk and CrkL adaptor proteins: Networks for physiological and pathological signaling. Cell Commun. Signal. 2009, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Rufer, A.C.; Rumpf, J.; von Holleben, M.; Beer, S.; Rittinger, K.; Groemping, Y. Isoform-selective interaction of the adaptor protein Tks5/FISH with Sos1 and dynamins. J. Mol. Biol. 2009, 390, 939–950. [Google Scholar] [CrossRef]
- Sini, P.; Cannas, A.; Koleske, A.J.; Di Fiore, P.P.; Scita, G. Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nat. Cell Biol. 2004, 6, 268–274. [Google Scholar] [CrossRef]
- Wilkinson, B.; Li, J.; Coba, M.P. Synaptic GAP and GEF complexes cluster proteins essential for GTP signaling. Sci. Rep. 2017, 7, 5272. [Google Scholar] [CrossRef]
- Chau, J.E.; Vish, K.J.; Boggon, T.J.; Stiegler, A.L. SH3 domain regulation of RhoGAP activity: Crosstalk between p120RasGAP and DLC1 RhoGAP. Nat. Commun. 2022, 13, 4788. [Google Scholar] [CrossRef]
- Berry, D.M.; Nash, P.; Liu, S.K.; Pawson, T.; McGlade, C.J. A high-affinity Arg-X-X-Lys SH3 binding motif confers specificity for the interaction between Gads and SLP-76 in T cell signaling. Curr. Biol. 2002, 12, 1336–1341. [Google Scholar] [CrossRef] [PubMed]
- Kazemein Jasemi, N.S.; Ahmadian, M.R. Allosteric regulation of GRB2 modulates RAS activation. Small GTPases 2022, 13, 282–286. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazemein Jasemi, N.S.; Mehrabipour, M.; Magdalena Estirado, E.; Brunsveld, L.; Dvorsky, R.; Ahmadian, M.R. Functional Classification and Interaction Selectivity Landscape of the Human SH3 Domain Superfamily. Cells 2024, 13, 195. https://doi.org/10.3390/cells13020195
Kazemein Jasemi NS, Mehrabipour M, Magdalena Estirado E, Brunsveld L, Dvorsky R, Ahmadian MR. Functional Classification and Interaction Selectivity Landscape of the Human SH3 Domain Superfamily. Cells. 2024; 13(2):195. https://doi.org/10.3390/cells13020195
Chicago/Turabian StyleKazemein Jasemi, Neda S., Mehrnaz Mehrabipour, Eva Magdalena Estirado, Luc Brunsveld, Radovan Dvorsky, and Mohammad R. Ahmadian. 2024. "Functional Classification and Interaction Selectivity Landscape of the Human SH3 Domain Superfamily" Cells 13, no. 2: 195. https://doi.org/10.3390/cells13020195
APA StyleKazemein Jasemi, N. S., Mehrabipour, M., Magdalena Estirado, E., Brunsveld, L., Dvorsky, R., & Ahmadian, M. R. (2024). Functional Classification and Interaction Selectivity Landscape of the Human SH3 Domain Superfamily. Cells, 13(2), 195. https://doi.org/10.3390/cells13020195







