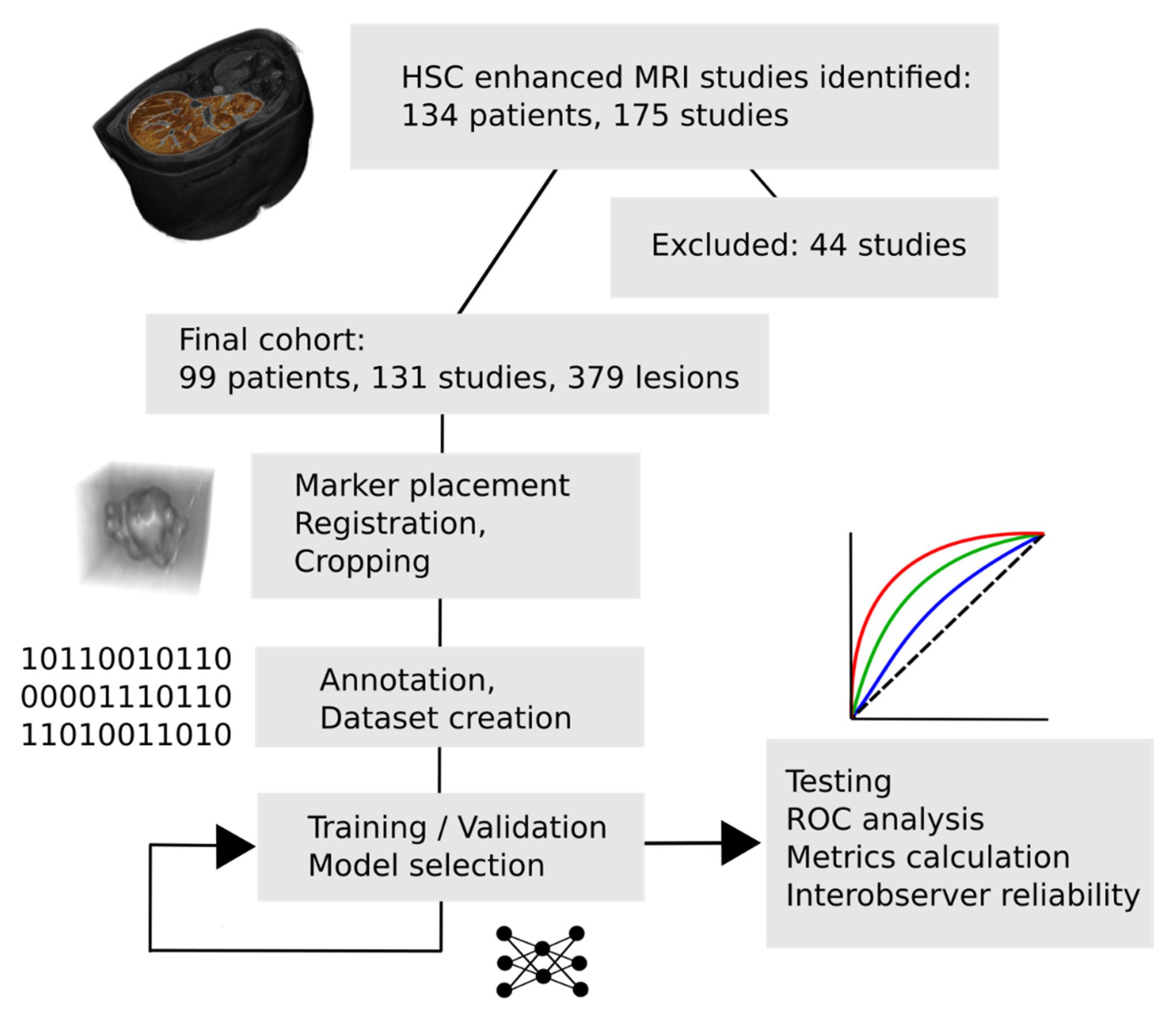
1. Introduction
The number of focal liver lesions (FLLs) detected with imaging studies is steadily growing worldwide, and accurate diagnosis is crucial not only for avoiding treatment delay but also for sparing patients and health care providers from unnecessary procedures and lowering costs. Today, magnetic resonance imaging (MRI) offers the most comprehensive non-invasive characterization of FLLs, and, contrary to computer tomography (CT), it does not expose patients to radiation. MRI imaging, performed using a selection of pulse sequences and often with intravenous contrast agents, provides excellent soft-tissue resolution and results in a highly accurate diagnosis, with a sensitivity of 94% and a specificity of 82–89% [
1].
