Prime Editing and DNA Repair System: Balancing Efficiency with Safety
Abstract
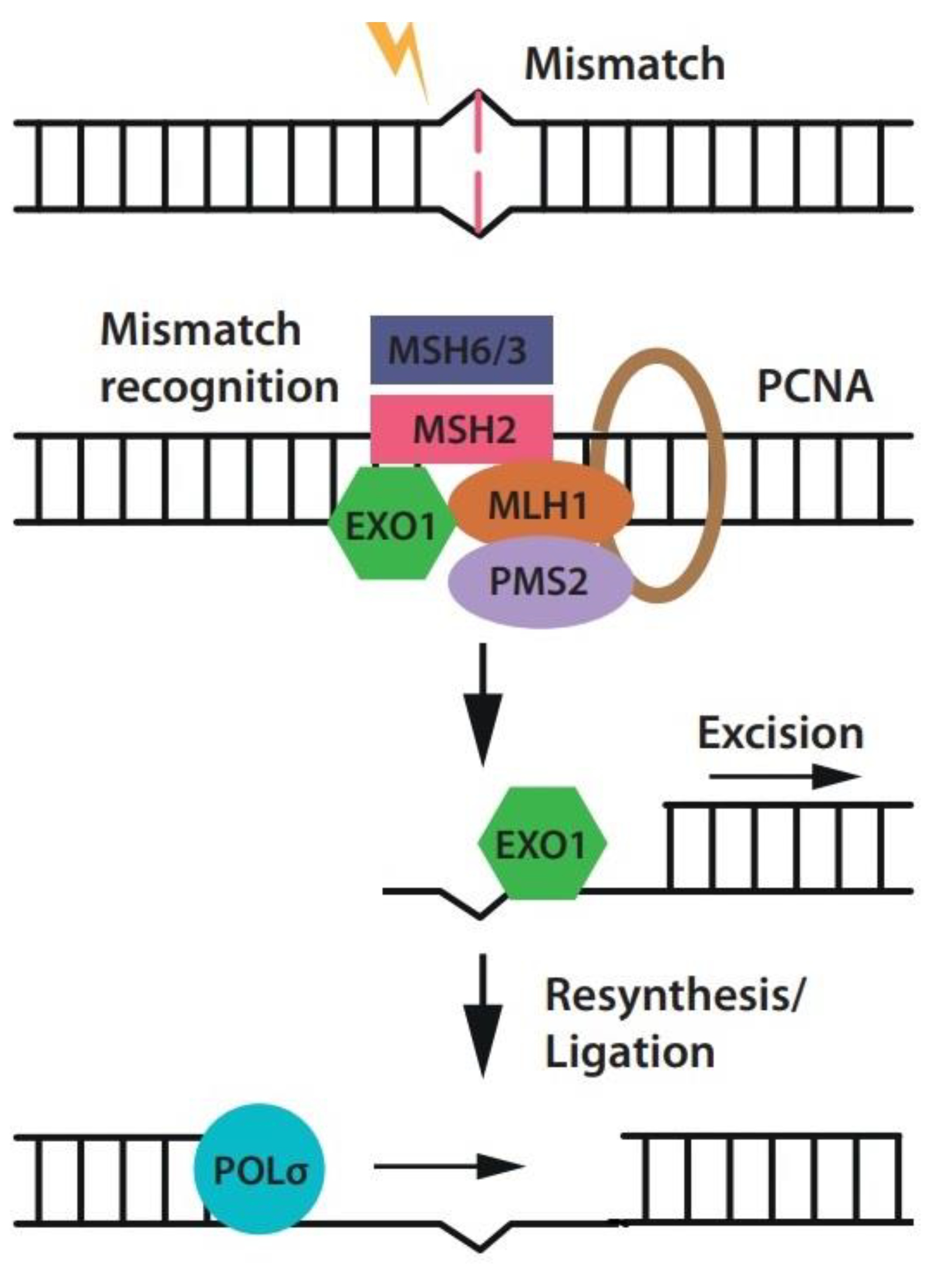
1. Introduction
2. Potential Molecular and Cellular Risks Associated with MLH1 Disruption
2.1. Mitochondria
2.2. Autophagy
2.3. Variability in Cellular Responses to MMR Defects across Different Tissues
2.4. Folate
2.5. microRNA
2.6. Wnt Signaling Pathway
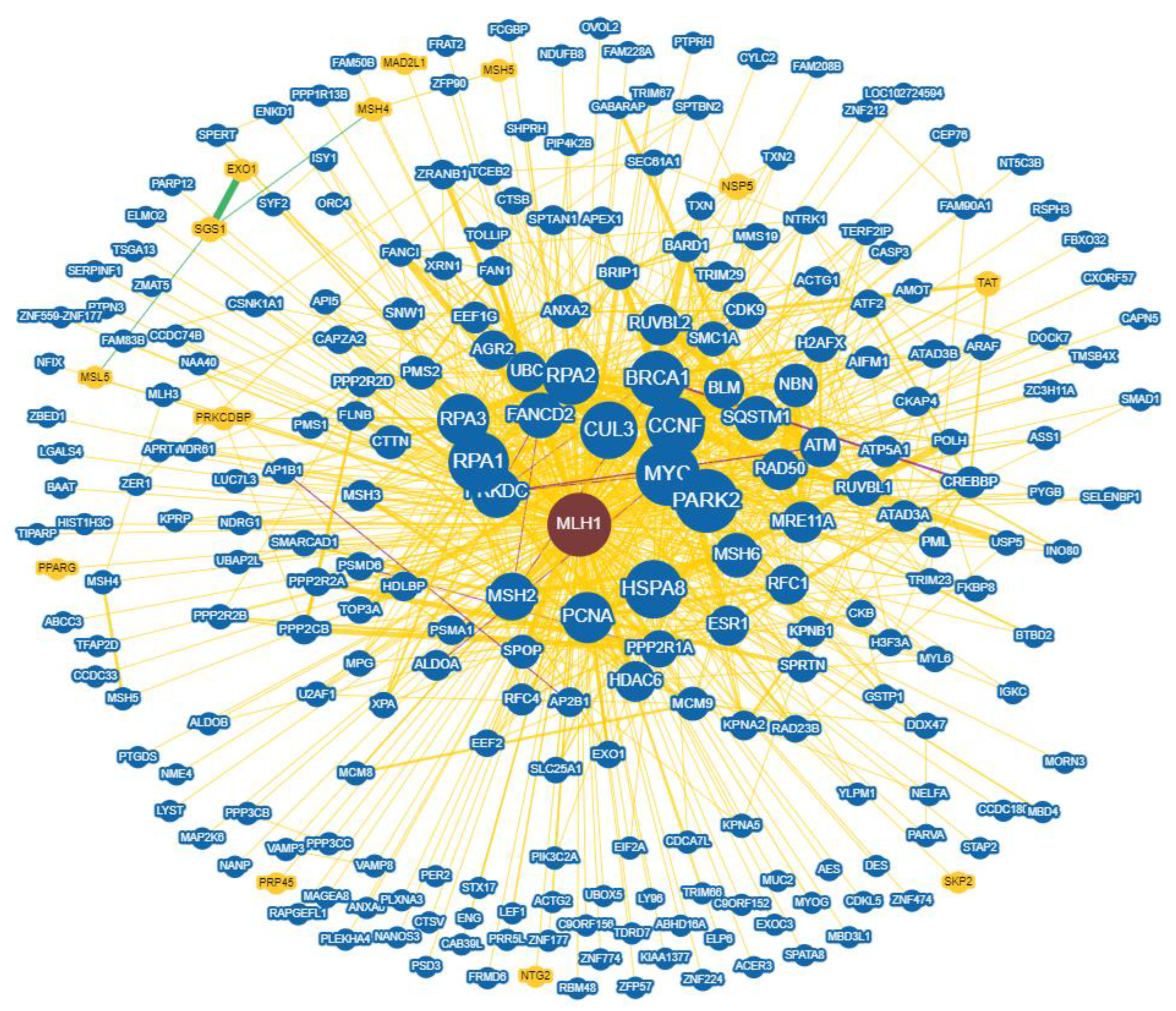
2.7. Interaction Networks
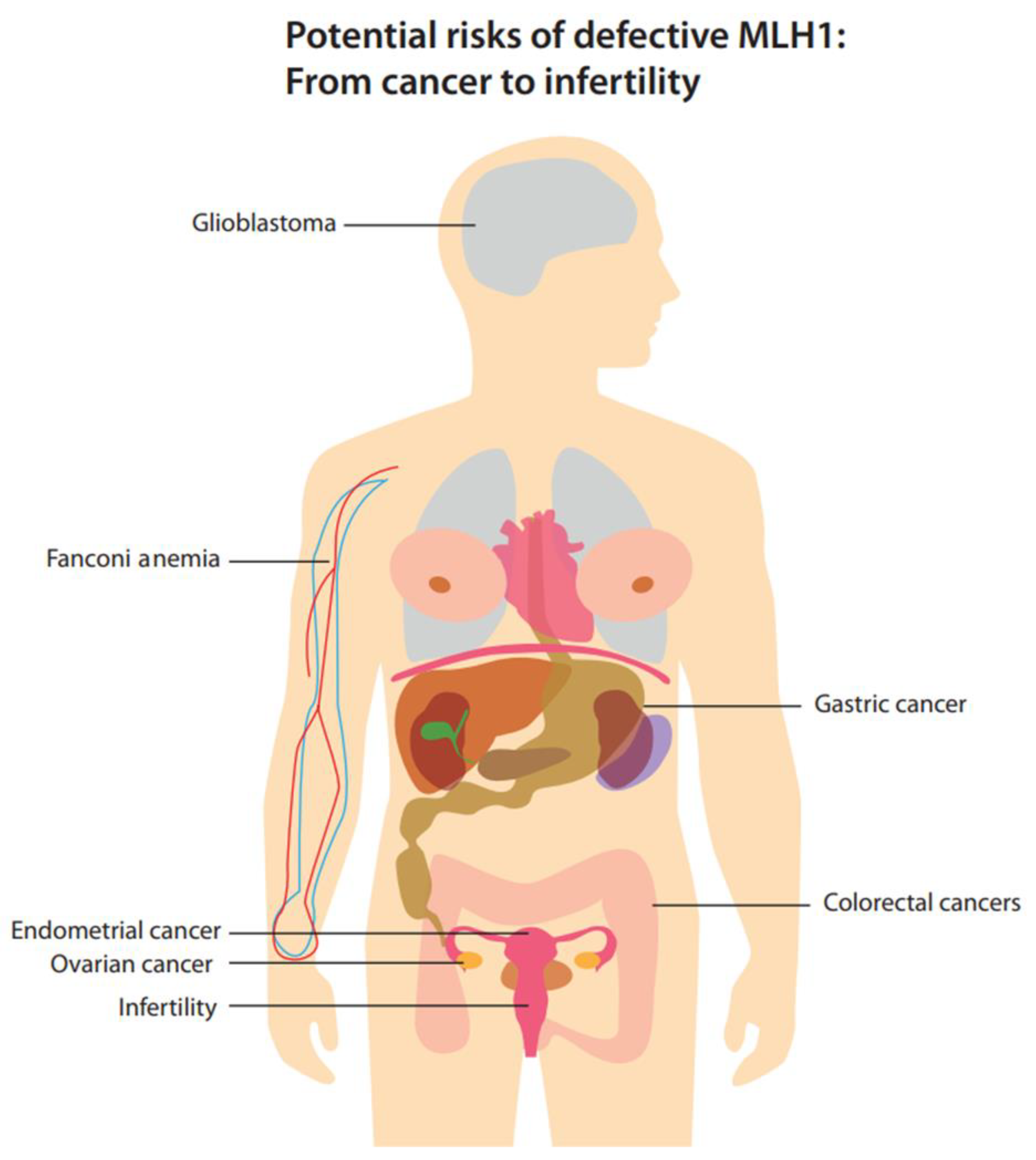
3. The Clinical Impact of Defective MLH1
3.1. Colorectal Cancer
3.2. Gastric Cancer
3.3. Glioblastoma
3.4. Endometrial Cancer
3.5. Ovarian Cancer
3.6. Fanconi Anemia
3.7. Fertility
4. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Chylinski, K.; Makarova, K.S.; Charpentier, E.; Koonin, E. V Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014, 42, 6091–6105. [Google Scholar] [CrossRef] [PubMed]
- Chylinski, K.; Le Rhun, A.; Charpentier, E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013, 10, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; He, X.; Zhao, T.; Gu, L.; Liu, Y.; Liu, X.; Liu, H.; Yang, F.; Tu, M.; Tang, L. Engineering the direct repeat sequence of crRNA for optimization of FnCpf1-mediated genome editing in human cells. Mol. Ther. 2018, 26, 2650–2657. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xu, D. Repair pathway choice for double-strand breaks. Essays Biochem. 2020, 64, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Karl, L.A.; Peritore, M.; Galanti, L.; Pfander, B. DNA double strand break repair and its control by nucleosome remodeling. Front. Genet. 2022, 12, 2833. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Ferreira da Silva, J.; Oliveira, G.P.; Arasa-Verge, E.A.; Kagiou, C.; Moretton, A.; Timelthaler, G.; Jiricny, J.; Loizou, J.I. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair. Nat. Commun. 2022, 13, 760. [Google Scholar] [CrossRef]
- Chen, P.J.; Hussmann, J.A.; Yan, J.; Knipping, F.; Ravisankar, P.; Chen, P.-F.; Chen, C.; Nelson, J.W.; Newby, G.A.; Sahin, M. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell 2021, 184, 5635–5652. [Google Scholar] [CrossRef]
- Jiricny, J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006, 7, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Bozek, G.; Lo, J.C.; Storb, U. Different mismatch repair deficiencies all have the same effects on somatic hypermutation: Intact primary mechanism accompanied by secondary modifications. J. Exp. Med. 1999, 190, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Mensenkamp, A.R.; Vogelaar, I.P.; van Zelst-Stams, W.A.G.; Goossens, M.; Ouchene, H.; Hendriks-Cornelissen, S.J.B.; Kwint, M.P.; Hoogerbrugge, N.; Nagtegaal, I.D.; Ligtenberg, M.J.L. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology 2014, 146, 643–646. [Google Scholar] [CrossRef] [PubMed]
