Tissue-Predisposition to Cancer Driver Mutations
Abstract
1. Introduction
2. Lessons from Normal Tissue
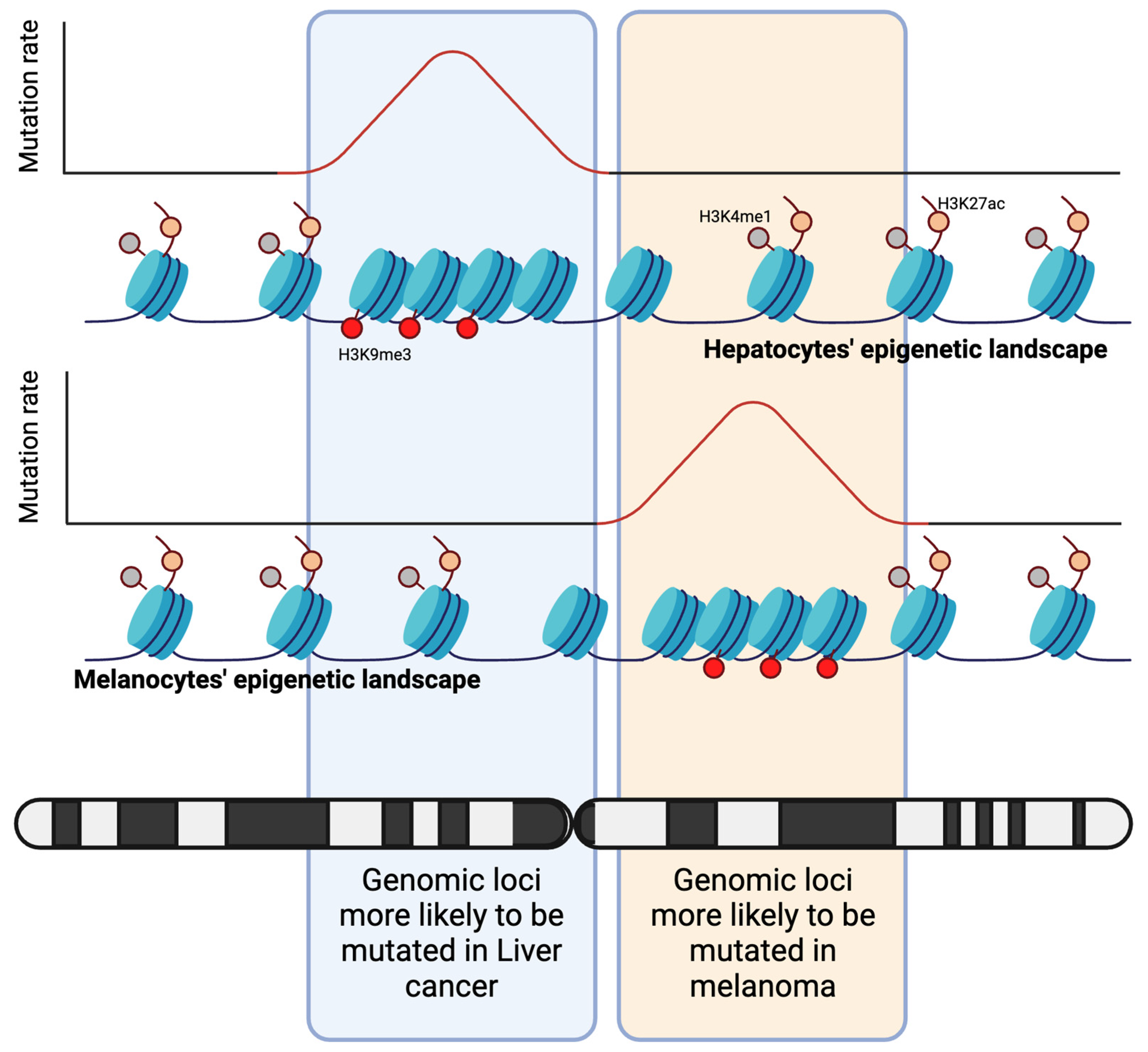
2.1. Mutations Are Encoded by the Epigenetic Landscape
2.2. Gene Expression or Function
2.3. Developmental Origins of Cancer
3. The Molecular Context of Mutations in Cancerous Tissue
3.1. The Paralogue Paradox
3.2. Synthetic Lethality
3.3. To Deal or Not to Deal with Senescence
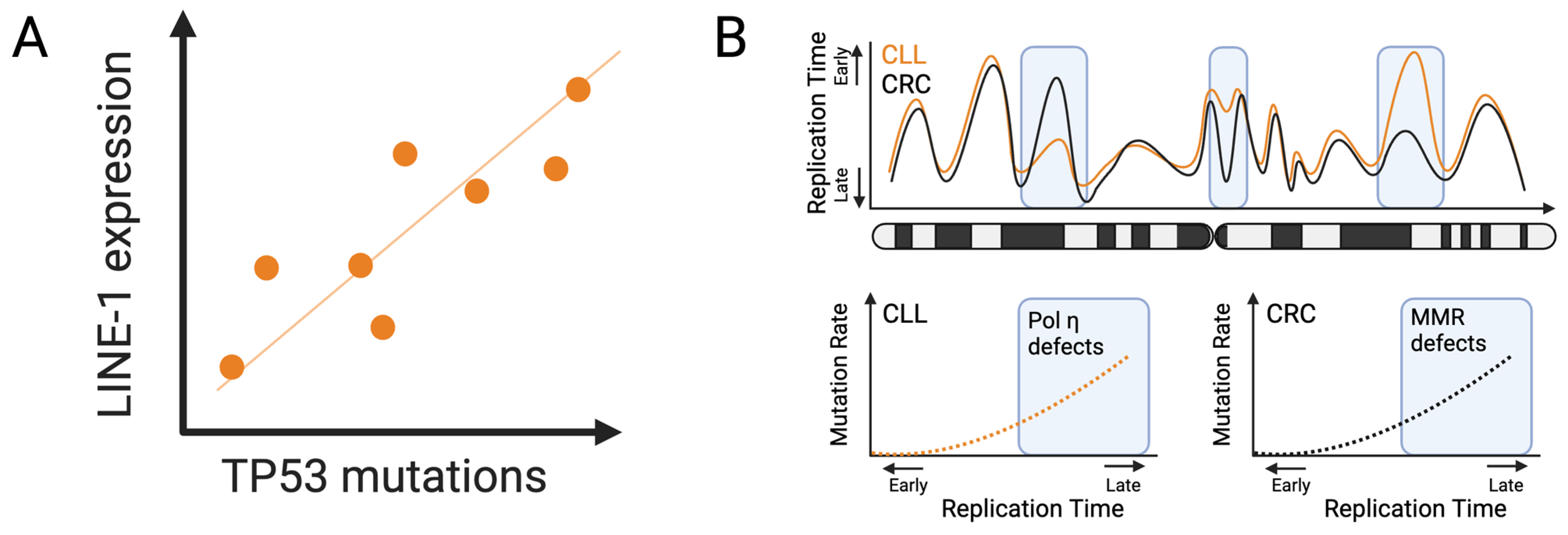
3.4. LINE-1 Transposable Elements
3.5. Replication Timing Dynamics
4. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Stoltzfus, A.; Cable, K. Mendelian-mutationism: The forgotten evolutionary synthesis. J. Hist. Biol. 2014, 47, 501–546. [Google Scholar] [CrossRef]
- Allen, G.E. Hugo de Vries and the reception of the “mutation theory”. J. Hist. Biol. 1969, 2, 55–87. [Google Scholar] [CrossRef]
- Koonin, E.V. The Origin at 150: Is a new evolutionary synthesis in sight? Trends Genet. 2009, 25, 473–475. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zhou, P.K. DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 254. [Google Scholar] [CrossRef] [PubMed]
- Hanley, G.E.; McAlpine, J.N.; Miller, D.; Huntsman, D.; Schrader, K.A.; Blake Gilks, C.; Mitchell, G. A population-based analysis of germline BRCA1 and BRCA2 testing among ovarian cancer patients in an era of histotype-specific approaches to ovarian cancer prevention. BMC Cancer 2018, 18, 254. [Google Scholar] [CrossRef] [PubMed]
- Plawski, A.; Banasiewicz, T.; Borun, P.; Kubaszewski, L.; Krokowicz, P.; Skrzypczak-Zielinska, M.; Lubinski, J. Familial adenomatous polyposis of the colon. Hered. Cancer Clin. Pract. 2013, 11, 15. [Google Scholar] [CrossRef][Green Version]
- Kennedy, M.C.; Lowe, S.W. Mutant p53: It’s not all one and the same. Cell Death Differ. 2022, 29, 983–987. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- De Braekeleer, E.; Douet-Guilbert, N.; Rowe, D.; Bown, N.; Morel, F.; Berthou, C.; Ferec, C.; De Braekeleer, M. ABL1 fusion genes in hematological malignancies: A review. Eur. J. Haematol. 2011, 86, 361–371. [Google Scholar] [CrossRef]
- Basu, A.; Tiwari, V.K. Epigenetic reprogramming of cell identity: Lessons from development for regenerative medicine. Clin. Epigenet. 2021, 13, 144. [Google Scholar] [CrossRef]