Hepatocyte-specific contrast agents (HSCs) have been proved highly useful in lesion detection and differentiation between benign and malignant foci, and they have been increasingly used for imaging of FLLs. These contrast agents, including gadoxetate disodium and gadobenate dimeglumine, are taken up by functioning hepatocytes and are excreted into the bile [
2]. Thus, HSCs can be used to differentiate between liver lesions consisting of normal hepatocytes and those containing poorly differentiated hepatocytes or cells with non-hepatocytic origin. It is also easier to detect small lesions because there is a strong contrast between the enhancing parenchyma and foci without normal hepatocytes in the hepatobiliary phase (HBP) [
2].
Some common FLLs, such as hepatocellular carcinoma (HCC) and liver metastasis (MET), constitute a significant diagnostic challenge, as these patients can only be cured with surgical resection or image-guided ablation; thus, the significance of early detection is paramount. METs are the most common malignancies of the liver. It has been reported that up to 70% of all patients with colorectal cancer would develop METs at some point in their lifetime [
3]. HSC-enhanced MRI has been shown to have the highest sensitivity (73.3%) for METs smaller than 10 mm in diameter among all imaging modalities, which also translates to a significantly better survival rate of patients imaged with MRI (70.8%) compared to those imaged with CT (48.1%) [
4]. HCC is the fifth most common solid malignancy worldwide, and the mortality rate from HCC is predicted to rise in the coming decades [
5]. HCC typically develops in the background of decades of chronic liver disease (CLD). MRI findings of an arterial-enhancing mass with subsequent washout and enhancing capsule on delayed interstitial phase images are diagnostic for HCC [
6]. Focal nodular hyperplasia (FNH) is the second most common benign solid FLL after hemangioma. FNH is a common incidental finding in imaging studies, and it is a frequent source of differential diagnostic dilemmas of malignant lesions. A definitive diagnosis of FNH can be established in patients who do not have CLD when typical features such as arterial phase and HBP hyperenhancement and a central scar are detected with HSC MRI [
7]. Standardized data collection and reporting systems have also been developed, such as the Liver Imaging Reporting and Data System (LI-RADS), to improve CT and MRI diagnosis by reducing variability in the interpretation of imaging studies [
8]. However, due to the complex nature of these systems, their integration into the clinical workflow can be cumbersome.
Artificial intelligence (AI) techniques have been introduced in growing numbers to facilitate lesion detection and classification, assess the patients’ prognosis, or identify risk factors of FLLs based on imaging studies [
9]. Some of these studies extracted large numbers of image features from dynamic contrast-enhanced MRI to build mathematical models for the automatic classification of FLLs [
10]. Deep learning models (DLMs) are state-of-the-art image processing algorithms predominantly based on convolutional neural networks (CNN). DLMs have been tested for analysis of all known imaging modalities and achieved excellent results in image-based detection and classification of various diseases [
11]. A handful of studies have also applied DLMs to classify FLLs in MRI images and demonstrated that the performance of the DLMs is excellent and comparable to the human observers’ diagnostic rate [
12,
13,
14,
15]. Among different DLM architectures, models using 3D convolutions could be efficiently trained on a relatively small number of cases for differentiating between the most common types of FLLs [
14,
15].
