Chronic Inflammation Disrupts Circadian Rhythms in Splenic CD4+ and CD8+ T Cells in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Immunization
2.2. Activity Analysis
2.3. Tissue Collection and Preparation
2.4. Real-Time Quantitative Reverse Transcription PCR (qPCR)
2.5. Statistical Analysis
3. Results
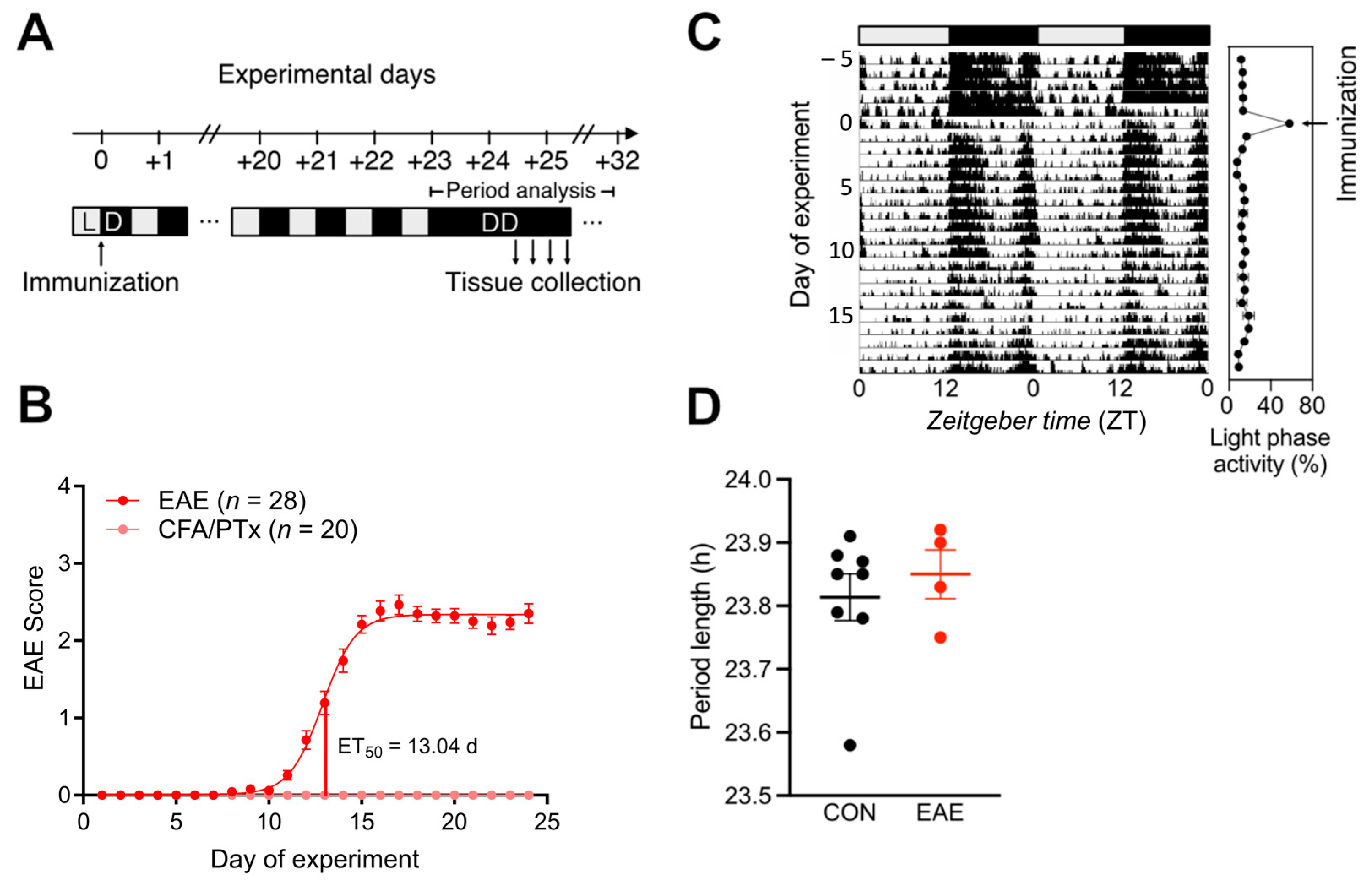
3.1. Central Circadian Rhythms Are Largely Unaffected in EAE
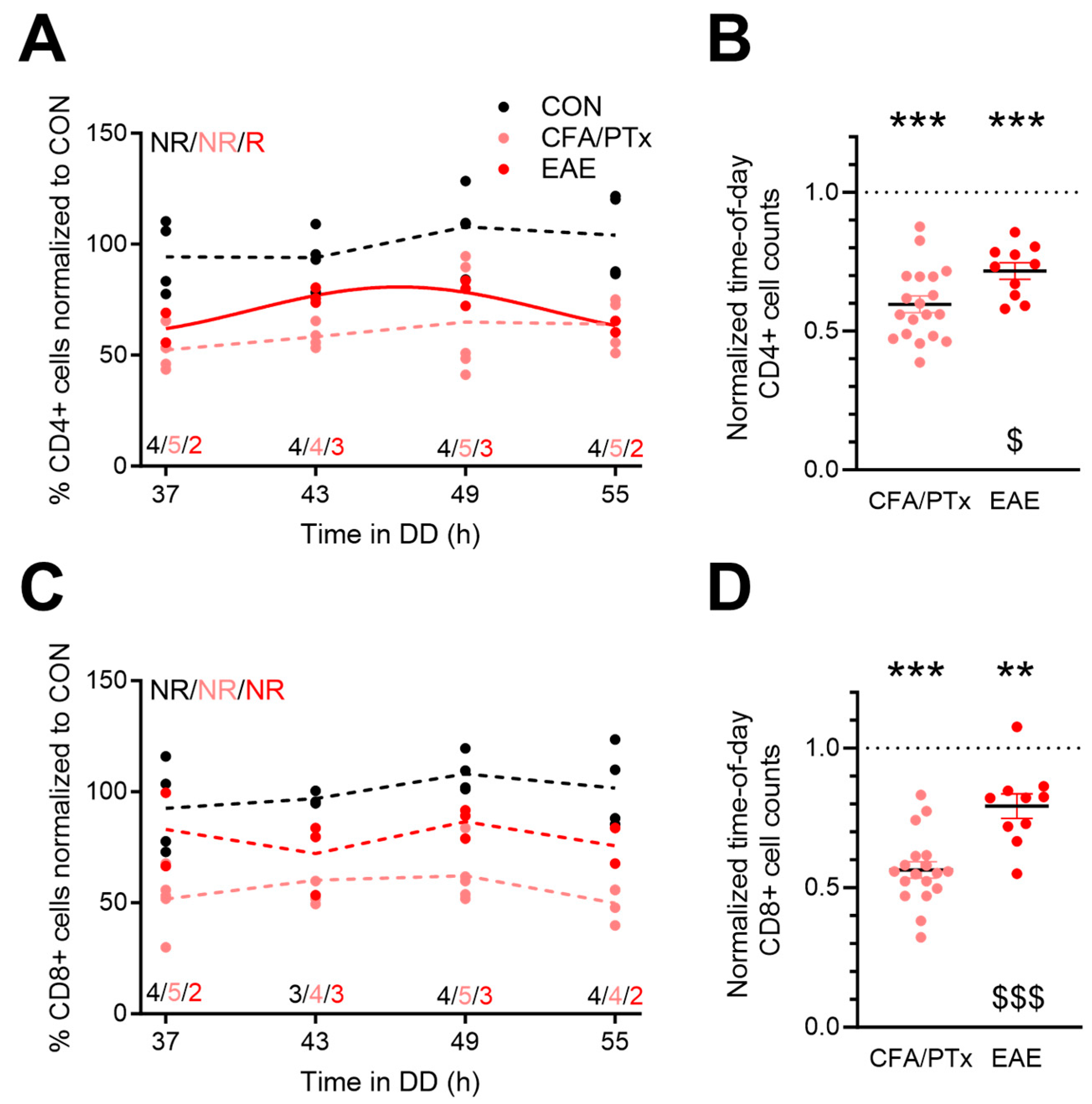
3.2. Splenic T Cell Numbers Are Reduced in Response to CFA/PTx Treatment and during Chronic EAE
3.3. Persistent Disruption of Circadian T Cell Clock Gene Rhythms in Response to CFA/PTx Treatment and EAE
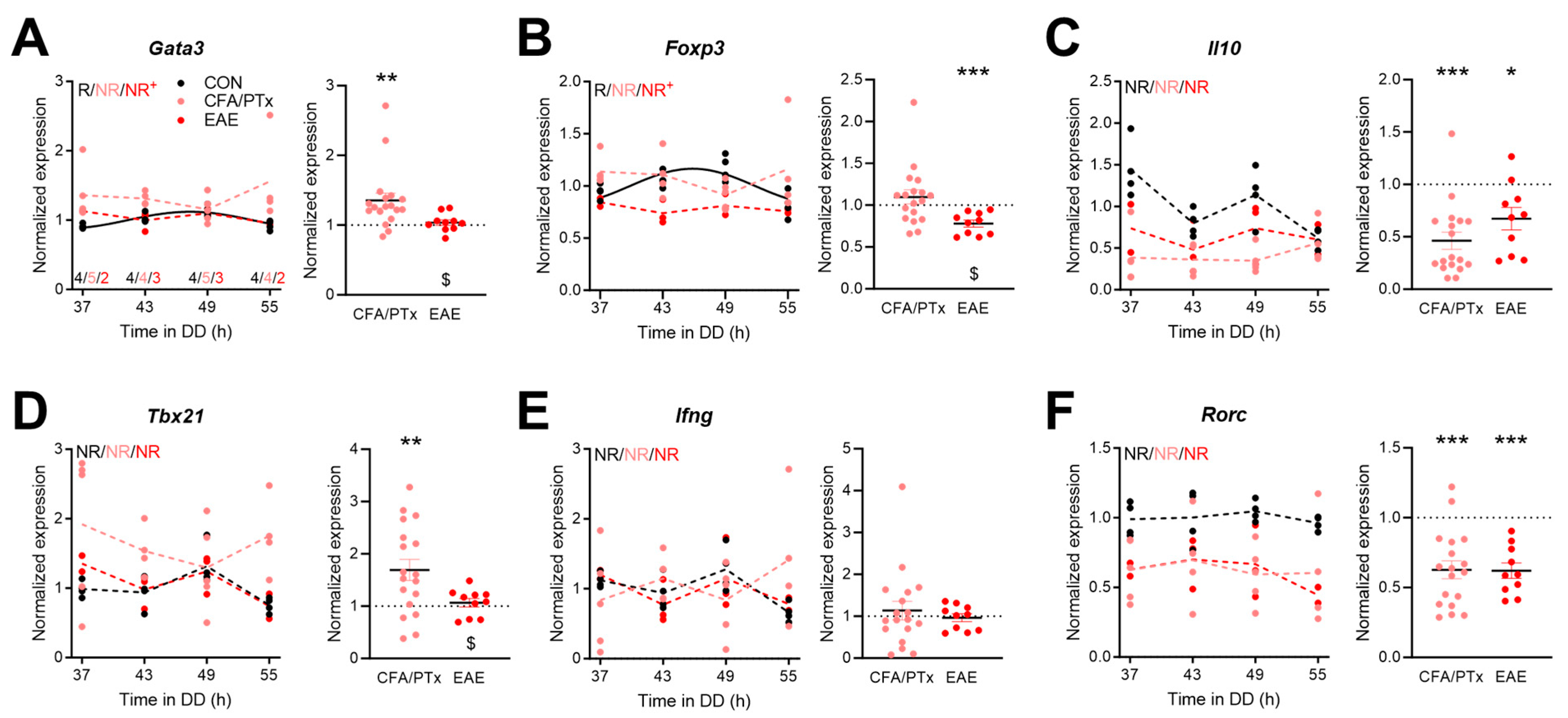
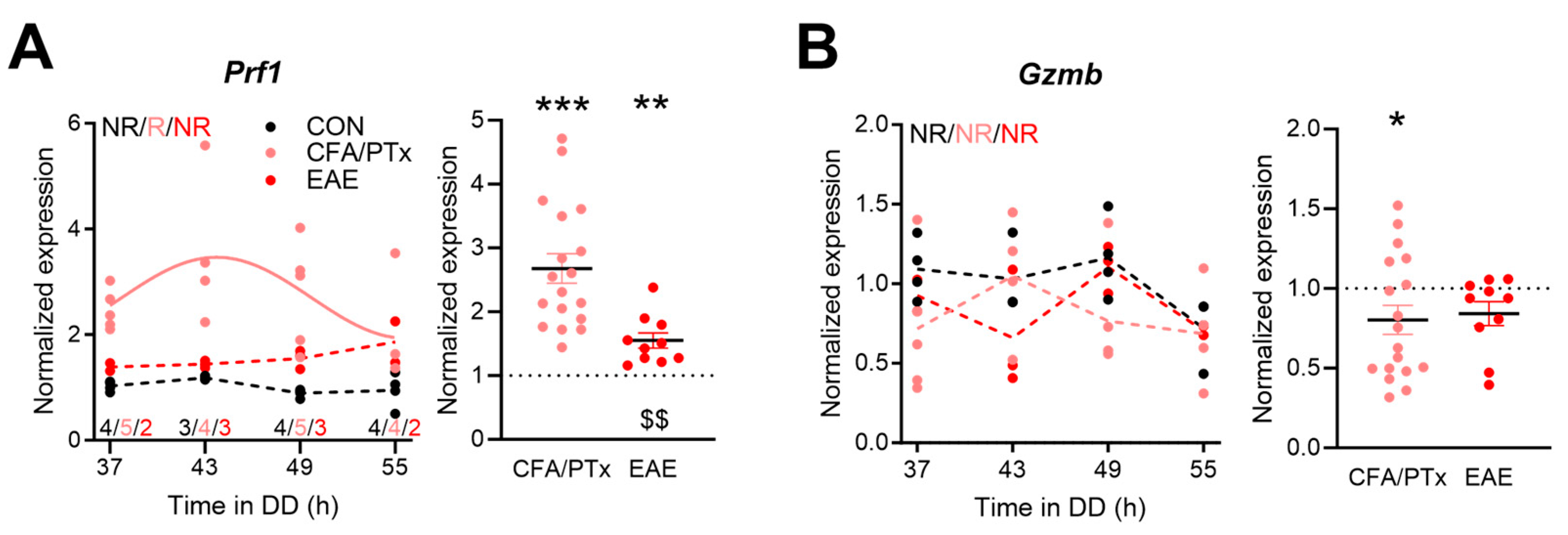
3.4. Circadian Expression of Immune-Function-Related Genes in Peripheral T Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Takahashi, J.S. Molecular Components of the Circadian Clock in Mammals. Diabetes Obes. Metab. 2015, 17, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Partch, C.L.; Green, C.B.; Takahashi, J.S. Molecular Architecture of the Mammalian Circadian Clock. Trends Cell Biol. 2014, 24, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, B.; Nakahata, Y.; Sahar, S.; Kaluzova, M.; Gauthier, D.; Pham, K.; Patel, N.; Hirayama, J.; Sassone-Corsi, P. Chromatin Remodeling and Circadian Control: Master Regulator CLOCK Is an Enzyme. Cold Spring Harb. Symp. Quant. Biol. 2007, 72, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Gallego, M.; Virshup, D.M. Post-Translational Modifications Regulate the Ticking of the Circadian Clock. Nat. Rev. Mol. Cell Biol. 2007, 8, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Shingle, D.L.; Green, C.B. Post-Transcriptional Control of Circadian Rhythms. J. Cell Sci. 2011, 124, 311–320. [Google Scholar] [CrossRef]
- Moore, R.Y.; Eichler, V.B. Loss of a Circadian Adrenal Corticosterone Rhythm Following Suprachiasmatic Lesions in the Rat. Brain Res. 1972, 42, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Stephan, F.K.; Zucker, I. Circadian Rhythms in Drinking Behavior and Locomotor Activity of Rats Are Eliminated by Hypothalamic Lesions. Proc. Natl. Acad. Sci. USA 1972, 69, 1583–1586. [Google Scholar] [CrossRef]