- Barrero, M.J.; Boue, S.; Izpisua Belmonte, J.C. Epigenetic mechanisms that regulate cell identity. Cell Stem Cell 2010, 7, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Beagan, J.A.; Phillips-Cremins, J.E. On the existence and functionality of topologically associating domains. Nat. Genet. 2020, 52, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Szalaj, P.; Plewczynski, D. Three-dimensional organization and dynamics of the genome. Cell Biol. Toxicol. 2018, 34, 381–404. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Sharp, P.M.; Li, W.H. Mutation rates differ among regions of the mammalian genome. Nature 1989, 337, 283–285. [Google Scholar] [CrossRef] [PubMed]
- Schuster-Bockler, B.; Lehner, B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 2012, 488, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Makova, K.D.; Hardison, R.C. The effects of chromatin organization on variation in mutation rates in the genome. Nat. Rev. Genet. 2015, 16, 213–223. [Google Scholar] [CrossRef]
- Polak, P.; Karlic, R.; Koren, A.; Thurman, R.; Sandstrom, R.; Lawrence, M.; Reynolds, A.; Rynes, E.; Vlahovicek, K.; Stamatoyannopoulos, J.A.; et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 2015, 518, 360–364. [Google Scholar] [CrossRef] [PubMed]
- Giroux, V.; Rustgi, A.K. Metaplasia: Tissue injury adaptation and a precursor to the dysplasia-cancer sequence. Nat. Rev. Cancer 2017, 17, 594–604. [Google Scholar] [CrossRef]
- Ha, K.; Kim, H.G.; Lee, H. Chromatin marks shape mutation landscape at early stage of cancer progression. NPJ Genom. Med. 2017, 2, 9. [Google Scholar] [CrossRef]
- Abdolhosseini, F.; Azarkhalili, B.; Maazallahi, A.; Kamal, A.; Motahari, S.A.; Sharifi-Zarchi, A.; Chitsaz, H. Cell Identity Codes: Understanding Cell Identity from Gene Expression Profiles using Deep Neural Networks. Sci. Rep. 2019, 9, 2342. [Google Scholar] [CrossRef]
- Efroni, I.; Ip, P.L.; Nawy, T.; Mello, A.; Birnbaum, K.D. Quantification of cell identity from single-cell gene expression profiles. Genome Biol. 2015, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Svejstrup, J.Q. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 2002, 3, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Tiong, K.L.; Yeang, C.H. Explaining cancer type specific mutations with transcriptomic and epigenomic features in normal tissues. Sci. Rep. 2018, 8, 11456. [Google Scholar] [CrossRef] [PubMed]
- Perrimon, N.; Pitsouli, C.; Shilo, B.Z. Signaling mechanisms controlling cell fate and embryonic patterning. Cold Spring Harb. Perspect. Biol. 2012, 4, a005975. [Google Scholar] [CrossRef] [PubMed]
- Plasschaert, R.N.; Bartolomei, M.S. Genomic imprinting in development, growth, behavior and stem cells. Development 2014, 141, 1805–1813. [Google Scholar] [CrossRef] [PubMed]
