NAD+ Modulates the Proliferation and Differentiation of Adult Neural Stem/Progenitor Cells via Akt Signaling Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Isolation and Culturing of Adult Neural Stem/Progenitor Cells and Hippocampal Neurons
2.2. NAD+ Exposure and the Proliferation and Differentiation Assays of aNSPCs
2.3. Neurospheres Formation Assay
2.4. Transfection of Neuronal Cells and Sholl Analysis
2.5. ATP, NADP+ and NADPH Level Measurement
2.6. Western Blot Assay
2.7. Immunostaining and Cell Quantification
2.8. Total RNA Isolation, Reverse Transcription and Quantitative Real-Time PCR
2.9. RNA-Seq and Data Analysis
2.10. Statistical Analysis
3. Results
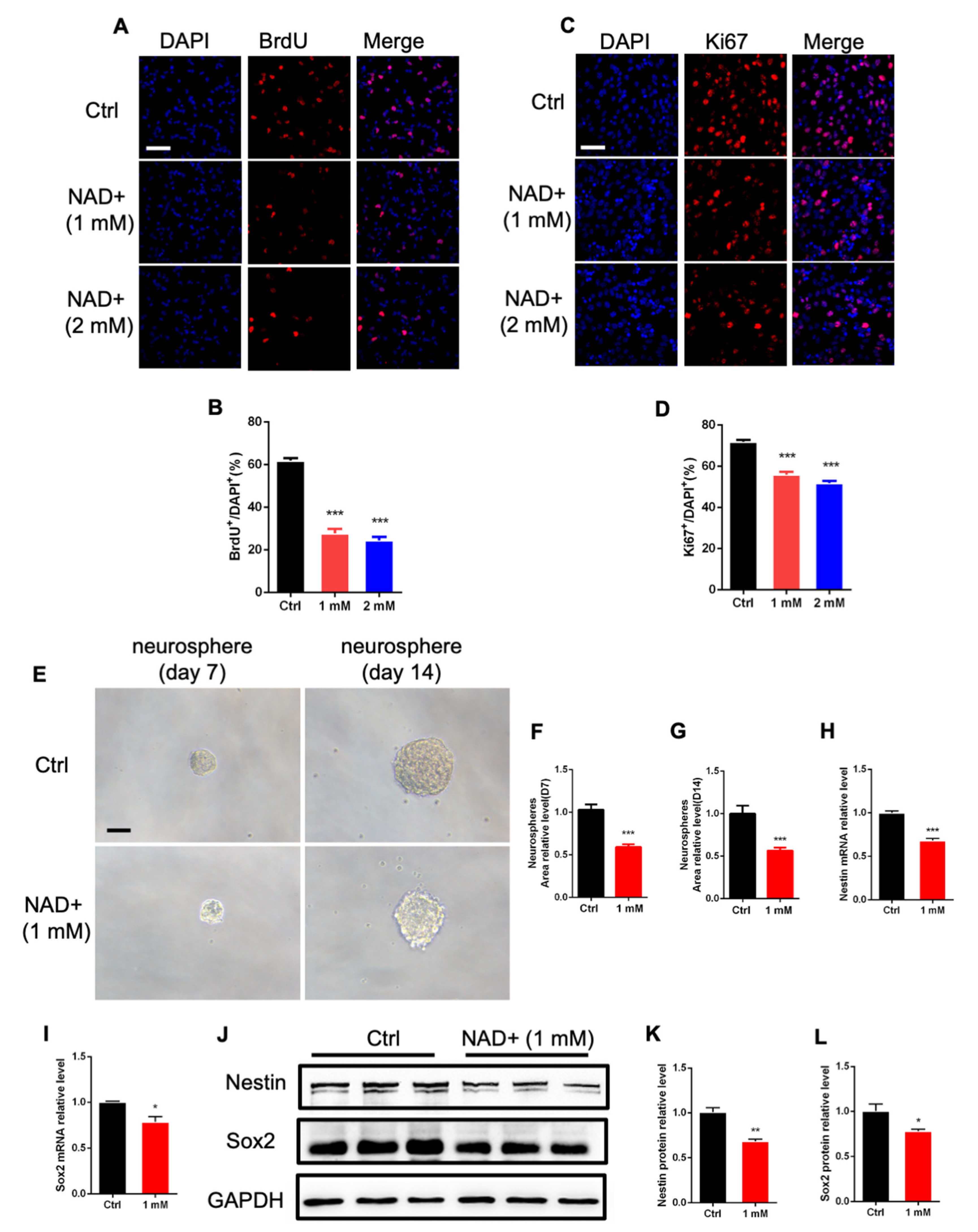
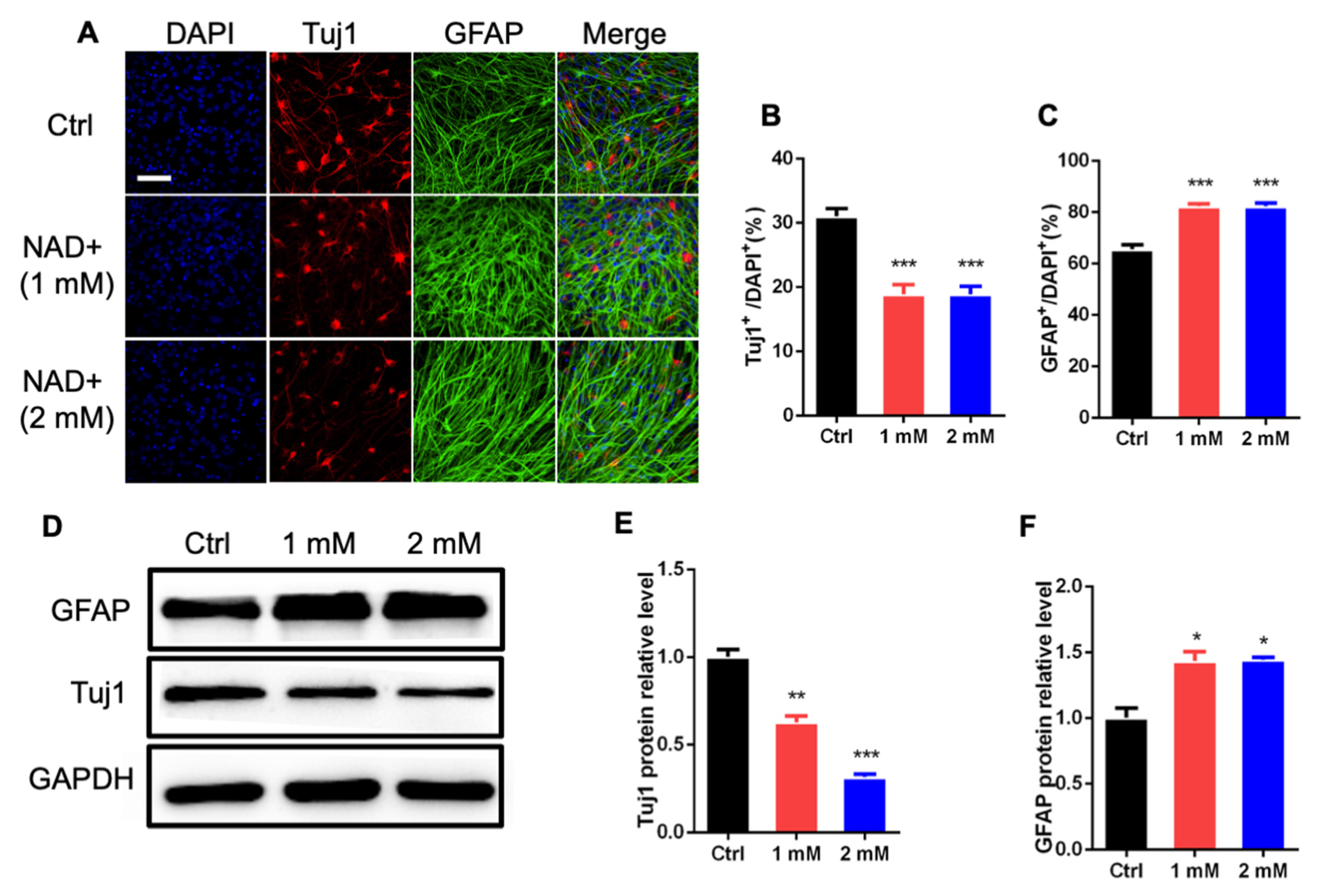
3.1. NAD+ Regulates the Proliferation and Differentiation of Adult Neural Stem/Progenitor Cells
3.2. NAD Regulates the Morphological Development of the aNSPCs-Derived Neuron and Hippocampal Neuron
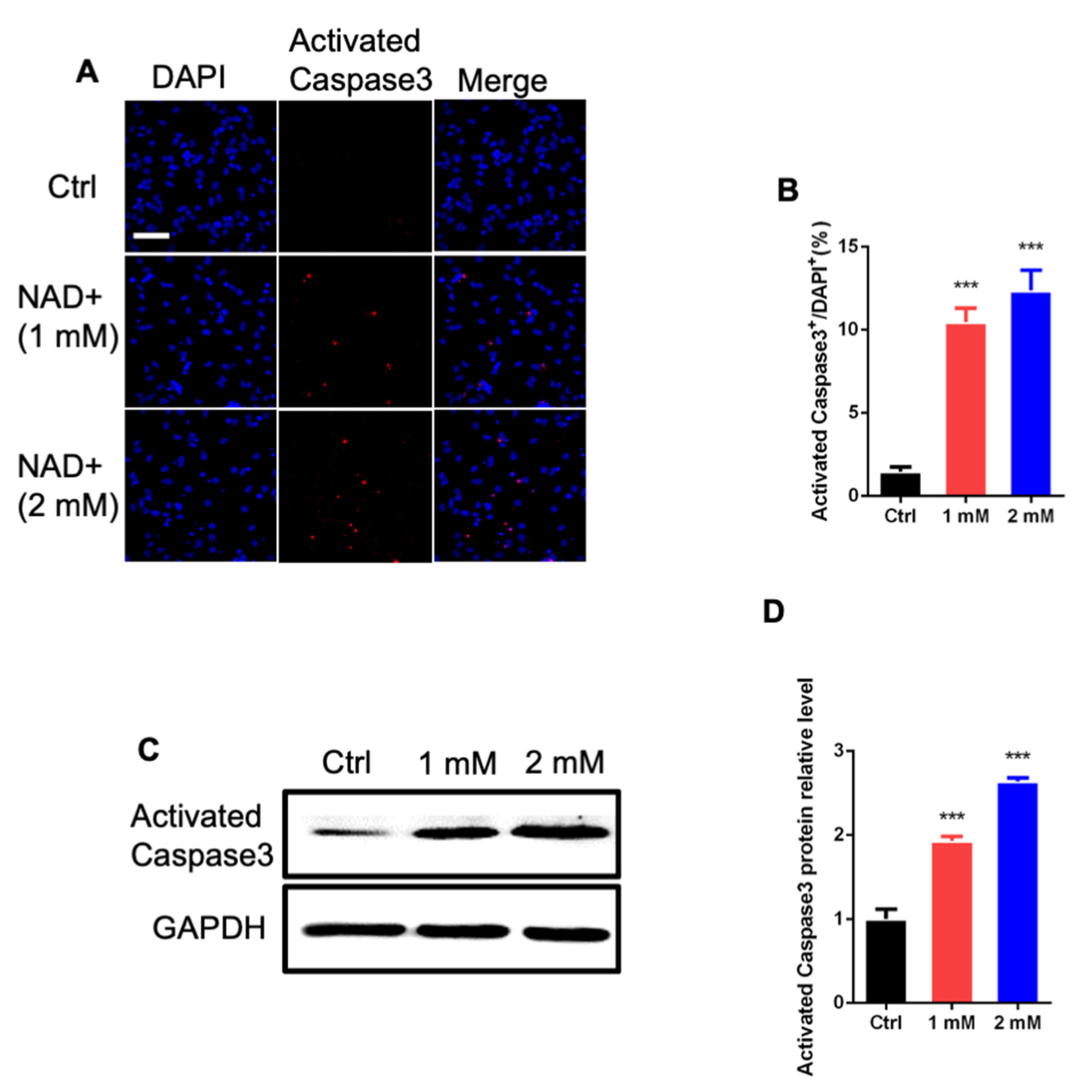
3.3. NAD+ Exposure Induces Apoptosis of aNSPCs
3.4. NAD+ Exposure Alters the Transcriptome of aNSPCs under Proliferating and Differentiation Conditions
3.5. NAD+ Exposure Alters Akt Signaling Pathway
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sahay, A.; Scobie, K.N.; Hill, A.S.; O’Carroll, C.M.; Kheirbek, M.A.; Burghardt, N.S.; Fenton, A.A.; Dranovsky, A.; Hen, R. Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature 2011, 472, 466–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Chen, J.; Liang, F.; Zhang, J.; Qu, W.; Huang, X.; Cheng, X.; Zhao, X.; Yang, Z.; Xu, S.; et al. RYBP modulates embryonic neurogenesis involving the Notch signaling pathway in a PRC1-independent pattern. Stem Cell Rep. 2021, 16, 2988–3004. [Google Scholar] [CrossRef]