Meanwhile, current AI classification models have limited value in clinical practice as these have been trained to diagnose only a handful of liver pathologies based only on a small set of MRI images. Current DLMs cannot recognize many FLLs belonging to less common diagnoses and lesions with atypical image features or with post-treatment changes [
16]. A clinically useful DLM must be able to analyze multiple image sequences to identify a comprehensive set of image features that can be used for the characterization of FLLs and the generation of a differential diagnosis [
17]. For the transparency of the AI-driven classification process, it is essential to know how many of the detected image features support the diagnosis and to validate the localization of these features via network visualization techniques, such as activation and occlusion sensitivity maps. Such sophisticated models are better suited for the systematic evaluation of FLLs and can increase the efficiency and the reproducibility of the imaging diagnosis.
Previously, we have shown that DLMs using multi-sequence HSC MRI images can accurately differentiate between FNHs, HCCs, and METs [
15]. In the present study, we aim to demonstrate that 3D CNNs can be applied for comprehensive evaluation and visualization of diagnostic image features in the same three types of FLLs in HSC MRI. We also investigate the consistency between the detected image features and the predicted diagnosis in each class, and the agreement between the DLM and radiologists.
3. Results
After training each model with multiple hyperparameter setups, the highest validation mean AUC (0.9147) was achieved by the EfficientNetB0 model after 480 epochs. In this setting, the network was trained with a batch size of 32. We provide the training results of the other model architectures as well in decreasing order, based on validation mean AUC: EfficientNetB6 (0.9033), EfficientNetB2 (0.9033), EfficientNetB3 (0.902), EfficientNetB4 (0.8988), EfficientNetB1 (0.8922), EfficientNetB5 (0.8922), DenseNet121 (0.8807), DenseNet169 (0.8792), DenseNet201 (0.8733), DenseNet264 (0.8682), EfficientNetB7 (0.856). The final EfficientNetB0 model could identify most features with excellent metrics when tested on the independent test dataset.
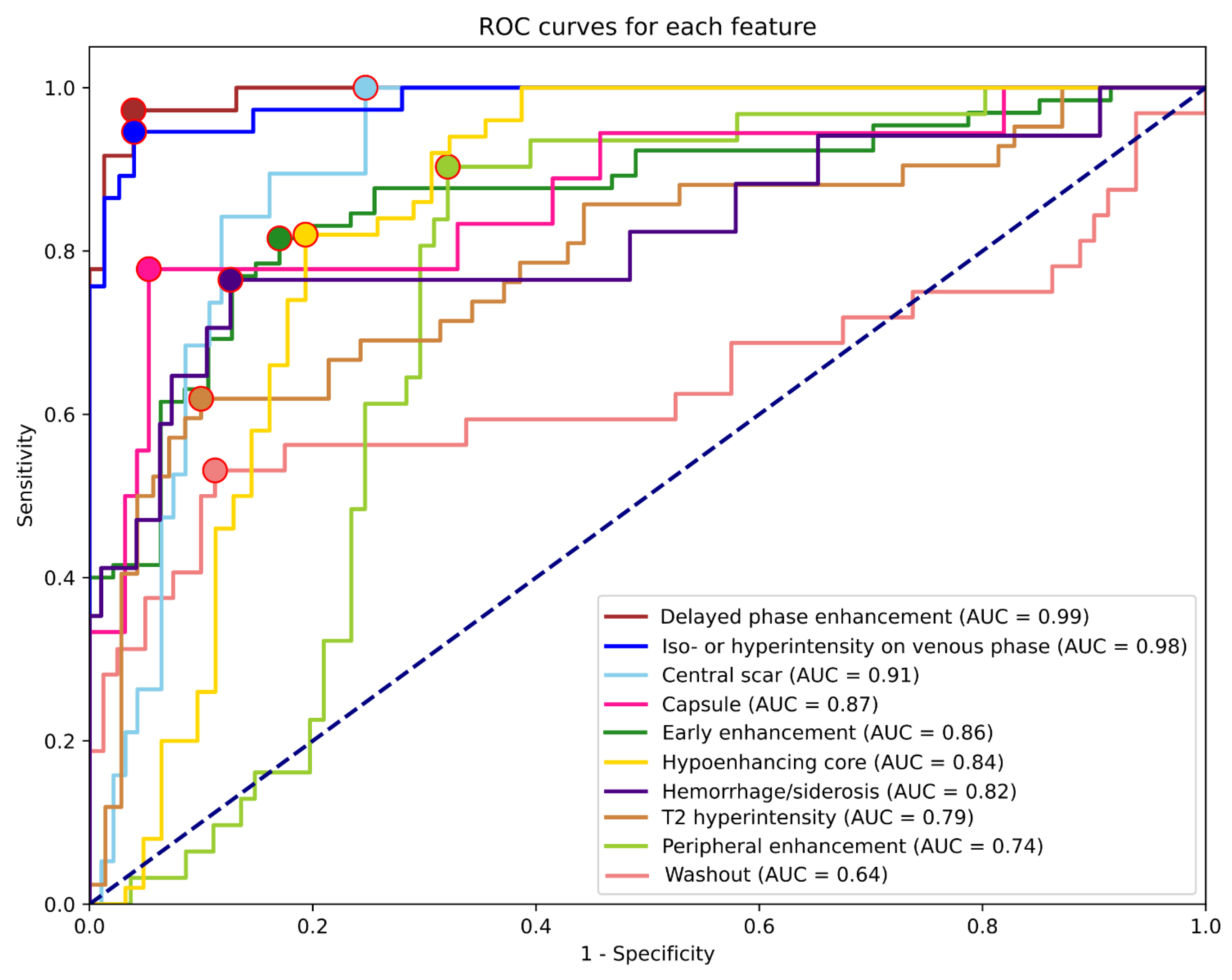
Table 3 summarizes the results for each feature, including all lesion types. The highest AUCs were reached for the detection of delayed phase enhancement (0.99) and iso- or hyperintensity on the venous phase (0.98). These features were only rarely detected as false positives or remained undetected. The least predictable features based on AUC were T2 hyperintensity (0.79), peripheral enhancement (0.74), and washout (0.64). ROC curves and corresponding AUC values are shown in
Figure 2.
The highest and lowest PPVs were reached for delayed phase enhancement (0.92) and central scar (0.44) detection, while the best and worst sensitivities were for central scar (0.95), delayed phase enhancement (0.94), and iso- or hyperintensity on venous phase (0.92) vs. T2 hyperintensity (0.60) and washout (0.50). NPVs and specificities were higher on average (0.89, 0.86) than PPVs and sensitivities (0.71, 0,78). Apart from early enhancement (0.75) and T2 hyperintensity (0.79), all other NPVs were above 0.8. The feature with the lowest specificity was peripheral enhancement (0.68), while the most specific was delayed phase enhancement (0.96). As shown in
Table 4, almost all feature AUCs were calculated with power reaching 0.98; therefore, the number of samples is more than sufficient to support these results.
To be able to explore the differences in predictions between the different lesion types, results are reported for FNHs, HCCs, and METs separately as well (
Table 4,
Table 5 and
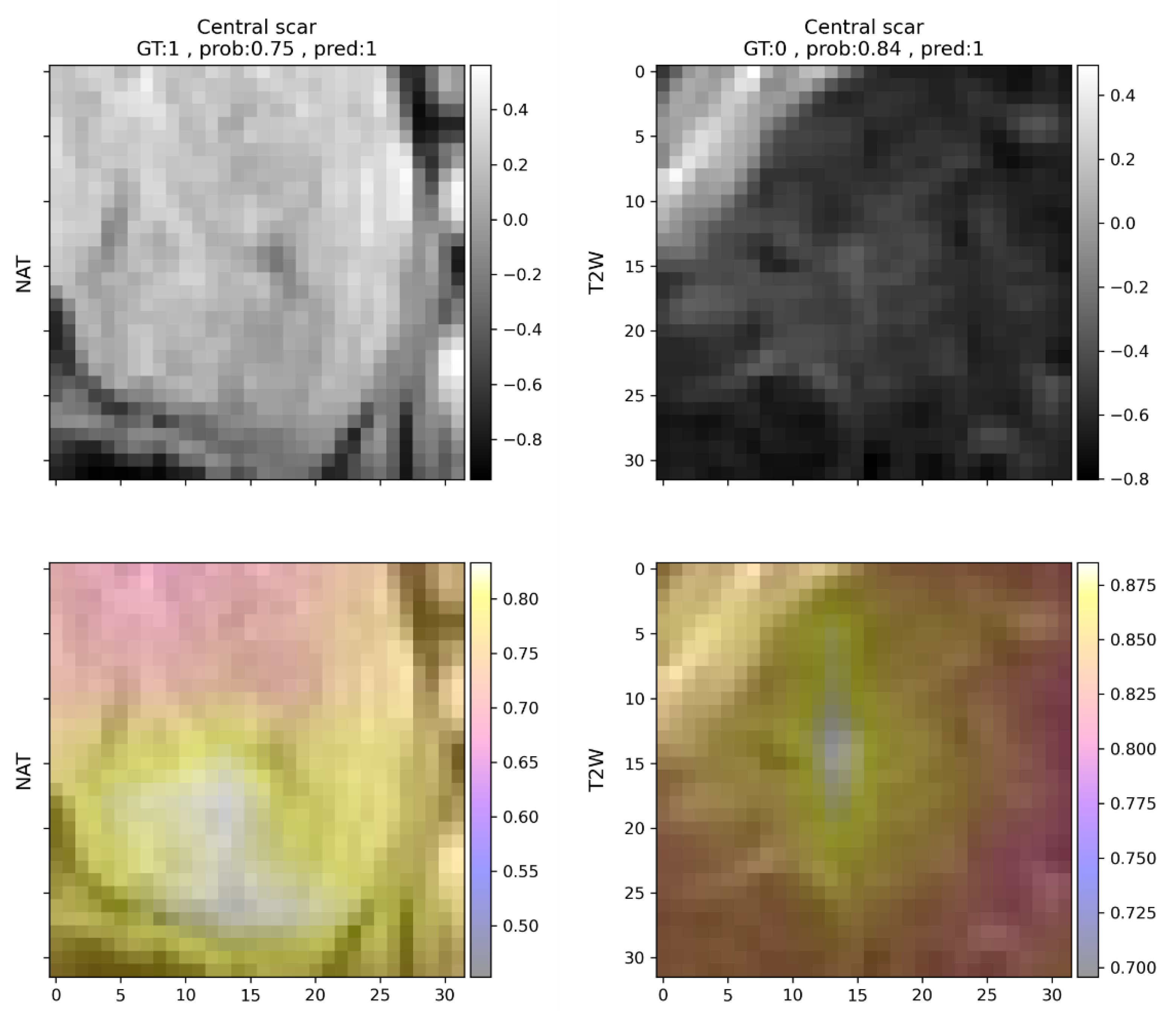