- Monk, M.; Holding, C. Human embryonic genes re-expressed in cancer cells. Oncogene 2001, 20, 8085–8091. [Google Scholar] [CrossRef] [PubMed]
- Siebel, C.; Lendahl, U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol. Rev. 2017, 97, 1235–1294. [Google Scholar] [CrossRef]
- Nishikawa, Y.; Kodama, Y.; Shiokawa, M.; Matsumori, T.; Marui, S.; Kuriyama, K.; Kuwada, T.; Sogabe, Y.; Kakiuchi, N.; Tomono, T.; et al. Hes1 plays an essential role in Kras-driven pancreatic tumorigenesis. Oncogene 2019, 38, 4283–4296. [Google Scholar] [CrossRef]
- Eser, S.; Reiff, N.; Messer, M.; Seidler, B.; Gottschalk, K.; Dobler, M.; Hieber, M.; Arbeiter, A.; Klein, S.; Kong, B.; et al. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013, 23, 406–420. [Google Scholar] [CrossRef]
- Deftos, M.L.; Bevan, M.J. Notch signaling in T cell development. Curr. Opin. Immunol. 2000, 12, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Radtke, F.; Wilson, A.; Stark, G.; Bauer, M.; van Meerwijk, J.; MacDonald, H.R.; Aguet, M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 1999, 10, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; MacDonald, H.R.; Radtke, F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J. Exp. Med. 2001, 194, 1003–1012. [Google Scholar] [CrossRef]
- Demarest, R.M.; Ratti, F.; Capobianco, A.J. It’s T-ALL about Notch. Oncogene 2008, 27, 5082–5091. [Google Scholar] [CrossRef] [PubMed]
- Zweidler-McKay, P.A.; He, Y.; Xu, L.; Rodriguez, C.G.; Karnell, F.G.; Carpenter, A.C.; Aster, J.C.; Allman, D.; Pear, W.S. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood 2005, 106, 3898–3906. [Google Scholar] [CrossRef]
- Clevers, H. Wnt/beta-catenin signaling in development and disease. Cell 2006, 127, 469–480. [Google Scholar] [CrossRef]
- Farin, H.F.; Van Es, J.H.; Clevers, H. Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 2012, 143, 1518–1529.e1517. [Google Scholar] [CrossRef]
- Li, L.; Neaves, W.B. Normal stem cells and cancer stem cells: The niche matters. Cancer Res. 2006, 66, 4553–4557. [Google Scholar] [CrossRef]
- Rowan, A.J.; Lamlum, H.; Ilyas, M.; Wheeler, J.; Straub, J.; Papadopoulou, A.; Bicknell, D.; Bodmer, W.F.; Tomlinson, I.P. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc. Natl. Acad. Sci. USA 2000, 97, 3352–3357. [Google Scholar] [CrossRef]
- Spirio, L.; Olschwang, S.; Groden, J.; Robertson, M.; Samowitz, W.; Joslyn, G.; Gelbert, L.; Thliveris, A.; Carlson, M.; Otterud, B.; et al. Alleles of the APC gene: An attenuated form of familial polyposis. Cell 1993, 75, 951–957. [Google Scholar] [CrossRef]
