Multiscale Modeling of Bistability in the Yeast Polarity Circuit
Abstract
1. Introduction
2. Methods
2.1. Experimental Datasets
2.2. Dose Response Curves Derived from Experimental Data
2.3. Reaction–Diffusion Equation Simulations
2.4. Particle-Based Simulations
3. Results
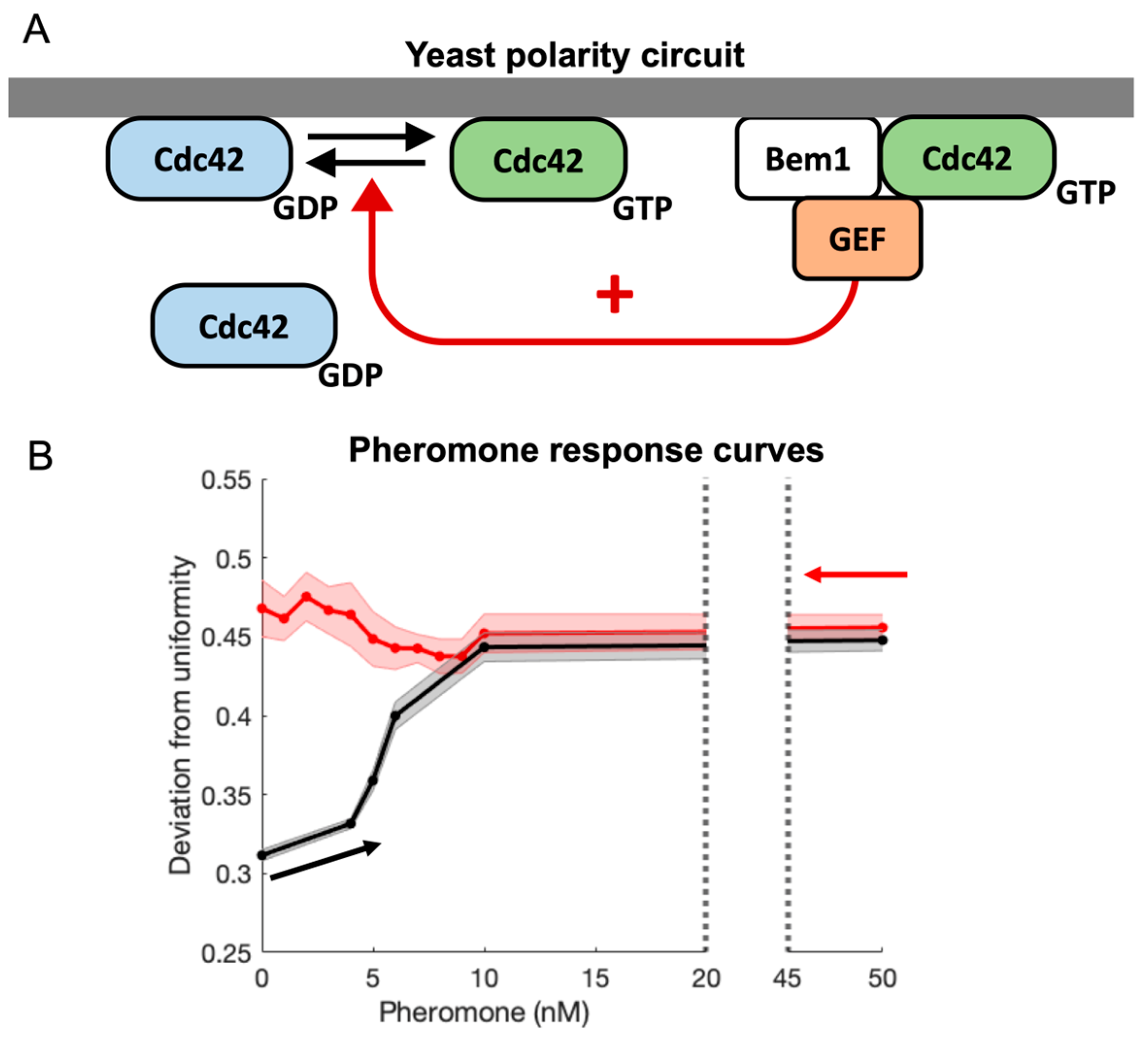
3.1. Experimental Observations
3.2. Mechanistic Considerations for Model Development
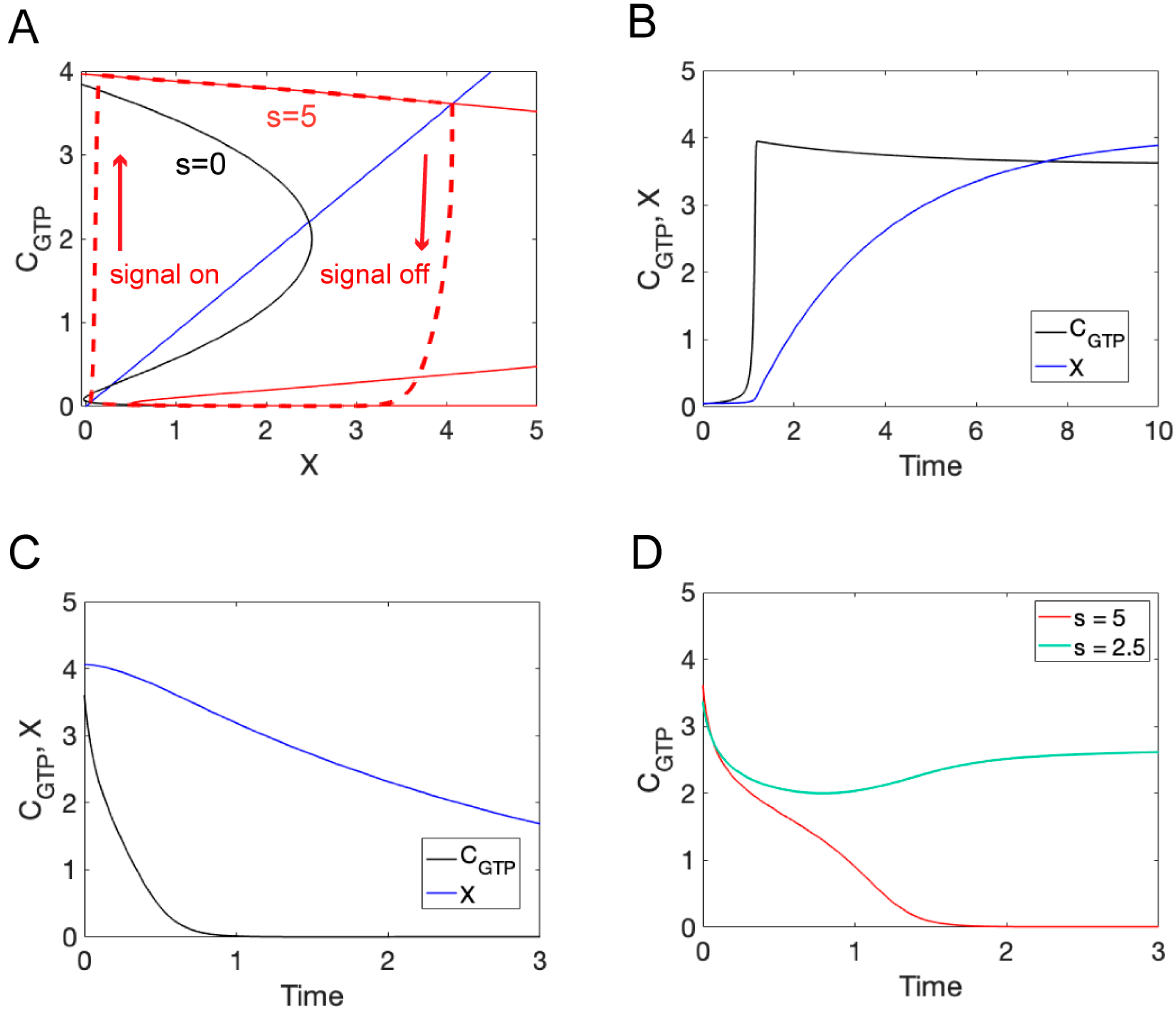
3.3. Ordinary Differential Equation Model
3.4. Reaction–Diffusion Equation Models
3.5. Detailed RDE Model
3.6. Particle-Based Model
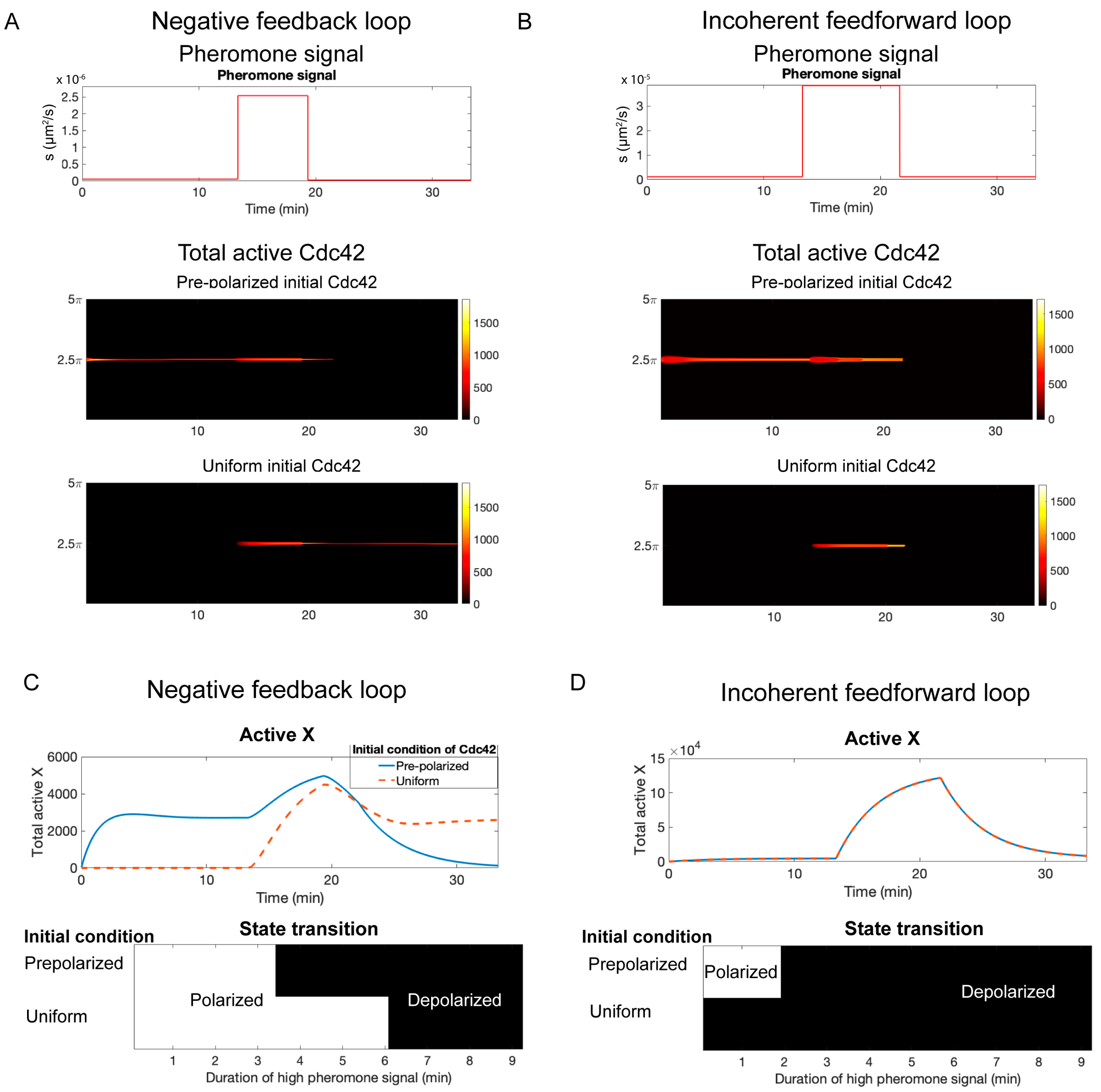
3.7. Negative Feedback Versus Incoherent Feedforward Regulation
4. Discussion
4.1. Multiscale Models of the Polarity Circuit
4.2. Negative Feedback Versus Incoherent Feedforward Regulation
4.3. Potential Benefits of Negative Regulation of a Bistable Circuit
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DfU | deviation from uniformity |
| GAP | GTPase activating protein |
| GEF | guanine nucleotide exchange factor |
| ODE | ordinary differential equation |
| RDE | reaction–diffusion equation |
| NFB | negative feedback |
| IFF | incoherent feedforward |
References
- Cretel, E.; Pierres, A.; Benoliel, A.-M.; Bongrand, P. How Cells feel their environment: A focus on early dynamic events. Cell. Mol. Bioeng. 2008, 1, 5–14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ruprecht, V.; Monzo, P.; Ravasio, A.; Yue, Z.; Makhija, E.; Strale, P.O.; Gauthier, N.; Shivashankar, G.V.; Studer, V.; Albiges-Rizo, C.; et al. How cells respond to environmental cues—Insights from bio-functionalized substrates. J. Cell Sci. 2017, 130, 51–61. [Google Scholar] [CrossRef]
- Vogel, V.; Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, K.C.; Lew, D.J. Pheromone Guidance of Polarity Site Movement in Yeast. Biomolecules 2022, 12, 502. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, S.; Barrowman, J. Biogenesis of the Saccharomyces cerevisiae pheromone a-factor, from yeast mating to human disease. Microbiol. Mol. Biol. Rev. 2012, 76, 626–651. [Google Scholar] [CrossRef]
