Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. In Vitro Mutagenesis and Generation of TAS2R46-Expressing HeLa Cells
2.3. Calcium Imaging
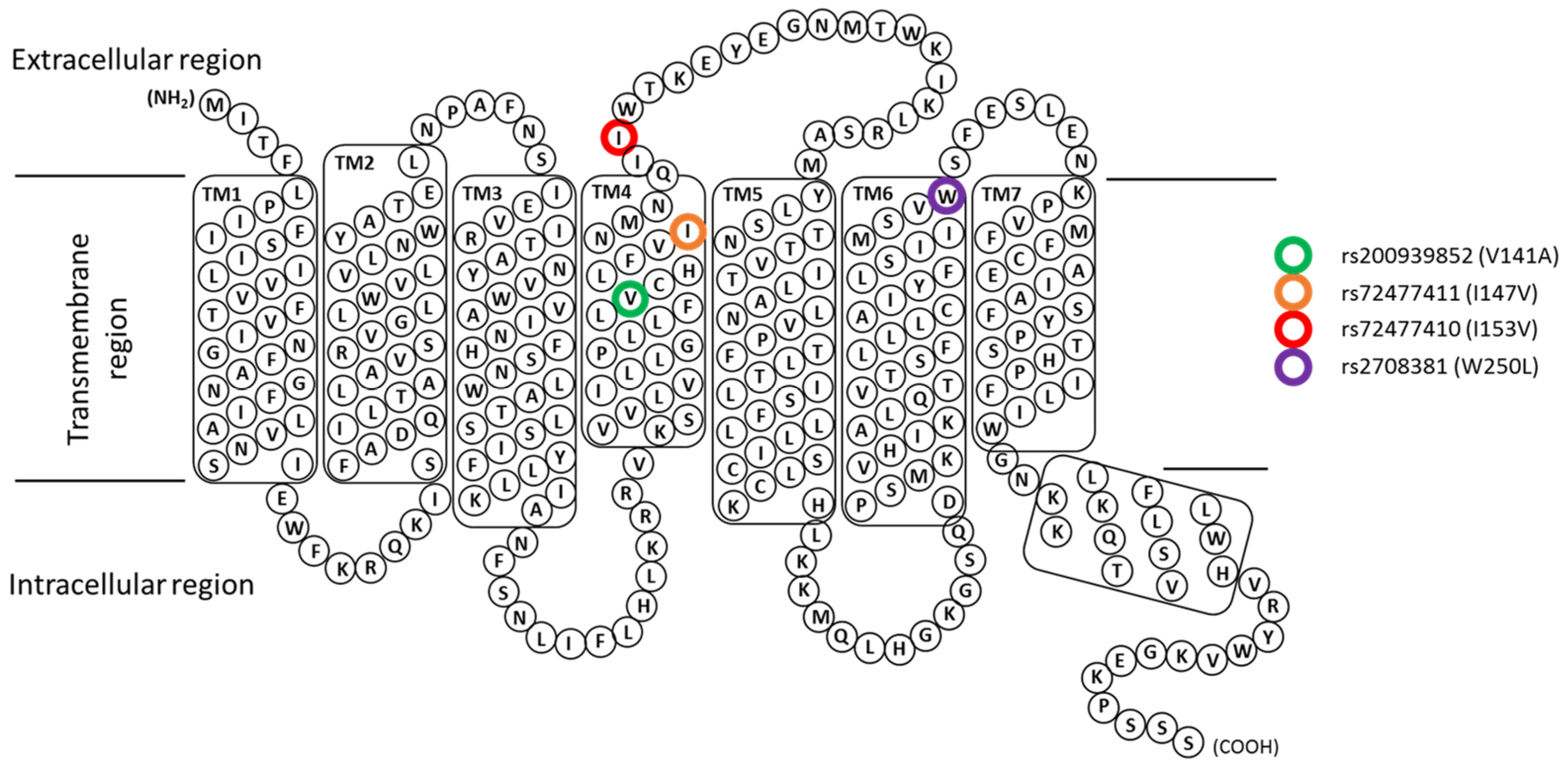
2.4. Model Building and Validations of Wild-Type and Mutant TAS2R46 Models
- 7XP4: TAS2R46 in apo state;
- 7XP5: TAS2R46 in ligand-free state;
- 7XP6: TAS2R46 in active state.