- Chen, J.; Dong, X.; Cheng, X.; Zhu, Q.; Zhang, J.; Li, Q.; Huang, X.; Wang, M.; Li, L.; Guo, W.; et al. Ogt controls neural stem/progenitor cell pool and adult neurogenesis through modulating Notch signaling. Cell Rep. 2021, 34, 108905. [Google Scholar] [CrossRef]
- Shen, H.; Zhao, X.; Chen, J.; Qu, W.; Huang, X.; Wang, M.; Shao, Z.; Shu, Q.; Li, X. O-GlcNAc transferase Ogt regulates embryonic neuronal development through modulating Wnt/beta-catenin signaling. Hum. Mol. Genet. 2021, 31, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yao, B.; Chen, L.; Kang, Y.; Li, Y.; Cheng, Y.; Li, L.; Lin, L.; Wang, Z.; Wang, M.; et al. Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat. Commun. 2017, 8, 15903. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhuang, Y.; Chen, J.; Xu, W.; Shou, Y.; Huang, X.; Shu, Q.; Li, X. Dynamic effects of Fto in regulating the proliferation and differentiation of adult neural stem cells of mice. Hum. Mol. Genet. 2020, 29, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, Y.-C.; Huang, C.; Shen, H.; Sun, B.; Cheng, X.; Zhang, Y.-J.; Yang, Y.-G.; Shu, Q.; Yang, Y.; et al. m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genom. Proteom. Bioinform. 2019, 17, 154–168. [Google Scholar] [CrossRef]
- Gao, H.; Cheng, X.; Chen, J.; Ji, C.; Guo, H.; Qu, W.; Dong, X.; Chen, Y.; Ma, L.; Shu, Q.; et al. Fto-modulated lipid niche regulates adult neurogenesis through modulating adenosine metabolism. Hum. Mol. Genet. 2020, 29, 2775–2787. [Google Scholar] [CrossRef]
- Li, L.; Zang, L.; Zhang, F.; Chen, J.; Shen, H.; Shu, L.; Liang, F.; Feng, C.; Chen, D.; Tao, H.; et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 2017, 26, 2398–2411. [Google Scholar] [CrossRef] [PubMed]
- Zocher, S.; Schilling, S.; Grzyb, A.N.; Adusumilli, V.S.; Lopes, J.B.; Günther, S.; Overall, R.W.; Winter, Y.; Kempermann, G. Early-life environmental enrichment generates persistent individualized behavior in mice. Sci. Adv. 2020, 6, eabb1478. [Google Scholar] [CrossRef]