- Husse, J.; Leliavski, A.; Tsang, A.H.; Oster, H.; Eichele, G. The Light-Dark Cycle Controls Peripheral Rhythmicity in Mice with a Genetically Ablated Suprachiasmatic Nucleus Clock. FASEB J. 2014, 28, 4950–4960. [Google Scholar] [CrossRef]
- Labrecque, N.; Cermakian, N. Circadian Clocks in the Immune System. J. Biol. Rhythm. 2015, 30, 277–290. [Google Scholar] [CrossRef]
- Cermakian, N.; Stegeman, S.K.; Tekade, K.; Labrecque, N. Circadian Rhythms in Adaptive Immunity and Vaccination. Semin. Immunopathol. 2022, 44, 193–207. [Google Scholar] [CrossRef]
- Scheiermann, C.; Gibbs, J.; Ince, L.; Loudon, A. Clocking in to Immunity. Nat. Rev. Immunol. 2018, 18, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lutes, L.K.; Barnoud, C.; Scheiermann, C. The Circadian Immune System. Sci. Immunol. 2022, 7, eabm2465. [Google Scholar] [CrossRef] [PubMed]
- Lange, T.; Dimitrov, S.; Born, J. Effects of Sleep and Circadian Rhythm on the Human Immune System. Ann. N. Y. Acad. Sci. 2010, 1193, 48–59. [Google Scholar] [CrossRef]
- Fortier, E.E.; Rooney, J.; Dardente, H.; Hardy, M.-P.; Labrecque, N.; Cermakian, N. Circadian Variation of the Response of T Cells to Antigen. J. Immunol. 2011, 187, 6291–6300. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, S.; Thijssen, S.; Alarcon Salvador, S.; Heine, G.H.; van Bentum, K.; Fliser, D.; Sester, M.; Sester, U. T-Cell Numbers and Antigen-Specific T-Cell Function Follow Different Circadian Rhythms. J. Clin. Immunol. 2012, 32, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Bollinger, T.; Leutz, A.; Leliavski, A.; Skrum, L.; Kovac, J.; Bonacina, L.; Benedict, C.; Lange, T.; Westermann, J.; Oster, H.; et al. Circadian Clocks in Mouse and Human CD4+ T Cells. PLoS ONE 2011, 6, e29801. [Google Scholar] [CrossRef] [PubMed]
- Hemmers, S.; Rudensky, A.Y. The Cell-Intrinsic Circadian Clock Is Dispensable for Lymphocyte Differentiation and Function. Cell Rep. 2015, 11, 1339–1349. [Google Scholar] [CrossRef]
- Nobis, C.C.; Dubeau Laramée, G.; Kervezee, L.; Maurice De Sousa, D.; Labrecque, N.; Cermakian, N. The Circadian Clock of CD8 T Cells Modulates Their Early Response to Vaccination and the Rhythmicity of Related Signaling Pathways. Proc. Natl. Acad. Sci. USA 2019, 116, 20077–20086. [Google Scholar] [CrossRef]