2.5. Molecular Docking Simulations
2.6. Statistical Analysis
3. Results
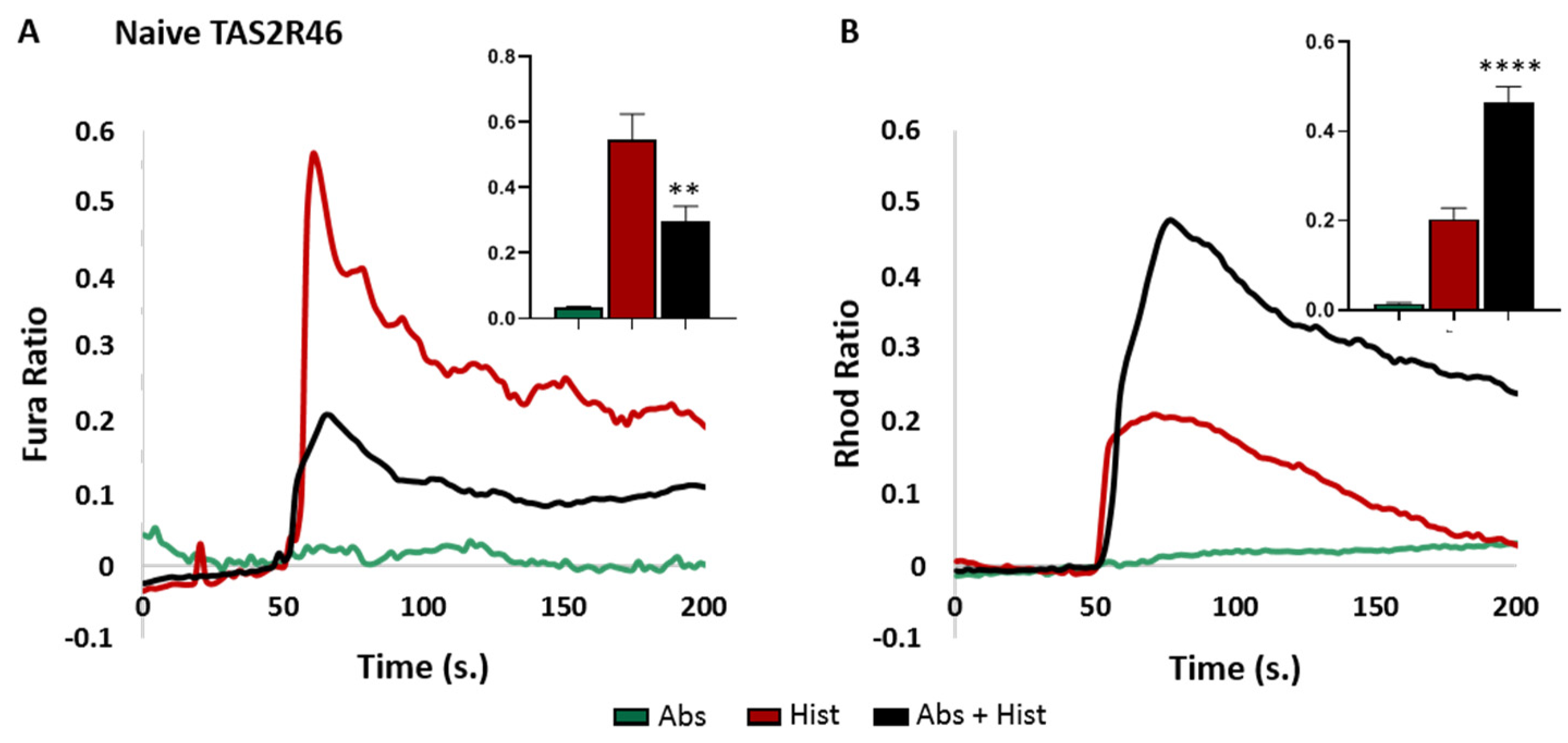
3.1. TAS2R46-HeLa Cells Efficiently Recapitulated the Calcium Movements Downstream of Receptor Activation
3.2. Effects of the Polymorphism on Downstream Signalling
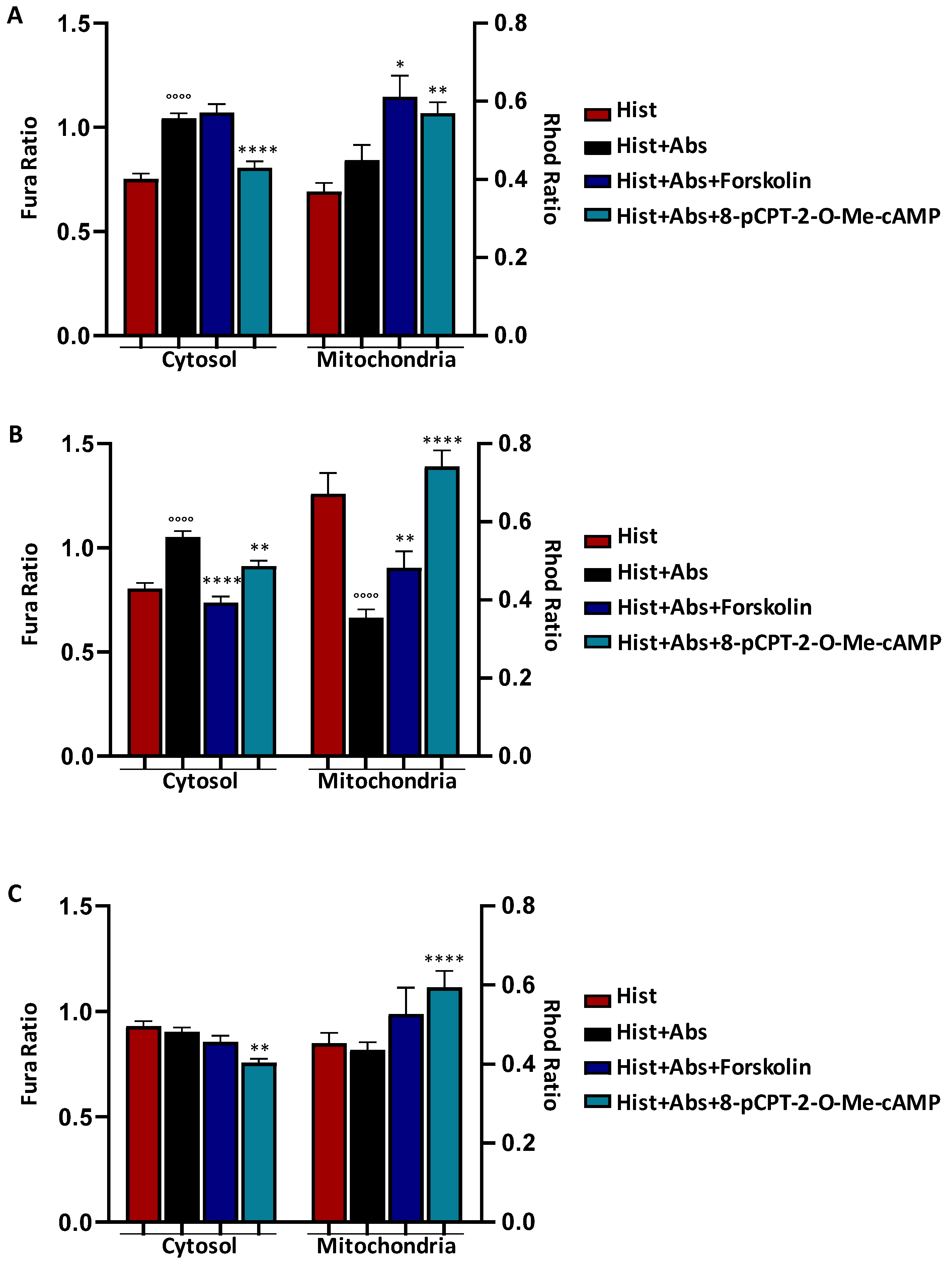
3.3. EPAC Activation Rescued the Absinthin Effect Downstream of Mutated Receptors
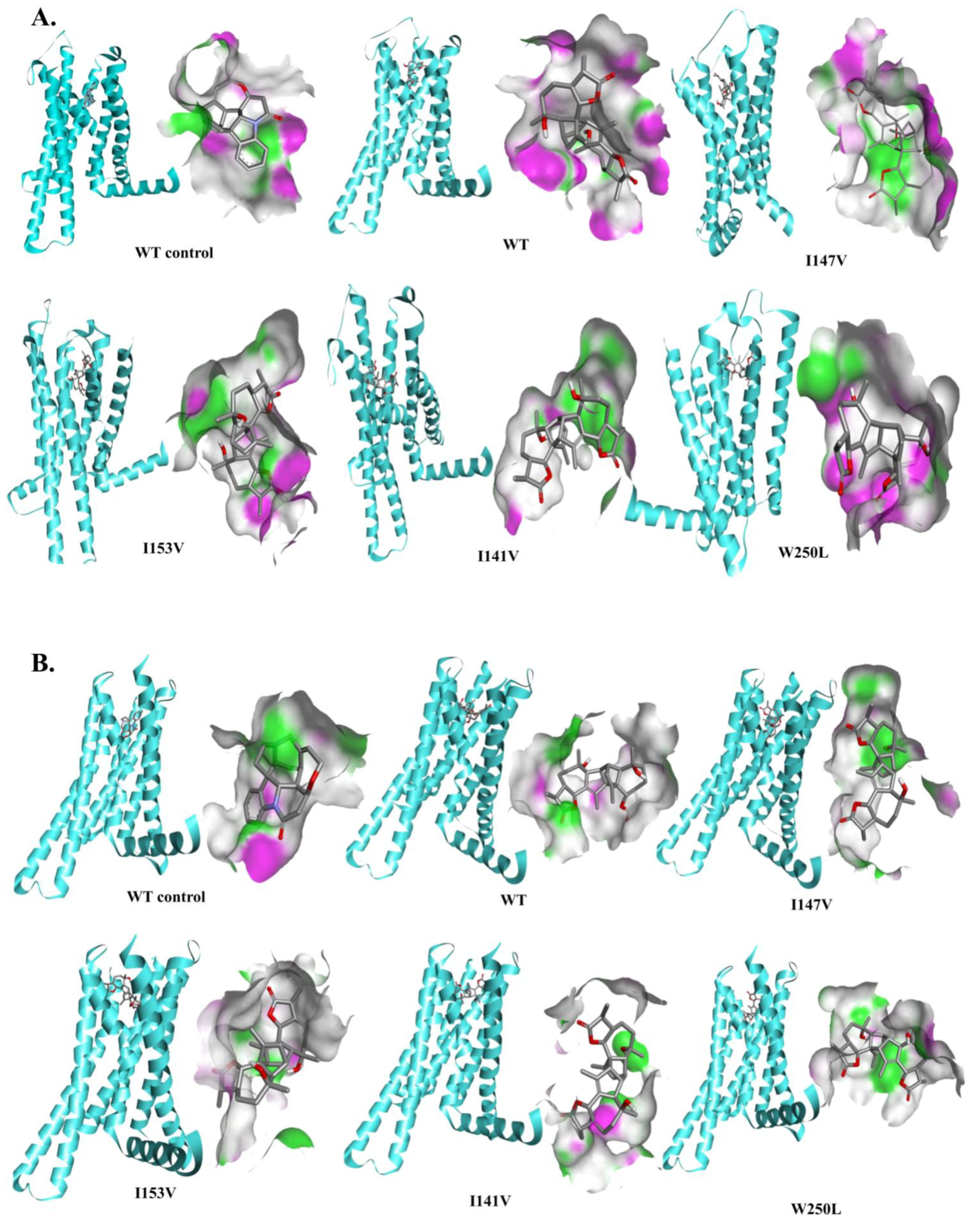
3.4. Structural Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lush, I.E.; Hornigold, N.; King, P.; Stoye, J.P. The Genetics of Tasting in Mice VII. Glycine Revisited, and the Chromosomal Location of Sac and Soa. Genet. Res. 1995, 66, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Adler, E.; Hoon, M.A.; Mueller, K.L.; Chandrashekar, J.; Ryba, N.J.P.; Zuker, C.S. A Novel Family of Mammalian Taste Receptors. Cell 2000, 100, 693–702. [Google Scholar] [CrossRef]
- Bachmanov, A.A.; Beauchamp, G.K. Taste Receptor Genes. Annu. Rev. Nutr. 2007, 27, 389–414. [Google Scholar] [CrossRef]
- Wooding, S.P.; Ramirez, V.A.; Behrens, M. Bitter Taste Receptors. Evol. Med. Public Health 2021, 9, 431–447. [Google Scholar] [CrossRef]
- Talmon, M.; Pollastro, F.; Fresu, L.G. The Complex Journey of the Calcium Regulation Downstream of TAS2R Activation. Cells 2022, 11, 3638. [Google Scholar] [CrossRef]
- Nayak, A.P.; Shah, S.D.; Michael, J.V.; Deshpande, D.A. Bitter Taste Receptors for Asthma Therapeutics. Front. Physiol. 2019, 10, 884. [Google Scholar] [CrossRef]
- An, S.S.; Liggett, S.B. Taste and Smell GPCRs in the Lung: Evidence for a Previously Unrecognized Widespread Chemosensory System. Cell. Signal. 2018, 41, 82–88. [Google Scholar] [CrossRef]
- Dalesio, N.M.; Barreto Ortiz, S.F.; Pluznick, J.L.; Berkowitz, D.E. Olfactory, Taste, and Photo Sensory Receptors in Non-Sensory Organs: It Just Makes Sense. Front. Physiol. 2018, 9, 1673. [Google Scholar] [CrossRef]