- de’Angelis Gian, L.; Lorena, B.; Cinzia, A.; Gioacchino, L.; Federica, G.; Francesca, N. Microsatellite instability in colorectal cancer. Acta Bio Medica Atenei Parm. 2018, 89, 97. [Google Scholar]
- Hemminki, A.; Peltomäki, P.; Mecklin, J.-P.; Järvinen, H.; Salovaara, R.; Nyström-Lahti, M.; de la Chapelle, A.; Aaltonen, L.A. Loss of the wild type MLH1 gene is a feature of hereditary nonpolyposis colorectal cancer. Nat. Genet. 1994, 8, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.-X.; Rabinowitz, J.D.; White, E. Mitochondria and cancer. Mol. Cell 2016, 61, 667–676. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, Y. Mitochondria as a target in cancer treatment. MedComm 2020, 1, 129–139. [Google Scholar] [CrossRef]
- Rashid, S.; Freitas, M.O.; Cucchi, D.; Bridge, G.; Yao, Z.; Gay, L.; Williams, M.; Wang, J.; Suraweera, N.; Silver, A. MLH1 deficiency leads to deregulated mitochondrial metabolism. Cell Death Dis. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Martin, S.A.; Hewish, M.; Sims, D.; Lord, C.J.; Ashworth, A. Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair–deficient cancers. Cancer Res. 2011, 71, 1836–1848. [Google Scholar] [CrossRef]
- Mootha, V.K.; Bunkenborg, J.; Olsen, J.V.; Hjerrild, M.; Wisniewski, J.R.; Stahl, E.; Bolouri, M.S.; Ray, H.N.; Sihag, S.; Kamal, M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 2003, 115, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S. Targeting the mitochondria for the treatment of MLH1-deficient disease 2017. J. Clin. Oncol. 2016, 34 (Suppl. 15), e23182. [Google Scholar] [CrossRef]
- Mishra, M.; Kowluru, R.A. Retinal mitochondrial DNA mismatch repair in the development of diabetic retinopathy, and its continued progression after termination of hyperglycemia. Invest. Ophthalmol. Vis. Sci. 2014, 55, 6960–6967. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Zhang, S.; Mizushima, N. Autophagy genes in biology and disease. Nat. Rev. Genet. 2023, 24, 382–400. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xiao, Z.; Lin, Z.; Zhou, J.; Chen, W.; Jie, W.; Cao, X.; Yin, Z.; Cheng, J. Autophagy influences the low-dose hyper-radiosensitivity of human lung adenocarcinoma cells by regulating MLH1. Int. J. Radiat. Biol. 2017, 93, 600–606. [Google Scholar] [CrossRef] [PubMed]
- McDaid, J.R.; Loughery, J.; Dunne, P.; Boyer, J.C.; Downes, C.S.; Farber, R.A.; Walsh, C.P. MLH1 mediates PARP-dependent cell death in response to the methylating agent N-methyl-N-nitrosourea. Br. J. Cancer 2009, 101, 441–451. [Google Scholar] [CrossRef]