- Druzd, D.; Matveeva, O.; Ince, L.; Harrison, U.; He, W.; Schmal, C.; Herzel, H.; Tsang, A.H.; Kawakami, N.; Leliavski, A.; et al. Lymphocyte Circadian Clocks Control Lymph Node Trafficking and Adaptive Immune Responses. Immunity 2017, 46, 120–132. [Google Scholar] [CrossRef]
- Yu, X.; Rollins, D.; Ruhn, K.A.; Stubblefield, J.J.; Green, C.B.; Kashiwada, M.; Rothman, P.B.; Takahashi, J.S.; Hooper, L.V. TH17 Cell Differentiation Is Regulated by the Circadian Clock. Science 2013, 342, 727–730. [Google Scholar] [CrossRef]
- Sutton, C.E.; Finlay, C.M.; Raverdeau, M.; Early, J.O.; DeCourcey, J.; Zaslona, Z.; O’Neill, L.A.J.; Mills, K.H.G.; Curtis, A.M. Loss of the Molecular Clock in Myeloid Cells Exacerbates T Cell-Mediated CNS Autoimmune Disease. Nat. Commun. 2017, 8, 1923. [Google Scholar] [CrossRef]
- Cervantes-Silva, M.P.; Carroll, R.G.; Wilk, M.M.; Moreira, D.; Payet, C.A.; O’Siorain, J.R.; Cox, S.L.; Fagan, L.E.; Klavina, P.A.; He, Y.; et al. The Circadian Clock Influences T Cell Responses to Vaccination by Regulating Dendritic Cell Antigen Processing. Nat. Commun. 2022, 13, 7217. [Google Scholar] [CrossRef]
- Sun, Y.; Yang, Z.; Niu, Z.; Peng, J.; Li, Q.; Xiong, W.; Langnas, A.N.; Ma, M.Y.; Zhao, Y. MOP3, a Component of the Molecular Clock, Regulates the Development of B Cells. Immunology 2006, 119, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Linthicum, D.S.; Munoz, J.J.; Blaskett, A. Acute Experimental Autoimmune Encephalomyelitis in Mice. I. Adjuvant Action of Bordetella Pertussis Is Due to Vasoactive Amine Sensitization and Increased Vascular Permeability of the Central Nervous System. Cell Immunol. 1982, 73, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Brückener, K.E.; el Bayâ, A.; Galla, H.-J.; Schmidt, M.A. Permeabilization in a Cerebral Endothelial Barrier Model by Pertussis Toxin Involves the PKC Effector Pathway and Is Abolished by Elevated Levels of cAMP. J. Cell Sci. 2003, 116, 1837–1846. [Google Scholar] [CrossRef]
- Raghavendra, V.; Tanga, F.Y.; DeLeo, J.A. Complete Freunds Adjuvant-Induced Peripheral Inflammation Evokes Glial Activation and Proinflammatory Cytokine Expression in the CNS. Eur. J. Neurosci. 2004, 20, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Rafiyan, M.; Sadeghmousavi, S.; Akbarzadehmoallemkolaei, M.; Rezaei, N. Experimental Animal Models of Chronic Inflammation. Curr. Res. Immunol. 2023, 4, 100063. [Google Scholar] [CrossRef]