- Merlini, L.; Dudin, O.; Martin, S.G. Mate and fuse: How yeast cells do it. Open Biol. 2013, 3, 130008. [Google Scholar] [CrossRef]
- Clark-Cotton, M.R.; Jacobs, K.C.; Lew, D.J. Chemotropism and Cell-Cell Fusion in Fungi. Microbiol. Mol. Biol. Rev. 2022, 86, e0016521. [Google Scholar] [CrossRef]
- Miller, K.E.; Kang, P.J.; Park, H.O. Regulation of Cdc42 for polarized growth in budding yeast. Microb. Cell 2020, 7, 175–189. [Google Scholar] [CrossRef]
- Witte, K.; Strickland, D.; Glotzer, M. Cell cycle entry triggers a switch between two modes of Cdc42 activation during yeast polarization. Elife 2017, 6, e26722. [Google Scholar] [CrossRef]
- Woods, B.; Lew, D.J. Polarity establishment by Cdc42: Key roles for positive feedback and differential mobility. Small GTPases 2019, 10, 130–137. [Google Scholar] [CrossRef]
- Goryachev, A.B.; Leda, M. Many roads to symmetry breaking: Molecular mechanisms and theoretical models of yeast cell polarity. Mol. Biol. Cell 2017, 28, 370–380. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Mosaddeghzadeh, N.; Ahmadian, M.R. The RHO Family GTPases: Mechanisms of Regulation and Signaling. Cells 2021, 10, 1831. [Google Scholar] [CrossRef] [PubMed]
- Perez, P.; Rincon, S.A. Rho GTPases: Regulation of cell polarity and growth in yeasts. Biochem. J. 2010, 426, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Turing, A.M. The chemical basis of morphogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1952, 237, 37–72. [Google Scholar]
- Goryachev, A.B.; Pokhilko, A.V. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008, 582, 1437–1443. [Google Scholar] [CrossRef] [PubMed]
- Levchenko, A.; Iglesias, P.A. Models of eukaryotic gradient sensing: Application to chemotaxis of amoebae and neutrophils. Biophys. J. 2002, 82 Pt 1, 50–63. [Google Scholar] [CrossRef] [PubMed]