- Groves, C.J.; Saunders, B.P.; Spigelman, A.D.; Phillips, R.K. Duodenal cancer in patients with familial adenomatous polyposis (FAP): Results of a 10 year prospective study. Gut 2002, 50, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Scurrah, C.R.; McKinley, E.T.; Simmons, A.J.; Ramirez-Solano, M.A.; Zhu, X.; Markham, N.O.; Heiser, C.N.; Vega, P.N.; Rolong, A.; et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 2021, 184, 6262–6280. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Goswami, S.; Hu, Y.; Tang, F.; Zafra, M.P.; Murphy, C.; Cao, Z.; Poirier, J.T.; Khurana, E.; Elemento, O.; et al. Lineage Reversion Drives WNT Independence in Intestinal Cancer. Cancer Discov. 2020, 10, 1590–1609. [Google Scholar] [CrossRef] [PubMed]
- Hartman, J.L.t.; Garvik, B.; Hartwell, L. Principles for the buffering of genetic variation. Science 2001, 291, 1001–1004. [Google Scholar] [CrossRef] [PubMed]
- To, M.D.; Wong, C.E.; Karnezis, A.N.; Del Rosario, R.; Di Lauro, R.; Balmain, A. Kras regulatory elements and exon 4A determine mutation specificity in lung cancer. Nat. Genet. 2008, 40, 1240–1244. [Google Scholar] [CrossRef] [PubMed]
- Cereda, M.; Mourikis, T.P.; Ciccarelli, F.D. Genetic Redundancy, Functional Compensation, and Cancer Vulnerability. Trends Cancer 2016, 2, 160–162. [Google Scholar] [CrossRef]
- Ciriello, G.; Cerami, E.; Sander, C.; Schultz, N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012, 22, 398–406. [Google Scholar] [CrossRef]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef]
- Meylan, E.; Dooley, A.L.; Feldser, D.M.; Shen, L.; Turk, E.; Ouyang, C.; Jacks, T. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature 2009, 462, 104–107. [Google Scholar] [CrossRef]
- Dajee, M.; Lazarov, M.; Zhang, J.Y.; Cai, T.; Green, C.L.; Russell, A.J.; Marinkovich, M.P.; Tao, S.; Lin, Q.; Kubo, Y.; et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature 2003, 421, 639–643. [Google Scholar] [CrossRef]
- Cheng, K.; Nair, N.U.; Lee, J.S.; Ruppin, E. Synthetic lethality across normal tissues is strongly associated with cancer risk, onset, and tumor suppressor specificity. Sci. Adv. 2021, 7, eabc2100. [Google Scholar] [CrossRef] [PubMed]
- Mijit, M.; Caracciolo, V.; Melillo, A.; Amicarelli, F.; Giordano, A. Role of p53 in the Regulation of Cellular Senescence. Biomolecules 2020, 10, 420. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, C.A.; Wang, B.; Demaria, M. Senescence and cancer - role and therapeutic opportunities. Nat. Rev. Clin. Oncol. 2022, 19, 619–636. [Google Scholar] [CrossRef] [PubMed]