- Shirani, K.; Iranshahi, M.; Askari, V.R.; Gholizadeh, Z.; Zadeh, A.A.; Zeinali, M.; Hassani, F.V.; Taherzadeh, Z. Comparative Evaluation of the Protective Effects of Oral Administration of Auraptene and Umbelliprenin against CFA-Induced Chronic Inflammation with Polyarthritis in Rats. Biomed. Pharmacother. 2021, 139, 111635. [Google Scholar] [CrossRef]
- Barthelmes, J.; Tafferner, N.; Kurz, J.; de Bruin, N.; Parnham, M.J.; Geisslinger, G.; Schiffmann, S. Induction of Experimental Autoimmune Encephalomyelitis in Mice and Evaluation of the Disease-Dependent Distribution of Immune Cells in Various Tissues. J. Vis. Exp. 2016. [Google Scholar] [CrossRef]
- Wang, X.-L.; Li, L. Circadian Clock Regulates Inflammation and the Development of Neurodegeneration. Front. Cell Infect. Microbiol. 2021, 11, 696554. [Google Scholar] [CrossRef]
- Morena da Silva, F.; Esser, K.A.; Murach, K.A.; Greene, N.P. Inflammation o’clock: Interactions of Circadian Rhythms with Inflammation-Induced Skeletal Muscle Atrophy. J. Physiol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Hashiramoto, A.; Yamane, T.; Tsumiyama, K.; Yoshida, K.; Komai, K.; Yamada, H.; Yamazaki, F.; Doi, M.; Okamura, H.; Shiozawa, S. Mammalian Clock Gene Cryptochrome Regulates Arthritis via Proinflammatory Cytokine TNF-α. J. Immunol. 2009, 184, 1560–1565. [Google Scholar] [CrossRef]
- Gibbs, J.E.; Ray, D.W. The Role of the Circadian Clock in Rheumatoid Arthritis. Arthritis Res. Ther. 2013, 15, 205. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Huang, L.; Zhao, J.; Chen, S.; Liu, J.; Li, G. The Circadian Clock and Inflammation: A New Insight. Clin. Chim. Acta 2021, 512, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Cermakian, N.; Westfall, S.; Kiessling, S. Circadian Clocks and Inflammation: Reciprocal Regulation and Shared Mediators. Arch. Immunol. Ther. Exp. 2014, 62, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Buenafe, A.C. Diurnal Rhythms Are Altered in a Mouse Model of Multiple Sclerosis. J. Neuroimmunol. 2012, 243, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Maury, E.; Ramsey, K.M.; Bass, J. Circadian Rhythms and Metabolic Syndrome: From Experimental Genetics to Human Disease. Circ. Res. 2010, 106, 447–462. [Google Scholar] [CrossRef]