- Zeng, X.; Kinsella, T.J. A novel role for DNA mismatch repair and the autophagic processing of chemotherapy drugs in human tumor cells. Autophagy 2007, 3, 368–370. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhao, Y. Autophagy and DNA damage repair. Genome Instab. Dis. 2020, 1, 172–183. [Google Scholar] [CrossRef]
- Gomes, L.R.; Menck, C.F.M.; Leandro, G.S. Autophagy roles in the modulation of DNA repair pathways. Int. J. Mol. Sci. 2017, 18, 2351. [Google Scholar] [CrossRef]
- Sonawane, A.R.; Platig, J.; Fagny, M.; Chen, C.-Y.; Paulson, J.N.; Lopes-Ramos, C.M.; DeMeo, D.L.; Quackenbush, J.; Glass, K.; Kuijjer, M.L. Understanding tissue-specific gene regulation. Cell Rep. 2017, 21, 1077–1088. [Google Scholar] [CrossRef]
- Shrestha, K.S.; Aska, E.-M.; Tuominen, M.M.; Kauppi, L. Tissue-specific reduction in MLH1 expression induces microsatellite instability in intestine of Mlh1+/− mice. DNA Repair (Amst) 2021, 106, 103178. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tsutsumi, S.; Kawaguchi, T.; Nagasaki, K.; Tatsuno, K.; Yamamoto, S.; Sang, F.; Sonoda, K.; Sugawara, M.; Saiura, A. Whole-exome sequencing of human pancreatic cancers and characterization of genomic instability caused by MLH1 haploinsufficiency and complete deficiency. Genome Res. 2012, 22, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Spies, M.; Fishel, R. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 2015, 7, a022657. [Google Scholar] [CrossRef]
- Elliott, B.; Jasin, M. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol. 2001, 21, 2671–2682. [Google Scholar] [CrossRef] [PubMed]
- Froese, D.S.; Fowler, B.; Baumgartner, M.R. Vitamin B12, folate, and the methionine remethylation cycle—Biochemistry, pathways, and regulation. J. Inherit. Metab. Dis. 2019, 42, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Waki, T.; Tamura, G.; Tsuchiya, T.; Sato, K.; Nishizuka, S.; Motoyama, T. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am. J. Pathol. 2002, 161, 399–403. [Google Scholar] [CrossRef]
- Gay, L.J.; Arends, M.J.; Mitrou, P.N.; Bowman, R.; Ibrahim, A.E.; Happerfield, L.; Luben, R.; McTaggart, A.; Ball, R.Y.; Rodwell, S.A. MLH1 promoter methylation, diet, and lifestyle factors in mismatch repair deficient colorectal cancer patients from EPIC-Norfolk. Nutr. Cancer 2011, 63, 1000–1010. [Google Scholar] [CrossRef]