Table 6). Since not all features are present in all lesion types, not all metrics can be calculated for all features in each case. To simplify this problem, feature predictions are ordered according to their respective f1 scores. To provide more details on false detections, non-abundant features are also listed for each lesion type. Features present in FNHs were generally well recognizable by the model. Features related to contrast enhancement that are representative of FNHs, such as early or delayed phase enhancement, had f1 scores above 0.95, while non-present features were rarely detected. Central scars were common false positive detections, but mostly if the lesion was FNH (
Figure 3). If the lesion analyzed was HCC (
Table 5) or MET (
Table 6), the model almost never predicted the presence of a central scar.
Among all lesion types, HCC feature prediction yielded the least desirable results. As reported in
Table 5, diagnostically important features, namely washout and early enhancement, were undetected in half and nearly half of all cases. Features that are present in both HCCs and METs, such as peripheral enhancement (MET), were common false positive findings in the HCC group, but not in the MET group. Capsule was less difficult to detect, but peripheral enhancement was falsely detected in half of the analyzed cases, possibly due to the similarity between the two. Although hemorrhage was reported only in HCCs by the expert annotator, the algorithm predicted it in four cases in FNHs as well, and two cases in METs. Hemorrhage in HCCs remained undetected in one-third of cases, similar to hypoenhancing core (
Table 5). Features related to contrast enhancement were detected less accurately in HCCs. Hypoenhancing core was missed in five cases and falsely detected in nine cases. The presence of other similar features such as early enhancement or hemorrhage might make the detection of a hypoenhancing core more difficult.