- Haas, S.; Straub, R.H. Disruption of Rhythms of Molecular Clocks in Primary Synovial Fibroblasts of Patients with Osteoarthritis and Rheumatoid Arthritis, Role of IL-1β/TNF. Arthritis Res. Ther. 2012, 14, R122. [Google Scholar] [CrossRef]
- Hand, L.E.; Gray, K.J.; Dickson, S.H.; Simpkins, D.A.; Ray, D.W.; Konkel, J.E.; Hepworth, M.R.; Gibbs, J.E. Regulatory T Cells Confer a Circadian Signature on Inflammatory Arthritis. Nat. Commun. 2020, 11, 1658. [Google Scholar] [CrossRef]
- Wang, S.; Lin, Y.; Yuan, X.; Li, F.; Guo, L.; Wu, B. REV-ERBα Integrates Colon Clock with Experimental Colitis through Regulation of NF-κB/NLRP3 Axis. Nat. Commun. 2018, 9, 4246. [Google Scholar] [CrossRef]
- Palmieri, O.; Mazzoccoli, G.; Bossa, F.; Maglietta, R.; Palumbo, O.; Ancona, N.; Corritore, G.; Latiano, T.; Martino, G.; Rubino, R.; et al. Systematic Analysis of Circadian Genes Using Genome-Wide cDNA Microarrays in the Inflammatory Bowel Disease Transcriptome. Chronobiol. Int. 2015, 32, 903–916. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, Y.; Cohen, S.; Chapnik, N.; Ben-Tov, A.; Yerushalmy-Feler, A.; Dotan, I.; Tauman, R.; Froy, O. Clock Gene Disruption Is an Initial Manifestation of Inflammatory Bowel Diseases. Clin. Gastroenterol. Hepatol. 2020, 18, 115–122.e1. [Google Scholar] [CrossRef]
- Liu, X.; Yu, R.; Zhu, L.; Hou, X.; Zou, K. Bidirectional Regulation of Circadian Disturbance and Inflammation in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 1741–1751. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucl. Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Parsons, R.; Parsons, R.; Garner, N.; Oster, H.; Rawashdeh, O. CircaCompare: A Method to Estimate and Statistically Support Differences in Mesor, Amplitude, and Phase, between Circadian Rhythms. Bioinformatics 2019. [Google Scholar] [CrossRef] [PubMed]