- Liggett, S.B. Bitter Taste Receptors on Airway Smooth Muscle as Targets for Novel Bronchodilators. Expert Opin. Ther. Targets 2013, 17, 721–731. [Google Scholar] [CrossRef]
- Brockhoff, A.; Behrens, M.; Massarotti, A.; Appendino, G.; Meyerhof, W. Broad Tuning of the Human Bitter Taste Receptor hTAS2R46 to Various Sesquiterpene Lactones, Clerodane and Labdane Diterpenoids, Strychnine, and Denatonium. J. Agric. Food Chem. 2007, 55, 6236–6243. [Google Scholar] [CrossRef]
- Talmon, M.; Bosso, L.; Quaregna, M.; Lopatriello, A.; Rossi, S.; Gavioli, D.; Marotta, P.; Caprioglio, D.; Boldorini, R.; Miggiano, R.; et al. Correction to Anti-Inflammatory Activity of Absinthin and Derivatives in Human Broncho-Epithelial Cells. J. Nat. Prod. 2020, 83, 2815–2816. [Google Scholar] [CrossRef]
- Talmon, M.; Rossi, S.; Lim, D.; Pollastro, F.; Palattella, G.; Ruffinatti, F.A.; Marotta, P.; Boldorini, R.; Genazzani, A.A.; Fresu, L.G. Absinthin, an Agonist of the Bitter Taste Receptor hTAS2R46, Uncovers an ER-to-Mitochondria Ca2+–Shuttling Event. J. Biol. Chem. 2019, 294, 12472–12482. [Google Scholar] [CrossRef] [PubMed]
- Kulbacka, J.; Choromańska, A.; Rossowska, J.; Weżgowiec, J.; Saczko, J.; Rols, M.-P. Cell Membrane Transport Mechanisms: Ion Channels and Electrical Properties of Cell Membranes. In Transport Across Natural and Modified Biological Membranes and its Implications in Physiology and Therapy; Kulbacka, J., Satkauskas, S., Eds.; Advances in Anatomy, Embryology and Cell Biology; Springer International Publishing: Cham, Switzerland, 2017; Volume 227, pp. 39–58. ISBN 978-3-319-56894-2. [Google Scholar]
- Di Pizio, A.; Levit, A.; Slutzki, M.; Behrens, M.; Karaman, R.; Niv, M.Y. Comparing Class A GPCRs to Bitter Taste Receptors. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 132, pp. 401–427. ISBN 978-0-12-803595-5. [Google Scholar]
- Pallante, L.; Cannariato, M.; Androutsos, L.; Zizzi, E.A.; Bompotas, A.; Hada, X.; Grasso, G.; Kalogeras, A.; Mavroudi, S.; Di Benedetto, G.; et al. VirtuousPocketome: A Computational Tool for Screening Protein–Ligand Complexes to Identify Similar Binding Sites. Sci. Rep. 2024, 14, 6296. [Google Scholar] [CrossRef] [PubMed]