- Rangamani, P.; Iyengar, R. Modelling cellular signalling systems. Essays Biochem. 2008, 45, 83–94. [Google Scholar]
- Tyson, J.J.; Laomettachit, T.; Kraikivski, P. Modeling the dynamic behavior of biochemical regulatory networks. J. Theor. Biol. 2019, 462, 514–527. [Google Scholar] [CrossRef]
- Voit, E.O.; Olivenca, D.V. Discrete Biochemical Systems Theory. Front. Mol. Biosci. 2022, 9, 874669. [Google Scholar] [CrossRef]
- Rappel, W.J.; Edelstein-Keshet, L. Mechanisms of Cell Polarization. Curr. Opin. Syst. Biol. 2017, 3, 43–53. [Google Scholar] [CrossRef]
- Edelstein-Keshet, L.; Holmes, W.R.; Zajac, M.; Dutot, M. From simple to detailed models for cell polarization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130003. [Google Scholar] [CrossRef]
- Shi, C.; Huang, C.-H.; Devreotes, P.N.; Iglesias, P.A. Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells. PLoS Comput. Biol. 2013, 9, e1003122. [Google Scholar] [CrossRef]
- Khalili, B.; Lovelace, H.D.; Rutkowski, D.M.; Holz, D.; Vavylonis, D. Fission Yeast Polarization: Modeling Cdc42 Oscillations, Symmetry Breaking, and Zones of Activation and Inhibition. Cells 2020, 9, 1769. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Miura, T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 2010, 329, 1616–1620. [Google Scholar] [CrossRef] [PubMed]
- Stefan, M.I.; Bartol, T.M.; Sejnowski, T.J.; Kennedy, M.B. Multi-state modeling of biomolecules. PLoS Comput. Biol. 2014, 10, e1003844. [Google Scholar] [CrossRef]
- Andrews, S.S.; Bray, D. Stochastic simulation of chemical reactions with spatial resolution and single molecule detail. Phys. Biol. 2004, 1, 137–151. [Google Scholar] [CrossRef]
- Pablo, M.; Ramirez, S.A.; Elston, T.C. Particle-based simulations of polarity establishment reveal stochastic promotion of turing pattern formation. PLoS Comput. Biol. 2018, 14, e1006016. [Google Scholar] [CrossRef] [PubMed]
- Chant, J. Cell polarity in yeast. Trends Genet. 1994, 10, 328–333. [Google Scholar] [CrossRef]