- Yeung, J.; Mermet, J.; Jouffe, C.; Marquis, J.; Charpagne, A.; Gachon, F.; Naef, F. Transcription Factor Activity Rhythms and Tissue-Specific Chromatin Interactions Explain Circadian Gene Expression across Organs. Genome Res. 2018, 28, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Pitzer, C.; Kuner, R.; Tappe-Theodor, A. Voluntary and Evoked Behavioral Correlates in Inflammatory Pain Conditions under Different Social Housing Conditions. Pain. Rep. 2016, 1, e564. [Google Scholar] [CrossRef]
- Buenafe, A.C.; Zwickey, H.; Moes, N.; Oken, B.; Jones, R.E. A Telemetric Study of Physiologic Changes in Mice with Induced Autoimmune Encephalomyelitis. Lab. Anim. 2008, 37, 361–368. [Google Scholar] [CrossRef]
- He, J.; Wang, Y.; Kastin, A.J.; Pan, W. Increased Sleep Fragmentation in Experimental Autoimmune Encephalomyelitis. Brain Behav. Immun. 2014, 38, 53–58. [Google Scholar] [CrossRef]
- Shimba, A.; Cui, G.; Tani-Ichi, S.; Ogawa, M.; Abe, S.; Okazaki, F.; Kitano, S.; Miyachi, H.; Yamada, H.; Hara, T.; et al. Glucocorticoids Drive Diurnal Oscillations in T Cell Distribution and Responses by Inducing Interleukin-7 Receptor and CXCR4. Immunity 2018, 48, 286–298.e6. [Google Scholar] [CrossRef]
- Mazzoccoli, G.; Cata, A.D.; Greco, A.; Damato, M.; Pinto, D.G.D.; Montrano, M.; Marzulli, N.; Dagostino, M.P.; Carughi, S. Opposing Circadian Rhythms of CD3+, CD4+ and CD3+, CD8+ Lymphocyte Subpopulations in Healthy Humans. Biol. Rhythm. Res. 2011, 42, 111–118. [Google Scholar] [CrossRef]
- Sonobe, Y.; Jin, S.; Wang, J.; Kawanokuchi, J.; Takeuchi, H.; Mizuno, T.; Suzumura, A. Chronological Changes of CD4(+) and CD8(+) T Cell Subsets in the Experimental Autoimmune Encephalomyelitis, a Mouse Model of Multiple Sclerosis. Tohoku J. Exp. Med. 2007, 213, 329–339. [Google Scholar] [CrossRef][Green Version]
- Silver, A.C.; Arjona, A.; Hughes, M.E.; Nitabach, M.N.; Fikrig, E. Circadian Expression of Clock Genes in Mouse Macrophages, Dendritic Cells, and B. Cells. Brain Behav. Immun. 2012, 26, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Adlanmerini, M.; Lazar, M.A. The REV-ERB Nuclear Receptors: Timekeepers for the Core Clock Period and Metabolism. Endocrinology 2023, 164, bqad069. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Bellet, M.M.; Sassone-Corsi, P.; O’Neill, L.A.J. Circadian Clock Proteins and Immunity. Immunity 2014, 40, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.J.; Gibbs, J.E. Adaptive Immunity, Chronic Inflammation and the Clock. Semin. Immunopathol. 2022, 44, 209–224. [Google Scholar] [CrossRef]