The most common mistake in the case of METs was the underdiagnosis of T2 hyperintensity (eight cases), which was most commonly marked in this group (
Table 6). Features mostly present in FNHs were almost perfectly predicted (
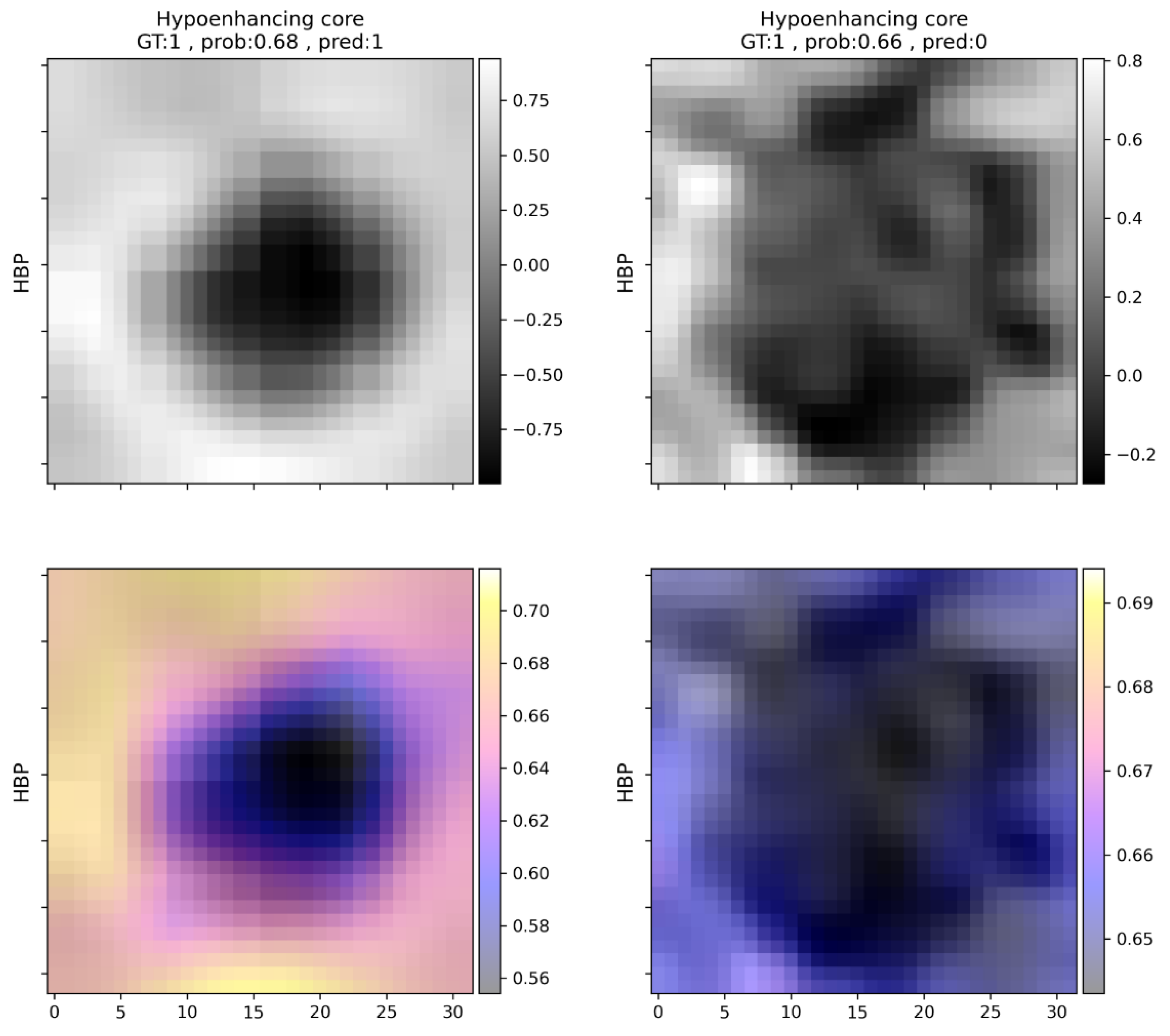
Table 4), while washout and early enhancement were the most common falsely detected features. Both peripheral enhancement and hypoenhancing core were identified with an f1 score above 0.9. For an example of hypoenhancing core prediction, see
Figure 4.