- Chiou, J.G.; Balasubramanian, M.K.; Lew, D.J. Cell Polarity in Yeast. Annu. Rev. Cell Dev. Biol. 2017, 33, 77–101. [Google Scholar] [CrossRef]
- Sieber, B.; Coronas-Serna, J.M.; Martin, S.G. A focus on yeast mating: From pheromone signaling to cell-cell fusion. Semin. Cell Dev. Biol. 2023, 133, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Duina, A.A.; Miller, M.E.; Keeney, J.B. Budding yeast for budding geneticists: A primer on the Saccharomyces cerevisiae model system. Genetics 2014, 197, 33–48. [Google Scholar] [CrossRef]
- Kiskowski, M.A.; Hancock, J.F.; Kenworthy, A.K. On the use of Ripley’s K-function and its derivatives to analyze domain size. Biophys. J. 2009, 97, 1095–1103. [Google Scholar] [CrossRef]
- Vered, L. Memory and Bistability in the Pheromone Response Pathway, in Chemistry. Ph.D. Thesis, University of North Carolina, Chapel Hill, NC, USA, 2018; p. 73. [Google Scholar] [CrossRef]
- Bennett, M.R.; Pang, W.L.; Ostroff, N.A.; Baumgartner, B.L.; Nayak, S.; Tsimring, L.S.; Hasty, J. Metabolic gene regulation in a dynamically changing environment. Nature 2008, 454, 1119–1122. [Google Scholar] [CrossRef]
- Pomeroy, A.E.; Peña, M.I.; Houser, J.R.; Dixit, G.; Dohlman, H.G.; Elston, T.C.; Errede, B. A predictive model of gene expression reveals the role of network motifs in the mating response of yeast. Sci. Signal. 2021, 14, eabb5235. [Google Scholar] [CrossRef] [PubMed]
- Shellhammer, J.P.; Pomeroy, A.E.; Li, Y.; Dujmusic, L.; Elston, T.C.; Hao, N.; Dohlman, H.G. Quantitative analysis of the yeast pheromone pathway. Yeast 2019, 36, 495–518. [Google Scholar] [CrossRef]
- Dyer, J.M.; Savage, N.S.; Jin, M.; Zyla, T.R.; Elston, T.C.; Lew, D.J. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 2013, 23, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.-G.; Ramirez, S.A.; Elston, T.C.; Witelski, T.P.; Schaeffer, D.G.; Lew, D.J. Principles that govern competition or co-existence in Rho-GTPase driven polarization. PLoS Comput. Biol. 2018, 14, e1006095. [Google Scholar] [CrossRef]