- Brockhoff, A.; Behrens, M.; Niv, M.Y.; Meyerhof, W. Structural Requirements of Bitter Taste Receptor Activation. Proc. Natl. Acad. Sci. USA 2010, 107, 11110–11115. [Google Scholar] [CrossRef] [PubMed]
- Sandal, M.; Behrens, M.; Brockhoff, A.; Musiani, F.; Giorgetti, A.; Carloni, P.; Meyerhof, W. Evidence for a Transient Additional Ligand Binding Site in the TAS2R46 Bitter Taste Receptor. J. Chem. Theory Comput. 2015, 11, 4439–4449. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wu, L.; Liu, S.; Liu, X.; Cao, X.; Zhou, C.; Zhang, J.; Fu, Y.; Guo, Y.; Wu, Y.; et al. Structural Basis for Strychnine Activation of Human Bitter Taste Receptor TAS2R46. Science 2022, 377, 1298–1304. [Google Scholar] [CrossRef]
- Ledda, M.; Kutalik, Z.; Souza Destito, M.C.; Souza, M.M.; Cirillo, C.A.; Zamboni, A.; Martin, N.; Morya, E.; Sameshima, K.; Beckmann, J.S.; et al. GWAS of Human Bitter Taste Perception Identifies New Loci and Reveals Additional Complexity of Bitter Taste Genetics. Hum. Mol. Genet. 2014, 23, 259–267. [Google Scholar] [CrossRef]
- Roudnitzky, N.; Behrens, M.; Engel, A.; Kohl, S.; Thalmann, S.; Hübner, S.; Lossow, K.; Wooding, S.P.; Meyerhof, W. Receptor Polymorphism and Genomic Structure Interact to Shape Bitter Taste Perception. PLoS Genet. 2015, 11, e1005530. [Google Scholar] [CrossRef] [PubMed]
- Chamoun, E.; Carroll, N.; Duizer, L.; Qi, W.; Feng, Z.; Darlington, G.; Duncan, A.; Haines, J.; Ma, D. The Guelph Family Health Study The Relationship between Single Nucleotide Polymorphisms in Taste Receptor Genes, Taste Function and Dietary Intake in Preschool-Aged Children and Adults in the Guelph Family Health Study. Nutrients 2018, 10, 990. [Google Scholar] [CrossRef]