- Li, X.; Yao, X.; Wang, Y.; Hu, F.; Wang, F.; Jiang, L.; Liu, Y.; Wang, D.; Sun, G.; Zhao, Y. MLH1 promoter methylation frequency in colorectal cancer patients and related clinicopathological and molecular features. PLoS ONE 2013, 8, e59064. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Chen, Y.; Petersen, I. Expression and promoter DNA methylation of MLH1 in colorectal cancer and lung cancer. Pathol. Pract. 2017, 213, 333–338. [Google Scholar] [CrossRef]
- Li, G.-M.; Presnell, S.R.; Gu, L. Folate deficiency, mismatch repair-dependent apoptosis, and human disease. J. Nutr. Biochem. 2003, 14, 568–575. [Google Scholar] [CrossRef]
- Murphy, L.E.; Mills, J.L.; Molloy, A.M.; Qian, C.; Carter, T.C.; Strevens, H.; Wide-Swensson, D.; Giwercman, A.; Levine, R.J. Folate and vitamin B12 in idiopathic male infertility. Asian J. Androl. 2011, 13, 856. [Google Scholar] [CrossRef] [PubMed]
- Altmäe, S.; Stavreus-Evers, A.; Ruiz, J.R.; Laanpere, M.; Syvänen, T.; Yngve, A.; Salumets, A.; Nilsson, T.K. Variations in folate pathway genes are associated with unexplained female infertility. Fertil. Steril. 2010, 94, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, L.A.; Dumas, B.; Milrod, C.J.; Hudspeth, J.C. Folate deficiency in an urban safety net population. Am. J. Med. 2021, 134, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Lee, S.; Ortega, J.; Gu, L.; Li, G.-M. Modulation of microRNA processing by mismatch repair protein MutLα. Cell Res. 2012, 22, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Çakmak, H.A.; Demir, M. MicroRNA and cardiovascular diseases. Balkan Med. J. 2020, 37, 60. [Google Scholar] [CrossRef] [PubMed]
- Hussen, B.M.; Hidayat, H.J.; Salihi, A.; Sabir, D.K.; Taheri, M.; Ghafouri-Fard, S. MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 2021, 138, 111528. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Pan, X.; Gu, L. Evidence that a mutation in the MLH1 3′-untranslated region confers a mutator phenotype and mismatch repair deficiency in patients with relapsed leukemia. J. Biol. Chem. 2008, 283, 3211–3216. [Google Scholar] [CrossRef]
- Rao, T.P.; Kühl, M. An updated overview on Wnt signaling pathways: A prelude for more. Circ. Res. 2010, 106, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef]
- Lugli, A.; Zlobec, I.; Minoo, P.; Baker, K.; Tornillo, L.; Terracciano, L.; Jass, J.R. Prognostic significance of the wnt signalling pathway molecules APC, β-catenin and E-cadherin in colorectal cancer—A tissue microarray-based analysis. Histopathology 2007, 50, 453–464. [Google Scholar] [CrossRef]
- Wang, J.; He, T.; Gao, Q.; Chang, H.; Dai, X.; Yang, J.; Liu, S.; Zhang, S.; Shan, C.; Zhang, C. The dysfunctional Wnt pathway down-regulates MLH1/SET expression and promotes microsatellite instability and immunotherapy response in colorectal cancer. Genes Dis. 2024, 11, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Bebek, G. Identifying gene interaction networks. Stat. Hum. Genet. Methods Protoc. 2012, 850, 483–494. [Google Scholar]