- Chou, C.S.; Nie, Q.; Yi, T.M. Modeling robustness tradeoffs in yeast cell polarization induced by spatial gradients. PLoS ONE 2008, 3, e3103. [Google Scholar] [CrossRef]
- Freisinger, T.; Klüender, B.; Johnson, J.; Müller, N.; Pichler, G.; Beck, G.; Costanzo, M.; Boone, C.; Cerione, R.A.; Frey, E.; et al. Establishment of a robust single axis of cell polarity by coupling multiple positive feedback loops. Nat. Commun. 2013, 4, 1807. [Google Scholar] [CrossRef]
- Jilkine, A.; Angenent, S.B.; Wu, L.F.; Altschuler, S.J. A density-dependent switch drives stochastic clustering and polarization of signaling molecules. PLoS Comput. Biol. 2011, 7, e1002271. [Google Scholar] [CrossRef]
- Klunder, B.; Freisinger, T.; Wedlich-Söldner, R.; Frey, E. GDI-mediated cell polarization in yeast provides precise spatial and temporal control of Cdc42 signaling. PLoS Comput. Biol. 2013, 9, e1003396. [Google Scholar] [CrossRef] [PubMed]
- Khalili, B.; Merlini, L.; Vincenzetti, V.; Martin, S.G.; Vavylonis, D. Exploration and stabilization of Ras1 mating zone: A mechanism with positive and negative feedbacks. PLoS Comput. Biol. 2018, 14, e1006317. [Google Scholar] [CrossRef]
- Mori, Y.; Jilkine, A.; Edelstein-Keshet, L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys. J. 2008, 94, 3684–3697. [Google Scholar] [CrossRef]
- Bernitt, E.; Döbereiner, H.-G.; Gov, N.S.; Yochelis, A. Fronts and waves of actin polymerization in a bistability-based mechanism of circular dorsal ruffles. Nat. Commun. 2017, 8, 15863. [Google Scholar] [CrossRef] [PubMed]
- Beta, C.; Edelstein-Keshet, L.; Gov, N.; Yochelis, A. From actin waves to mechanism and back: How theory aids biological understanding. Elife 2023, 12, e87181. [Google Scholar] [CrossRef] [PubMed]
- Michaud, A.; Leda, M.; Swider, Z.T.; Kim, S.; He, J.; Landino, J.; Valley, J.R.; Huisken, J.; Goryachev, A.B.; von Dassow, G.; et al. A versatile cortical pattern-forming circuit based on Rho, F-actin, Ect2, and RGA-3/4. J. Cell Biol. 2022, 221, e202203017. [Google Scholar] [CrossRef]