- Courtois-Cox, S.; Jones, S.L.; Cichowski, K. Many roads lead to oncogene-induced senescence. Oncogene 2008, 27, 2801–2809. [Google Scholar] [CrossRef] [PubMed]
- Pribluda, A.; Elyada, E.; Wiener, Z.; Hamza, H.; Goldstein, R.E.; Biton, M.; Burstain, I.; Morgenstern, Y.; Brachya, G.; Billauer, H.; et al. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell 2013, 24, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Sedic, M.; Skibinski, A.; Brown, N.; Gallardo, M.; Mulligan, P.; Martinez, P.; Keller, P.J.; Glover, E.; Richardson, A.L.; Cowan, J.; et al. Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat. Commun. 2015, 6, 7505. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.P.; Naler, L.B.; Ma, S.; Lu, C. Cell-type-specific epigenomic variations associated with BRCA1 mutation in pre-cancer human breast tissues. NAR Genom. Bioinform. 2022, 4, lqac006. [Google Scholar] [CrossRef]
- Falcomata, C.; Barthel, S.; Ulrich, A.; Diersch, S.; Veltkamp, C.; Rad, L.; Boniolo, F.; Solar, M.; Steiger, K.; Seidler, B.; et al. Genetic Screens Identify a Context-Specific PI3K/p27Kip1 Node Driving Extrahepatic Biliary Cancer. Cancer Discov. 2021, 11, 3158–3177. [Google Scholar] [CrossRef]
- Morton, J.P.; Timpson, P.; Karim, S.A.; Ridgway, R.A.; Athineos, D.; Doyle, B.; Jamieson, N.B.; Oien, K.A.; Lowy, A.M.; Brunton, V.G.; et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 246–251. [Google Scholar] [CrossRef]
- Burns, K.H. Transposable elements in cancer. Nat. Rev. Cancer 2017, 17, 415–424. [Google Scholar] [CrossRef]
- Turelli, P.; Castro-Diaz, N.; Marzetta, F.; Kapopoulou, A.; Raclot, C.; Duc, J.; Tieng, V.; Quenneville, S.; Trono, D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014, 24, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Ergun, S.; Buschmann, C.; Heukeshoven, J.; Dammann, K.; Schnieders, F.; Lauke, H.; Chalajour, F.; Kilic, N.; Stratling, W.H.; Schumann, G.G. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 2004, 279, 27753–27763. [Google Scholar] [CrossRef] [PubMed]
- Belancio, V.P.; Roy-Engel, A.M.; Pochampally, R.R.; Deininger, P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. 2010, 38, 3909–3922. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Babarinde, I.A.; Sun, L.; Xu, S.; Chen, R.; Shi, J.; Wei, Y.; Li, Y.; Ma, G.; Zhuang, Q.; et al. Identifying transposable element expression dynamics and heterogeneity during development at the single-cell level with a processing pipeline scTE. Nat. Commun. 2021, 12, 1456. [Google Scholar] [CrossRef] [PubMed]