Features reported in other lesion types were variably predictable (
Table 7). Peripheral enhancement might be confused with nodular enhancement, exhibited by hemangiomas, which was not explicitly analyzed, as only a low number of cases were available. Hypoenhancing core represents a similar case, as both cysts and hemangiomas may mislead the model predictions due to their enhancement characteristics.
To assess the agreement between the model, a radiology resident, and an expert radiologist with substantial experience in abdominal radiology, Cohen’s Kappas were calculated for each feature in each combination (
Table 8). The mean score was 0.60 for the agreement between the predictive model and the expert, similar to novice opinion compared to model predictions, indicating moderate reliability. In the case of delayed phase enhancement and venous phase iso- or hyperintensity, the agreement was almost perfect (>0.8). Even the worst feature predictions (central scar, peripheral enhancement, capsule) showed moderate agreement (>0.4) with the expert opinion. Features that were less accurately predicted by the network were also subject to disagreement between the two human observers. Central scar, for example, was more frequently identified by both the model and the radiology resident, while only moderate agreement was observable in the case of washout in all three comparisons.
Generally, the described model predictions are reliable, and they could provide descriptions of radiological features present in FLLs, putting more weight on the exclusion of a feature and allowing false positive predictions depending on the type of lesion and features present. It must also be mentioned that the listed mistakes may partly be due to human uncertainty or the lack of consensus among experts on the definition of a given radiological feature, not to mention various imaging artifacts and image processing errors that may make proper predictions more difficult.
4. Discussion
The current paper explores our findings on how DLMs may perform on a small, single-institutional dataset concerning a complex reporting task. HSC MRI-based approaches are not novel in abdominal radiomics. Several research groups have reported excellent results on the automatic, DLM-based classification of various types of FLLs, but these put more emphasis on predicting lesion class and less emphasis on mimicking the human observers [
12,
14,
15,
28]. More interpretable methods have been described in radiology in general, the most obvious one being chest X-ray reporting using deep learning methods, where multiple findings have to be identified in parallel by the AI [
29]. While chest X-ray interpretation is among the most advanced research areas in deep learning radiomics, other examination types and areas with less frequently performed studies and much more complex reporting tasks lack sufficient proof for the application of AI methods. Research on radiological feature descriptors is also of importance as many of the lesions are multifocal, many types may be found parallel, and histological confirmation cannot be acquired in all cases, thus, a certain diagnosis may not be possible (and necessary) for all lesions. Additionally, the described features allow a much broader extension of applications, since each may allow the user to draw different conclusions, such as whether tumor recurrence is observable (enhancement) or whether the malignant transformation of a regenerative nodule has occurred.
These are partly the reasons why the main emphasis of the current paper is on radiological feature identification. Although the classification of different FLLs based on the identified features could seem like a straightforward task, various challenges promote it to a research topic on its own. While the majority of the lesions evaluated in our study fell into three main lesion types, the liver is host to one of the largest varieties of focal pathologies; as such, it would be worth examining diagnostic algorithms built upon the present feature identifier in a more detailed manner. As such, they should be evaluated on a larger variety of pathologies. There are multiple lesion types, for example, cholangiocellular carcinoma, that are not present in the current dataset, but in future studies should be evaluated, considering their clinical importance. Apart from this, further evaluation in this direction could be carried out in multiple ways that did not fit within the scope of the current manuscript. A classifier model could be built solely for the diagnosis of FLLs, as well as by reusing the currently presented feature identifier, for example, via transfer learning. In this case, the training of the model would be guided to take into account the radiological features identifiable by human observers, apart from deep features. The diagnosis of the tumor could also be based on the probabilities of the predictions for each feature. In this case, the top-N features would be used to create an algorithmic approach for diagnosis making. Our current interpretation of the feature detector partly opposes this approach, as the predictions of the model are evaluated based on the calculated optimal cut-off values. Apart from these, there could be other ways to create a diagnostic model that integrates the feature identifier for better interpretability. Because of this, the automatic classification of focal liver lesions lies outside the scope of the current paper.