- Hladyshau, S.; Kho, M.; Nie, S.; Tsygankov, D. Spatiotemporal development of coexisting wave domains of Rho activity in the cell cortex. Sci. Rep. 2021, 11, 19512. [Google Scholar] [CrossRef]
- Herron, J.C.; Hu, S.; Liu, B.; Watanabe, T.; Hahn, K.M.; Elston, T.C. Spatial models of pattern formation during phagocytosis. PLoS Comput. Biol. 2022, 18, e1010092. [Google Scholar] [CrossRef]
- Howell, A.S.; Jin, M.; Wu, C.-F.; Zyla, T.R.; Elston, T.C.; Lew, D.J. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell 2012, 149, 322–333. [Google Scholar] [CrossRef]
- Ramirez, S.A.; Pablo, M.; Burk, S.; Lew, D.J.; Elston, T.C. A novel stochastic simulation approach enables exploration of mechanisms for regulating polarity site movement. PLoS Comput. Biol. 2021, 17, e1008525. [Google Scholar] [CrossRef]
- Kuo, C.C.; Savage, N.S.; Chen, H.; Wu, C.-F.; Zyla, T.R.; Lew, D.J. Inhibitory GEF phosphorylation provides negative feedback in the yeast polarity circuit. Curr. Biol. 2014, 24, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Caviston, J.P.; Longtine, M.; Pringle, J.R.; Bi, E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 2003, 14, 4051–4066. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.E.; Lo, W.-C.; Miller, K.E.; Chou, C.-S.; Park, H.-O. Regulation of Cdc42 polarization by the Rsr1 GTPase and Rga1, a Cdc42 GTPase-activating protein, in budding yeast. J. Cell Sci. 2015, 128, 2106–2117. [Google Scholar] [CrossRef][Green Version]
- Okada, S.; Leda, M.; Hanna, J.; Savage, N.S.; Bi, E.; Goryachev, A.B. Daughter cell identity emerges from the interplay of Cdc42, septins, and exocytosis. Dev. Cell 2013, 26, 148–161. [Google Scholar] [CrossRef]
- Esch, R.K.; Wang, Y.; Errede, B. Pheromone-induced degradation of Ste12 contributes to signal attenuation and the specificity of developmental fate. Eukaryot. Cell 2006, 5, 2147–2160. [Google Scholar] [CrossRef]