- Costanzo, M.; VanderSluis, B.; Koch, E.N.; Baryshnikova, A.; Pons, C.; Tan, G.; Wang, W.; Usaj, M.; Hanchard, J.; Lee, S.D. A global genetic interaction network maps a wiring diagram of cellular function. Science (80-) 2016, 353, aaf1420. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Oughtred, R.; Boucher, L.; Rust, J.; Chang, C.; Kolas, N.K. Biological general repository for interaction datasets (BioGRID). FAIRsharing.
- Wilkins, A.; Corbett, R.; Eeles, R. Age distribution and a multi-stage theory of carcinogenesis: 70 years on. Br. J. Cancer 2023, 128, 404–406. [Google Scholar] [CrossRef] [PubMed]
- Pećina-Šlaus, N.; Kafka, A.; Salamon, I.; Bukovac, A. Mismatch repair pathway, genome stability and cancer. Front. Mol. Biosci. 2020, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Centelles, J.J. General aspects of colorectal cancer. Int. Sch. Res. Not. 2012, 2012, 139268. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. Mech. Dis. 2011, 6, 479–507. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.V.; Yamada, H.Y. Genomic instability and colon carcinogenesis: From the perspective of genes. Front. Oncol. 2013, 3, 130. [Google Scholar] [CrossRef] [PubMed]
- Morán, A.; Ortega, P.; de Juan, C.; Fernández-Marcelo, T.; Frías, C.; Sánchez-Pernaute, A.; Torres, A.J.; Díaz-Rubio, E.; Iniesta, P.; Benito, M. Differential colorectal carcinogenesis: Molecular basis and clinical relevance. World J. Gastrointest. Oncol. 2010, 2, 151. [Google Scholar] [CrossRef]
- Lawes, D.A.; Pearson, T.; Sengupta, S.; Boulos, P.B. The role of MLH1, MSH2 and MSH6 in the development of multiple colorectal cancers. Br. J. Cancer 2005, 93, 472–477. [Google Scholar] [CrossRef]
- Mestrallet, G.; Brown, M.; Bozkus, C.C.; Bhardwaj, N. Immune escape and resistance to immunotherapy in mismatch repair deficient tumors. Front. Immunol. 2023, 14, 1210164. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Shen, H.; Kapesa, L.; Zeng, S. Lauren classification and individualized chemotherapy in gastric cancer. Oncol. Lett. 2016, 11, 2959–2964. [Google Scholar] [CrossRef] [PubMed]
- Tahara, E. Genetic pathways of two types of gastric cancer. IARC Sci. Publ. 2004, 157, 327–349. [Google Scholar]
- Vahidi, S.; Samadani, A.A. TERRA gene expression in gastric cancer: Role of hTERT. J. Gastrointest. Cancer 2021, 52, 431–447. [Google Scholar] [CrossRef] [PubMed]