Abdominal imaging studies, such as HSC MRI, are less frequently approached in a similar manner due to the higher cost of imaging, the complexity of the task, the smaller amount of available data, and the more variable agreement on radiological feature abundance among professionals, as well as the need for more time-consuming data preparation and analyses. Most papers use some form of deep learning interpretation method, such as attention maps, to try to find explanations for classification predictions, while direct feature predictions have rarely been the focus of research. Wang et al., in their 2019 study, were among the first to use CNNs for focal liver lesion feature identification [
17]. The reported model was able to correctly identify radiological features present in test lesions with 76.5% PPV and 82.9% sensitivity, which is similar to our results, though their method was built on a precious lesion classifier, from which feature predictions were derived. Our study deliberately avoided the diagnosis of lesions and focused solely on feature identification. Sheng et al. also used deep learning to predict radiological features based on gadoxetate disodium-enhanced MRI, dedicated to LI-RADS grading in an automated and semi-automatic manner. They reported AUCs of 0.941, 0.859, and 0.712 (internal testing) for arterial phase enhancement, washout, and capsule prediction. The model was also tested on an external test set, achieving AUCs of 0.792, 0.654, and 0.568, respectively [
30]. Though they evaluated fewer features, similarly to our findings, arterial phase enhancement was more accurately predictable than washout and capsule, both of which are challenging for the AI to predict. The results of Wang et al. also led to a similar conclusion, as arterial phase hyperenhancement and delayed phase hyperenhancement, among others, just as according to our results, were well predictable features, while others, such as central scar and washout, were especially difficult to accurately predict [
17]. Central scar and washout were also fairly difficult to identify and were quite often false positive findings; furthermore, in our experience, circle-like features such as peripheral enhancement, which might be confused with capsule by the model, were just as common false positive findings. The difficulty in the detection of these features is consistent with previous research on gadoxetate disodium, as HCC indicative features, such as capsule and washout, are less distinguishable using gadoxetate disodium than with extracellular contrast agents [
31]. Delayed phase enhancement, which is related to the hepatocyte-specific nature of gadoxetate disodium, was an accordingly straightforward prediction.
In the current study various occlusion sensitivity maps are shown that attempt to visually explain the decision-making process of the neural network classifier. The maps can be helpful in explaining the decision-making process even in a very complex task and can draw attention to erroneous decision-making that may be based on, for example, image artifacts or non-task-related image areas. The modification of the padding value from the image mean intensity to specific values depending on scan type and predicted radiological feature may be a promising direction for further investigation.
As mentioned previously, features on which there might be disagreement between expert radiologists as well (e.g., central scar) are more difficult to build a model upon. In the future, it is possible that more thorough curation of training data based on the opinion of multiple experts would be necessary to optimize these methods. A promising research direction would be a more detailed examination of how each image, their quality, and the reported expert consensus could be used to construct balanced, high-quality datasets that are more representative of radiological liver lesion features. The current study has additional limitations. It was retrospectively conducted within a single institute, and only a small number of patients were included. To mitigate the consequences of these problems, further multi-institutional studies are needed. Additional methods, such as transfer learning with other, similar, multi-modal datasets may be used in addition to the previously mentioned dataset reannotation. Further data augmentation methods, such as random cropping, also have to be evaluated. Splitting the model into multiple feature predictors based on conflicting features and corresponding scans may also be examined as a potential solution for inaccurate predictions (e.g., T2 hyperenhancement and hypoenhancing core). Apart from these, the tested methodology has the potential to aid less experienced radiologists or other clinicians in understanding and interpreting HSC MRI of FLLs in an automated, controllable manner by providing predictions of radiological features in a few seconds.