- Xiao-Jie, L.; Hui-Ying, X.; Qi, X.; Jiang, X.; Shi-Jie, M. LINE-1 in cancer: Multifaceted functions and potential clinical implications. Genet. Med. 2016, 18, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Philippe, C.; Vargas-Landin, D.B.; Doucet, A.J.; van Essen, D.; Vera-Otarola, J.; Kuciak, M.; Corbin, A.; Nigumann, P.; Cristofari, G. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife 2016, 5, e13926. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.C.; Gardner, E.J.; Masood, A.; Chuang, N.T.; Vertino, P.M.; Devine, S.E. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016, 26, 745–755. [Google Scholar] [CrossRef]
- Belgnaoui, S.M.; Gosden, R.G.; Semmes, O.J.; Haoudi, A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006, 6, 13. [Google Scholar] [CrossRef]
- McKerrow, W.; Wang, X.; Mendez-Dorantes, C.; Mita, P.; Cao, S.; Grivainis, M.; Ding, L.; LaCava, J.; Burns, K.H.; Boeke, J.D.; et al. LINE-1 expression in cancer correlates with p53 mutation, copy number alteration, and S phase checkpoint. Proc. Natl. Acad. Sci. USA 2022, 119, e2115999119. [Google Scholar] [CrossRef]
- Doucet-O’Hare, T.T.; Rodic, N.; Sharma, R.; Darbari, I.; Abril, G.; Choi, J.A.; Young Ahn, J.; Cheng, Y.; Anders, R.A.; Burns, K.H.; et al. LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc. Natl. Acad. Sci. USA 2015, 112, E4894–E4900. [Google Scholar] [CrossRef]
- Rhind, N.; Gilbert, D.M. DNA replication timing. Cold Spring Harb. Perspect. Biol. 2013, 5, a010132. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Baris, A.; Aladjem, M.I. Replication timing and nuclear structure. Curr. Opin. Cell Biol. 2018, 52, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Stamatoyannopoulos, J.A.; Adzhubei, I.; Thurman, R.E.; Kryukov, G.V.; Mirkin, S.M.; Sunyaev, S.R. Human mutation rate associated with DNA replication timing. Nat. Genet. 2009, 41, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Lehner, B. Differential DNA mismatch repair underlies mutation rate variation across the human genome. Nature 2015, 521, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Ryba, T.; Hiratani, I.; Lu, J.; Itoh, M.; Kulik, M.; Zhang, J.; Schulz, T.C.; Robins, A.J.; Dalton, S.; Gilbert, D.M. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 2010, 20, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Caballero, M.; Boos, D.; Koren, A. Cell-type specificity of the human mutation landscape with respect to DNA replication dynamics. Cell Genom. 2023, 3, 100315. [Google Scholar] [CrossRef]