- Egstrand, S.; Mace, M.L.; Morevati, M.; Nordholm, A.; Engelholm, L.H.; Thomsen, J.S.; Brüel, A.; Naveh-Many, T.; Guo, Y.; Olgaard, K.; et al. Hypomorphic Expression of Parathyroid Bmal1 Disrupts the Internal Parathyroid Circadian Clock and Increases Parathyroid Cell Proliferation in Response to Uremia. Kidney Int. 2022, 101, 1232–1250. [Google Scholar] [CrossRef]
- Chen, X.; Winkler-Pickett, R.T.; Carbonetti, N.H.; Ortaldo, J.R.; Oppenheim, J.J.; Zack Howard, O.M. Pertussis Toxin as an Adjuvant Suppresses the Number and Function of CD4+CD25+ T Regulatory Cells. Eur. J. Immunol. 2006, 36, 671–680. [Google Scholar] [CrossRef]
- van Hamburg, J.P.; Mus, A.-M.; de Bruijn, M.J.W.; de Vogel, L.; Boon, L.; Cornelissen, F.; Asmawidjaja, P.; Hendriks, R.W.; Lubberts, E. GATA-3 Protects against Severe Joint Inflammation and Bone Erosion and Reduces Differentiation of Th17 Cells during Experimental Arthritis. Arthritis Rheum. 2009, 60, 750–759. [Google Scholar] [CrossRef]
- Amir, M.; Chaudhari, S.; Wang, R.; Campbell, S.; Mosure, S.A.; Chopp, L.B.; Lu, Q.; Shang, J.; Pelletier, O.B.; He, Y.; et al. REV-ERBα Regulates TH17 Cell Development and Autoimmunity. Cell Rep. 2018, 25, 3733–3749.e8. [Google Scholar] [CrossRef]
- Eddy, J.L.; Krukowski, K.; Janusek, L.; Mathews, H.L. Glucocorticoids Regulate Natural Killer Cell Function Epigenetically. Cell. Immunol. 2014, 290, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Olsen, S.; Moldovan, J.; Fu, X.; Sarkar, F.H.; Moudgil, V.K.; Callewaert, D.M. Glucocorticoid Regulation of Natural Cytotoxicity: Effects of Cortisol on the Phenotype and Function of a Cloned Human Natural Killer Cell Line. Cell. Immunol. 1997, 178, 108–116. [Google Scholar] [CrossRef]
- Paul, M.J.; Indic, P.; Schwartz, W.J. Social Synchronization of Circadian Rhythmicity in Female Mice Depends on the Number of Cohabiting Animals. Biol. Lett. 2015, 11, 20150204. [Google Scholar] [CrossRef]
- Pollak, Y.; Ovadia, H.; Goshen, I.; Gurevich, R.; Monsa, K.; Avitsur, R.; Yirmiya, R. Behavioral Aspects of Experimental Autoimmune Encephalomyelitis. J. Neuroimmunol. 2000, 104, 31–36. [Google Scholar] [CrossRef]