- Risso, D.S.; Kozlitina, J.; Sainz, E.; Gutierrez, J.; Wooding, S.; Getachew, B.; Luiselli, D.; Berg, C.J.; Drayna, D. Genetic Variation in the TAS2R38 Bitter Taste Receptor and Smoking Behaviors. PLoS ONE 2016, 11, e0164157. [Google Scholar] [CrossRef]
- Risso, D.; Carmagnola, D.; Morini, G.; Pellegrini, G.; Canciani, E.; Antinucci, M.; Henin, D.; Dellavia, C. Distribution of TAS2R38 Bitter Taste Receptor Phenotype and Haplotypes among COVID-19 Patients. Sci. Rep. 2022, 12, 7381. [Google Scholar] [CrossRef] [PubMed]
- Purnell, P.R.; Addicks, B.L.; Zalzal, H.G.; Shapiro, S.; Wen, S.; Ramadan, H.H.; Setola, V.; Siderovski, D.P. Single Nucleotide Polymorphisms in Chemosensory Pathway Genes GNB3, TAS2R19, and TAS2R38 Are Associated with Chronic Rhinosinusitis. Int. Arch. Allergy Immunol. 2019, 180, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.J.; Xiong, G.; Kofonow, J.M.; Chen, B.; Lysenko, A.; Jiang, P.; Abraham, V.; Doghramji, L.; Adappa, N.D.; Palmer, J.N.; et al. T2R38 Taste Receptor Polymorphisms Underlie Susceptibility to Upper Respiratory Infection. J. Clin. Investig. 2012, 122, 4145–4159. [Google Scholar] [CrossRef]
- Ryu, C.S.; Sakong, J.H.; Ahn, E.H.; Kim, J.O.; Ko, D.; Kim, J.H.; Lee, W.S.; Kim, N.K. Association Study of the Three Functional Polymorphisms (TAS2R46G>A, OR4C16G>A, and OR4X1A>T) with Recurrent Pregnancy Loss. Genes Genom. 2019, 41, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Dotson, C.D.; Zhang, L.; Xu, H.; Shin, Y.-K.; Vigues, S.; Ott, S.H.; Elson, A.E.T.; Choi, H.J.; Shaw, H.; Egan, J.M.; et al. Bitter Taste Receptors Influence Glucose Homeostasis. PLoS ONE 2008, 3, e3974. [Google Scholar] [CrossRef] [PubMed]
- Eswar, N. Tools for Comparative Protein Structure Modeling and Analysis. Nucleic Acids Res. 2003, 31, 3375–3380. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. ModLoop: Automated Modeling of Loops in Protein Structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Fazal, L.; Laudette, M.; Paula-Gomes, S.; Pons, S.; Conte, C.; Tortosa, F.; Sicard, P.; Sainte-Marie, Y.; Bisserier, M.; Lairez, O.; et al. Multifunctional Mitochondrial Epac1 Controls Myocardial Cell Death. Circ. Res. 2017, 120, 645–657. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, D.; Varin, A.; Nicolas, V.; Courilleau, D.; Mateo, P.; Caubere, C.; Rouet, P.; Gomez, A.-M.; Vandecasteele, G.; et al. A Cardiac Mitochondrial cAMP Signaling Pathway Regulates Calcium Accumulation, Permeability Transition and Cell Death. Cell Death Dis. 2016, 7, e2198. [Google Scholar] [CrossRef]
- Talmon, M.; Massara, E.; Quaregna, M.; De Battisti, M.; Boccafoschi, F.; Lecchi, G.; Puppo, F.; Bettega Cajandab, M.A.; Salamone, S.; Bovio, E.; et al. Bitter Taste Receptor (TAS2R) 46 in Human Skeletal Muscle: Expression and Activity. Front. Pharmacol. 2023, 14, 1205651. [Google Scholar] [CrossRef]
- Kooistra, A.J.; Mordalski, S.; Pándy-Szekeres, G.; Esguerra, M.; Mamyrbekov, A.; Munk, C.; Keserű, G.M.; Gloriam, D.E. GPCRdb in 2021: Integrating GPCR Sequence, Structure and Function. Nucleic Acids Res. 2021, 49, D335–D343. [Google Scholar] [CrossRef] [PubMed]
- Horn, F. GPCRDB Information System for G Protein-Coupled Receptors. Nucleic Acids Res. 2003, 31, 294–297. [Google Scholar] [CrossRef]
- Fredriksson, R.; Lagerström, M.C.; Lundin, L.-G.; Schiöth, H.B. The G-Protein-Coupled Receptors in the Human Genome Form Five Main Families. Phylogenetic Analysis, Paralogon Groups, and Fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef] [PubMed]
- Cvicek, V.; Goddard, W.A.; Abrol, R. Structure-Based Sequence Alignment of the Transmembrane Domains of All Human GPCRs: Phylogenetic, Structural and Functional Implications. PLoS Comput. Biol. 2016, 12, e1004805. [Google Scholar] [CrossRef] [PubMed]
- Di Pizio, A.; Niv, M.Y. Promiscuity and Selectivity of Bitter Molecules and Their Receptors. Bioorg. Med. Chem. 2015, 23, 4082–4091. [Google Scholar] [CrossRef]