- Austin, S.H.L.; Gabarró-Solanas, R.; Rigo, P.; Paun, O.; Harris, L.; Guillemot, F.; Urbán, N. Wnt/beta-catenin signalling is dispensable for adult neural stem cell homeostasis and activation. Development 2021, 148, dev199629. [Google Scholar] [CrossRef] [PubMed]
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Yao, B.; Christian, K.M.; He, C.; Jin, B.Y.P.; Ming, G.-L.; Song, K.M.C.G.-L.M.H. Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 2016, 17, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, J.; Zhao, X. Genetics and Epigenetics in Adult Neurogenesis. Cold Spring Harb. Perspect. Biol. 2016, 8, a018911. [Google Scholar] [CrossRef]
- Snyder, J.S.; Soumier, A.; Brewer, M.; Pickel, J.; Cameron, H.A. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature 2011, 476, 458–461. [Google Scholar] [CrossRef]
- Szulwach, K.E.; Li, X.; Smrt, R.D.; Li, Y.; Luo, Y.; Lin, L.; Santistevan, N.J.; Li, W.; Zhao, X.; Jin, P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 2010, 189, 127–141. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Mei, Y.; He, Y.; Wang, D.; Wang, J.; Wei, X.; Yang, E.; Zhou, D.; Shen, H.; Peng, G.; et al. Ablating Adult Neural Stem Cells Improves Synaptic and Cognitive Functions in Alzheimer Models. Stem Cell Rep. 2021, 16, 89–105. [Google Scholar] [CrossRef]
- Lautrup, S.; Sinclair, D.A.; Mattson, M.P.; Fang, E.F. NAD+ in Brain Aging and Neurodegenerative Disorders. Cell Metab. 2019, 30, 630–655. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Huang, G.X.; Bonkowski, M.; Longchamp, A.; Li, C.; Schultz, M.B.; Kim, L.-J.; Osborne, B.; Joshi, S.; Lu, Y.; et al. Impairment of an Endothelial NAD+-H2S Signaling Network Is a Reversible Cause of Vascular Aging. Cell 2018, 173, 74–89.e20. [Google Scholar] [CrossRef] [Green Version]