- Rahi, S.J.; Larsch, J.; Pecani, K.; Katsov, A.Y.; Mansouri, N.; Tsaneva-Atanasova, K.; Sontag, E.D.; Cross, F.R. Oscillatory stimuli differentiate adapting circuit topologies. Nat. Methods 2017, 14, 1010–1016. [Google Scholar] [CrossRef]
- Sharma, R.; Roberts, E. Gradient sensing by a bistable regulatory motif enhances signal amplification but decreases accuracy in individual cells. Phys. Biol. 2016, 13, 036003. [Google Scholar] [CrossRef] [PubMed]
- Beta, C.; Amselem, G.; Bodenschatz, E. A bistable mechanism for directional sensing. New J. Phys. 2008, 10, 083015. [Google Scholar] [CrossRef]
- McClure, A.W.; Minakova, M.; Dyer, J.M.; Zyla, T.R.; Elston, T.C.; Lew, D.J. Role of Polarized G Protein Signaling in Tracking Pheromone Gradients. Dev. Cell 2015, 35, 471–482. [Google Scholar] [CrossRef]
- Watson, L.J.; Rossi, G.; Brennwald, P. Quantitative analysis of membrane trafficking in regulation of Cdc42 polarity. Traffic 2014, 15, 1330–1343. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hladyshau, S.; Guan, K.; Nivedita, N.; Errede, B.; Tsygankov, D.; Elston, T.C. Multiscale Modeling of Bistability in the Yeast Polarity Circuit. Cells 2024, 13, 1358. https://doi.org/10.3390/cells13161358
Hladyshau S, Guan K, Nivedita N, Errede B, Tsygankov D, Elston TC. Multiscale Modeling of Bistability in the Yeast Polarity Circuit. Cells. 2024; 13(16):1358. https://doi.org/10.3390/cells13161358
Chicago/Turabian StyleHladyshau, Siarhei, Kaiyun Guan, Nivedita Nivedita, Beverly Errede, Denis Tsygankov, and Timothy C. Elston. 2024. "Multiscale Modeling of Bistability in the Yeast Polarity Circuit" Cells 13, no. 16: 1358. https://doi.org/10.3390/cells13161358
APA StyleHladyshau, S., Guan, K., Nivedita, N., Errede, B., Tsygankov, D., & Elston, T. C. (2024). Multiscale Modeling of Bistability in the Yeast Polarity Circuit. Cells, 13(16), 1358. https://doi.org/10.3390/cells13161358