- Nagai, K.; Nishio, T.; Nakagawa, H.; Nakamura, S.; Fukuda, Y. Effect of Bilateral Lesions of the Suprachiasmatic Nuclei on the Circadian Rhythm of Food-Intake. Brain Res. 1978, 142, 384–389. [Google Scholar] [CrossRef]
- Tsang, A.H.; Koch, C.E.; Kiehn, J.-T.; Schmidt, C.X.; Oster, H. An Adipokine Feedback Regulating Diurnal Food Intake Rhythms in Mice. Elife 2020, 9, e55388. [Google Scholar] [CrossRef] [PubMed]
- Delezie, J.; Dumont, S.; Dardente, H.; Oudart, H.; Gréchez-Cassiau, A.; Klosen, P.; Teboul, M.; Delaunay, F.; Pévet, P.; Challet, E. The Nuclear Receptor REV-ERBα Is Required for the Daily Balance of Carbohydrate and Lipid Metabolism. FASEB J. 2012, 26, 3321–3335. [Google Scholar] [CrossRef]
- Huntemann, N.; Vogelsang, A.; Groeneweg, L.; Willison, A.; Herrmann, A.M.; Meuth, S.G.; Eichler, S. An Optimized and Validated Protocol for Inducing Chronic Experimental Autoimmune Encephalomyelitis in C57BL/6J Mice. J. Neurosci. Methods 2022, 367, 109443. [Google Scholar] [CrossRef]
- Stils, H.F., Jr. Adjuvants and Antibody Production: Dispelling the Myths Associated with Freund’s Complete and Other Adjuvants. ILAR J. 2005, 46, 280–293. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hirose, M.; Leliavski, A.; de Assis, L.V.M.; Matveeva, O.; Skrum, L.; Solbach, W.; Oster, H.; Heyde, I. Chronic Inflammation Disrupts Circadian Rhythms in Splenic CD4+ and CD8+ T Cells in Mice. Cells 2024, 13, 151. https://doi.org/10.3390/cells13020151
Hirose M, Leliavski A, de Assis LVM, Matveeva O, Skrum L, Solbach W, Oster H, Heyde I. Chronic Inflammation Disrupts Circadian Rhythms in Splenic CD4+ and CD8+ T Cells in Mice. Cells. 2024; 13(2):151. https://doi.org/10.3390/cells13020151
Chicago/Turabian StyleHirose, Misa, Alexei Leliavski, Leonardo Vinícius Monteiro de Assis, Olga Matveeva, Ludmila Skrum, Werner Solbach, Henrik Oster, and Isabel Heyde. 2024. "Chronic Inflammation Disrupts Circadian Rhythms in Splenic CD4+ and CD8+ T Cells in Mice" Cells 13, no. 2: 151. https://doi.org/10.3390/cells13020151
APA StyleHirose, M., Leliavski, A., de Assis, L. V. M., Matveeva, O., Skrum, L., Solbach, W., Oster, H., & Heyde, I. (2024). Chronic Inflammation Disrupts Circadian Rhythms in Splenic CD4+ and CD8+ T Cells in Mice. Cells, 13(2), 151. https://doi.org/10.3390/cells13020151