- Nordstrom, K.J.V.; Sallman Almen, M.; Edstam, M.M.; Fredriksson, R.; Schioth, H.B. Independent HHsearch, Needleman-Wunsch-Based, and Motif Analyses Reveal the Overall Hierarchy for Most of the G Protein-Coupled Receptor Families. Mol. Biol. Evol. 2011, 28, 2471–2480. [Google Scholar] [CrossRef]
- Yang, L.; Liu, D.; Wüthrich, K. GPCR Structural Characterization by NMR Spectroscopy in Solution. Acta Biochim. Biophys. Sin. 2022, 54, 1207–1212. [Google Scholar] [CrossRef]
- Brockhoff, A.; Behrens, M.; Roudnitzky, N.; Appendino, G.; Avonto, C.; Meyerhof, W. Receptor Agonism and Antagonism of Dietary Bitter Compounds. J. Neurosci. 2011, 31, 14775–14782. [Google Scholar] [CrossRef]
- Balasubramanian, S. Sequence Variation in G-Protein-Coupled Receptors: Analysis of Single Nucleotide Polymorphisms. Nucleic Acids Res. 2005, 33, 1710–1721. [Google Scholar] [CrossRef]





| Primer Name | Sequence | Tm (°C) |
|---|---|---|
| rs2708381 fw | 5′-TTTTCCAGACTCTCAAAACTCWAAACTGACATGATTATGGACAG-3′ | 78.8 |
| rs2708381 rev | 5′-CTGTCCATAATCATGTCAGTTTWGAGTTTTGAGAGTCTGGAAAA-3′ | 78.8 |
| rs72477410 fw | 5′-TTCATATTCTTTTGTCCATACAATCTGATTCATGTTTATCACAAAAAGATGACAAACC-3′ | 79.5 |
| rs72477410 rev | 5′-GGTTTGTCATCTTTTTGTGATAAACATGAATCAGATTGTATGGACAAAAGAATATGAA-3′ | 79.5 |
| rs72477411 fw | 5′-TTGTCCATATAATCTGATTCATGTTTACCACAAAAAGATGACAAACCAAAAATAG-3′ | 78.6 |
| rs72477411 rev | 5′-CTATTTTTGGTTTGTCATCTTTTTGTGGTAAACATGAATCAGATTATATGGACAA-3′ | 78.6 |
| rs200936852 fw | 5′-TGTTTATCACAAAAAGATGACAAGCCAAAAATAGCAAAGGCCCCAA-3′ | 78.9 |
| rs200936852 rev | 5′-TTGGGGCCTTTGCTATTTTTGGCTTGTCATCTTTTTGTGATAAACA-3′ | 78.9 |
| Orthosteric Site | ||
|---|---|---|
| Name of the Protein–Ligand Complex | Binding Free Energy (kcal/mol) | Inhibition Constant (µM) |
| Model_WT_strychnine | −8.68 | 0.43 |
| Model_WT_absinthin | −10.91 | 0.01 |
| Model_I147V_absinthin | −11.77 | 0.002 |
| Model_I153V_absinthin | −12.59 | 0.0005 |
| Model_V141A_absinthin | −12.54 | 0.0006 |
| Model_W250L_absinthin | −11.01 | 0.008 |
| 7XP6_WT_strychnine | −9.63 | 0.087 |
| 7XP6_WT_absinthin | −12.10 | 0.001 |
| 7XP6_I147V_absinthin | −12.18 | 0.001 |