- Busuttil, R.A.; Zapparoli, G.V.; Haupt, S.; Fennell, C.; Wong, S.Q.; Pang, J.-M.B.; Takeno, E.A.; Mitchell, C.; Di Costanzo, N.; Fox, S. Role of p53 in the progression of gastric cancer. Oncotarget 2014, 5, 12016. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.P.; Meltzer, S.J. A review of the genomics of gastric cancer. Clin. Gastroenterol. Hepatol. 2006, 4, 416–425. [Google Scholar] [CrossRef]
- Falchetti, M.; Saieva, C.; Lupi, R.; Masala, G.; Rizzolo, P.; Zanna, I.; Ceccarelli, K.; Sera, F.; Mariani-Costantini, R.; Nesi, G. Gastric cancer with high-level microsatellite instability: Target gene mutations, clinicopathologic features, and long-term survival. Hum. Pathol. 2008, 39, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Shokal, U.; Sharma, P.C. Implication of microsatellite instability in human gastric cancers. Indian J. Med. Res. 2012, 135, 599. [Google Scholar] [PubMed]
- Alexander, B.M.; Cloughesy, T.F. Adult glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef]
- Stark, A.M.; Doukas, A.; Hugo, H.-H.; Hedderich, J.; Hattermann, K.; Maximilian Mehdorn, H.; Held-Feindt, J. Expression of DNA mismatch repair proteins MLH1, MSH2, and MSH6 in recurrent glioblastoma. Neurol. Res. 2015, 37, 95–105. [Google Scholar] [CrossRef]
- Indraccolo, S.; Lombardi, G.; Fassan, M.; Pasqualini, L.; Giunco, S.; Marcato, R.; Gasparini, A.; Candiotto, C.; Nalio, S.; Fiduccia, P. Genetic, epigenetic, and immunologic profiling of MMR-deficient relapsed glioblastoma. Clin. Cancer Res. 2019, 25, 1828–1837. [Google Scholar] [CrossRef] [PubMed]
- Caccese, M.; Ius, T.; Simonelli, M.; Fassan, M.; Cesselli, D.; Dipasquale, A.; Cavallin, F.; Padovan, M.; Salvalaggio, A.; Gardiman, M.P. Mismatch-repair protein expression in high-grade gliomas: A large retrospective multicenter study. Int. J. Mol. Sci. 2020, 21, 6716. [Google Scholar] [CrossRef] [PubMed]
- Kataki, A.C.; Baruah, U.; Maheshwari, A.; Medhi, P.; Kataki, K.J. Endometrial Cancer. In Fundamentals in Gynaecologic Malignancy; Springer: Berlin/Heidelberg, Germany, 2023; pp. 247–278. [Google Scholar]
- Murali, R.; Soslow, R.A.; Weigelt, B. Classification of endometrial carcinoma: More than two types. Lancet Oncol. 2014, 15, e268–e278. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.L.; Mutter, G.L. Molecular and pathologic aspects of endometrial carcinogenesis. J. Clin. Oncol. 2006, 24, 4783–4791. [Google Scholar] [CrossRef] [PubMed]