- Vannini, N.; Campos, V.; Girotra, M.; Trachsel, V.; Rojas-Sutterlin, S.; Tratwal, J.; Ragusa, S.; Stefanidis, E.; Ryu, D.; Rainer, P.Y.; et al. The NAD-Booster Nicotinamide Riboside Potently Stimulates Hematopoiesis through Increased Mitochondrial Clearance. Cell Stem Cell 2019, 24, 405–418.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tobin, M.K.; Musaraca, K.; Disouky, A.; Shetti, A.; Bheri, A.; Honer, W.G.; Kim, N.; Dawe, R.J.; Bennett, D.A.; Arfanakis, K.; et al. Human Hippocampal Neurogenesis Persists in Aged Adults and Alzheimer’s Disease Patients. Cell Stem Cell 2019, 24, 974–982.e3. [Google Scholar] [CrossRef]
- Ortiz-Cordero, C.; Magli, A.; Dhoke, N.R.; Kuebler, T.; Selvaraj, S.; Oliveira, N.A.; Zhou, H.; Sham, Y.Y.; Bang, A.G.; Perlingeiro, R.C. NAD+ enhances ribitol and ribose rescue of alpha-dystroglycan functional glycosylation in human FKRP-mutant myotubes. eLife 2021, 10, e65443. [Google Scholar] [CrossRef]
- Fang, E.F.; Lautrup, S.H.; Hou, Y.; Demarest, T.G.; Croteau, D.L.; Mattson, M.P.; Bohr, V.A. NAD+ in Aging: Molecular Mechanisms and Translational Implications. Trends Mol. Med. 2017, 23, 899–916. [Google Scholar] [CrossRef]
- Hou, Y.; Lautrup, S.; Cordonnier, S.; Wang, Y.; Croteau, D.L.; Zavala, E.; Zhang, Y.; Moritoh, K.; O’Connell, J.F.; Baptiste, B.A.; et al. NAD+ supplementation normalizes key Alzheimer’s features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proc. Natl. Acad. Sci. USA 2018, 115, E1876–E1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ummarino, S.; Hausman, C.; Gaggi, G.; Rinaldi, L.; Bassal, M.A.; Zhang, Y.; Seelam, A.J.; Kobayashi, I.S.; Borchiellini, M.; Ebralidze, A.K.; et al. NAD Modulates DNA Methylation and Cell Differentiation. Cells 2021, 10, 2986. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, T.A.; Blackman, A.V.; Oyrer, J.; Jayabal, S.; Chung, A.J.; Watt, A.J.; Sjostrom, P.J.; van Meyel, D.J. Neuronal morphometry directly from bitmap images. Nat. Methods 2014, 11, 982–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.