| 7XP6_I153V_absinthin | −11.40 | 0.004 |
| 7XP6_V141A_absinthin | −12.01 | 0.001 |
| 7XP6_W250L_absinthin | −10.77 | 0.012 |
| Vestibular Site | ||
|---|---|---|
| Name of the Protein–Ligand Complex | Binding Free Energy (kcal/mol) | Inhibition Constant (µM) |
| 7XP6_WT_strychnine | −5.21 | 152.25 |
| 7XP6_WT_absinthin | −8.16 | 1.04 |
| 7XP6_I147V_absinthin | −7.58 | 2.80 |
| 7XP6_I153V_absinthin | −9.82 | 0.63 |
| 7XP6_V141A_absinthin | −8.10 | 1.15 |
| 7XP6_W250L_absinthin | −8.35 | 0.76 |
| Model_WT_strychnine | −8.01 | 1.34 |
| Model_WT_absinthin | −9.60 | 0.09 |
| Model_I147V_absinthin | −7.26 | 4.77 |
| Model_I153V_absinthin | −9.59 | 0.092 |
| Model_V141A_absinthin | −8.12 | 1.12 |
| Model_W250L_absinthin | −7.83 | 1.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lecchi, G.; Mocchetti, C.; Tunesi, D.; Berto, A.; Balasubramanian, H.B.; Biswas, S.; Bagchi, A.; Pollastro, F.; Fresu, L.G.; Talmon, M. Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells. Cells 2024, 13, 1204. https://doi.org/10.3390/cells13141204
Lecchi G, Mocchetti C, Tunesi D, Berto A, Balasubramanian HB, Biswas S, Bagchi A, Pollastro F, Fresu LG, Talmon M. Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells. Cells. 2024; 13(14):1204. https://doi.org/10.3390/cells13141204
Chicago/Turabian StyleLecchi, Giulia, Chiara Mocchetti, Davide Tunesi, Arianna Berto, Hari Baskar Balasubramanian, Sima Biswas, Angshuman Bagchi, Federica Pollastro, Luigia Grazia Fresu, and Maria Talmon. 2024. "Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells" Cells 13, no. 14: 1204. https://doi.org/10.3390/cells13141204
APA StyleLecchi, G., Mocchetti, C., Tunesi, D., Berto, A., Balasubramanian, H. B., Biswas, S., Bagchi, A., Pollastro, F., Fresu, L. G., & Talmon, M. (2024). Single-Nucleotide Polymorphisms of TAS2R46 Affect the Receptor Downstream Calcium Regulation in Histamine-Challenged Cells. Cells, 13(14), 1204. https://doi.org/10.3390/cells13141204