- Gordhandas, S.; Kahn, R.M.; Gamble, C.; Talukdar, N.; Maddy, B.; Nelson, B.B.; Askin, G.; Christos, P.J.; Holcomb, K.; Caputo, T.A. Clinicopathologic features of endometrial cancer with mismatch repair deficiency. Ecancermedicalscience 2020, 14, 1061. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, E.; Jansen, A.M.L.; Osse, E.M.; Nout, R.A.; Creutzberg, C.L.; Ruano, D.; Church, D.N.; Morreau, H.; Smit, V.; Van Wezel, T. Practical guidance for mismatch repair-deficiency testing in endometrial cancer. Ann. Oncol. 2017, 28, 96–102. [Google Scholar] [CrossRef]
- Cohn, D.E.; Frankel, W.L.; Resnick, K.E.; Zanagnolo, V.L.; Copeland, L.J.; Hampel, H.; Kelbick, N.; Morrison, C.D.; Fowler, J.M. Improved survival with an intact DNA mismatch repair system in endometrial cancer. Obstet. Gynecol. 2006, 108, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA. Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Toss, A.; Tomasello, C.; Razzaboni, E.; Contu, G.; Grandi, G.; Cagnacci, A.; Schilder, R.J.; Cortesi, L. Hereditary ovarian cancer: Not only BRCA 1 and 2 genes. Biomed Res. Int. 2015, 2015, 341723. [Google Scholar] [CrossRef]
- Samimi, G.; Fink, D.; Varki, N.M.; Husain, A.; Hoskins, W.J.; Alberts, D.S.; Howell, S.B. Analysis of MLH1 and MSH2 expression in ovarian cancer before and after platinum drug-based chemotherapy. Clin. cancer Res. 2000, 6, 1415–1421. [Google Scholar]
- Leskela, S.; Romero, I.; Cristobal, E.; Pérez-Mies, B.; Rosa-Rosa, J.M.; Gutierrez-Pecharroman, A.; Caniego-Casas, T.; Santón, A.; Ojeda, B.; López-Reig, R. Mismatch repair deficiency in ovarian carcinoma: Frequency, causes, and consequences. Am. J. Surg. Pathol. 2020, 44, 649–656. [Google Scholar] [CrossRef]
- Travaglino, A.; Raffone, A.; Gencarelli, A.; Saracinelli, S.; Riccardi, C.; Mollo, A.; Zullo, F.; Insabato, L. Clinico-pathological features associated with mismatch repair deficiency in endometrial undifferentiated/dedifferentiated carcinoma: A systematic review and meta-analysis. Gynecol. Oncol. 2021, 160, 579–585. [Google Scholar] [CrossRef]
- Ludford, K.; Ho, W.J.; Thomas, J.V.; Raghav, K.P.S.; Murphy, M.B.; Fleming, N.D.; Lee, M.S.; Smaglo, B.G.; You, Y.N.; Tillman, M.M. Neoadjuvant pembrolizumab in localized microsatellite instability high/deficient mismatch repair solid tumors. J. Clin. Oncol. 2023, 41, 2181–2190. [Google Scholar] [CrossRef] [PubMed]
- Bhamidipati, D.; Subbiah, V. Tumor-agnostic drug development in dMMR/MSI-H solid tumors. Trends Cancer 2023, 9, 828–839. [Google Scholar] [CrossRef] [PubMed]