-J.; Guan, F.-L.; Li, Q.; Dai, G.; Li, H.-F.; Li, X.-K. AlCl3 exposure regulates neuronal development by modulating DNA modification. World J. Stem Cells 2020, 12, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-H.; Lu, M.; Lee, B.-Y.; Ugurbil, K.; Chen, W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc. Natl. Acad. Sci. USA 2015, 112, 2876–2881. [Google Scholar] [CrossRef] [Green Version]
- Fang, E.F.; Scheibye-Knudsen, M.; Brace, L.E.; Kassahun, H.; SenGupta, T.; Nilsen, H.; Mitchell, J.R.; Croteau, D.L.; Bohr, V.A. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 2014, 157, 882–896. [Google Scholar] [CrossRef] [Green Version]
- Fang, E.F.; Kassahun, H.; Croteau, D.L.; Scheibye-Knudsen, M.; Marosi, K.; Lu, H.; Shamanna, R.A.; Kalyanasundaram, S.; Bollineni, R.C.; Wilson, M.A.; et al. NAD(+) Replenishment Improves Lifespan and Healthspan in Ataxia Telangiectasia Models via Mitophagy and DNA Repair. Cell Metab 2016, 24, 566–581. [Google Scholar] [CrossRef] [Green Version]
- Stein, L.R.; Imai, S. Specific ablation of Nampt in adult neural stem cells recapitulates their functional defects during aging. EMBO J. 2014, 33, 1321–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnay, F.; Veloso, A.; Steinmann, V.; Köcher, T.; Abdusselamoglu, M.D.; Bajaj, S.; Rivelles, E.; Landskron, L.; Esterbauer, H.; Zinzen, R.P.; et al. Oxidative Metabolism Drives Immortalization of Neural Stem Cells during Tumorigenesis. Cell 2020, 182, 1490–1507.e19. [Google Scholar] [CrossRef] [PubMed]