- Yaacov, A.; Rosenberg, S.; Simon, I. Mutational signatures association with replication timing in normal cells reveals similarities and differences with matched cancer tissues. Sci. Rep. 2023, 13, 7833. [Google Scholar] [CrossRef]
- Williams, B.O.; Remington, L.; Albert, D.M.; Mukai, S.; Bronson, R.T.; Jacks, T. Cooperative tumorigenic effects of germline mutations in Rb and p53. Nat. Genet. 1994, 7, 480–484. [Google Scholar] [CrossRef]
- Bianchi, J.J.; Zhao, X.; Mays, J.C.; Davoli, T. Not all cancers are created equal: Tissue specificity in cancer genes and pathways. Curr. Opin. Cell Biol. 2020, 63, 135–143. [Google Scholar] [CrossRef]
- Foster, D.S.; Griffin, M.; Januszyk, M.; Delitto, D.; Norton, J.A.; Longaker, M.T. Optimized Nuclei Isolation from Fresh and Frozen Solid Tumor Specimens for Multiome Sequencing. J. Vis. Exp. 2023, 200, e65831. [Google Scholar] [CrossRef]
- Muyas, F.; Sauer, C.M.; Valle-Inclan, J.E.; Li, R.; Rahbari, R.; Mitchell, T.J.; Hormoz, S.; Cortes-Ciriano, I. De novo detection of somatic mutations in high-throughput single-cell profiling data sets. Nat. Biotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, A.; Pikarsky, E.; Ben-Neriah, Y. Putative homeostatic role of cancer driver mutations. Trends Cell Biol. 2022, 32, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Hyman, D.M.; Puzanov, I.; Subbiah, V.; Faris, J.E.; Chau, I.; Blay, J.Y.; Wolf, J.; Raje, N.S.; Diamond, E.L.; Hollebecque, A.; et al. Vemurafenib in Multiple Nonmelanoma Cancers with BRAF V600 Mutations. N. Engl. J. Med. 2015, 373, 726–736. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peters, L.; Venkatachalam, A.; Ben-Neriah, Y. Tissue-Predisposition to Cancer Driver Mutations. Cells 2024, 13, 106. https://doi.org/10.3390/cells13020106
Peters L, Venkatachalam A, Ben-Neriah Y. Tissue-Predisposition to Cancer Driver Mutations. Cells. 2024; 13(2):106. https://doi.org/10.3390/cells13020106
Chicago/Turabian StylePeters, Luriano, Avanthika Venkatachalam, and Yinon Ben-Neriah. 2024. "Tissue-Predisposition to Cancer Driver Mutations" Cells 13, no. 2: 106. https://doi.org/10.3390/cells13020106
APA StylePeters, L., Venkatachalam, A., & Ben-Neriah, Y. (2024). Tissue-Predisposition to Cancer Driver Mutations. Cells, 13(2), 106. https://doi.org/10.3390/cells13020106