- Moreno, O.M.; Paredes, A.C.; Suarez-Obando, F.; Rojas, A. An update on Fanconi anemia: Clinical, cytogenetic and molecular approaches. Biomed. Rep. 2021, 15, 74. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Xie, J.; Ucher, A.; Stavnezer, J.; Cantor, S.B. Crosstalk between BRCA-F anconi anemia and mismatch repair pathways prevents MSH 2-dependent aberrant DNA damage responses. EMBO J. 2014, 33, 1698–1712. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Guillemette, S.; Peng, M.; Gilbert, C.; Buermeyer, A.; Cantor, S.B. An MLH1 mutation links BACH1/FANCJ to colon cancer, signaling, and insight toward directed therapy. Cancer Prev. Res. 2010, 3, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Wilson, J.B.; Thomashevski, A.; Jones, N.J.; Kupfer, G. A Role for Mismatch Repair Factors in the FA-BRCA Pathway. Blood 2009, 114, 1095. [Google Scholar] [CrossRef]
- Xie, J.X. Regulation of BACH1/FANCJ Function in DNA Damage Repair: A Dissertation. Ph.D. Thesis, UMass Chan Medical School, Worcester, MA, USA, 2009. [Google Scholar]
- Cantor, S.B.; Xie, J. Assessing the link between BACH1/FANCJ and MLH1 in DNA crosslink repair. Environ. Mol. Mutagen. 2010, 51, 500–507. [Google Scholar] [CrossRef]
- Horan, T.S.; Ascenção, C.F.R.; Mellor, C.; Wang, M.; Smolka, M.B.; Cohen, P.E. The DNA helicase FANCJ (BRIP1) functions in double strand break repair processing, but not crossover formation during prophase I of meiosis in male mice. PLoS Genet. 2024, 20, e1011175. [Google Scholar] [CrossRef]
- Singh, P.; Fragoza, R.; Blengini, C.S.; Tran, T.N.; Pannafino, G.; Al-Sweel, N.; Schimenti, K.J.; Schindler, K.; Alani, E.A.; Yu, H. Human MLH1/3 variants causing aneuploidy, pregnancy loss, and premature reproductive aging. Nat. Commun. 2021, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Feitsma, H.; Leal, M.C.; Moens, P.B.; Cuppen, E.; Schulz, R.W. Mlh1 deficiency in zebrafish results in male sterility and aneuploid as well as triploid progeny in females. Genetics 2007, 175, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Hume, S.; Dianov, G.L.; Ramadan, K. A unified model for the G1/S cell cycle transition. Nucleic Acids Res. 2020, 48, 12483–12501. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daliri, K.; Hescheler, J.; Pfannkuche, K.P. Prime Editing and DNA Repair System: Balancing Efficiency with Safety. Cells 2024, 13, 858. https://doi.org/10.3390/cells13100858
Daliri K, Hescheler J, Pfannkuche KP. Prime Editing and DNA Repair System: Balancing Efficiency with Safety. Cells. 2024; 13(10):858. https://doi.org/10.3390/cells13100858
Chicago/Turabian StyleDaliri, Karim, Jürgen Hescheler, and Kurt Paul Pfannkuche. 2024. "Prime Editing and DNA Repair System: Balancing Efficiency with Safety" Cells 13, no. 10: 858. https://doi.org/10.3390/cells13100858
APA StyleDaliri, K., Hescheler, J., & Pfannkuche, K. P. (2024). Prime Editing and DNA Repair System: Balancing Efficiency with Safety. Cells, 13(10), 858. https://doi.org/10.3390/cells13100858