- Wiley, C.; Campisi, J. NAD+ controls neural stem cell fate in the aging brain. EMBO J. 2014, 33, 1289–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okun, E.; Marton, D.; Cohen, D.; Griffioen, K.; Kanfi, Y.; Illouz, T.; Madar, R.; Cohen, H.Y. Sirt6 alters adult hippocampal neurogenesis. PLoS ONE 2017, 12, e0179681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Ryu, D.; Wu, Y.; Gariani, K.; Wang, X.; Luan, P.; D’Amico, D.; Ropelle, E.R.; Lutolf, M.P.; Aebersold, R.; et al. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 2016, 352, 1436–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ear, P.H.; Chadda, A.; Gumusoglu, S.B.; Schmidt, M.; Vogeler, S.; Malicoat, J.; Kadel, J.; Moore, M.M.; Migaud, M.E.; Stevens, H.; et al. Maternal Nicotinamide Riboside Enhances Postpartum Weight Loss, Juvenile Offspring Development, and Neurogenesis of Adult Offspring. Cell Rep. 2019, 26, 969–983.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Cao, B.; Naval-Sanchez, M.; Pham, T.; Sun, Y.B.Y.; Williams, B.; Heazlewood, S.Y.; Deshpande, N.; Li, J.; Kraus, F.; et al. Nicotinamide riboside attenuates age-associated metabolic and functional changes in hematopoietic stem cells. Nat. Commun. 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Hayakawa, K.; Esposito, E.; Wang, X.; Terasaki, Y.; Liu, Y.; Xing, C.; Ji, X.; Lo, E.H. Transfer of mitochondria from astrocytes to neurons after stroke. Nature 2016, 535, 551–555. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Guo, H.; Cheng, X.; Zhang, J.; Qu, W.; Ding, Q.; Sun, Q.; Shu, Q.; Li, X. NAD+ Modulates the Proliferation and Differentiation of Adult Neural Stem/Progenitor Cells via Akt Signaling Pathway. Cells 2022, 11, 1283. https://doi.org/10.3390/cells11081283
Huang X, Guo H, Cheng X, Zhang J, Qu W, Ding Q, Sun Q, Shu Q, Li X. NAD+ Modulates the Proliferation and Differentiation of Adult Neural Stem/Progenitor Cells via Akt Signaling Pathway. Cells. 2022; 11(8):1283. https://doi.org/10.3390/cells11081283
Chicago/Turabian StyleHuang, Xiaoli, Hongfeng Guo, Xuejun Cheng, Jinyu Zhang, Wenzheng Qu, Qianyun Ding, Qihang Sun, Qiang Shu, and Xuekun Li. 2022. "NAD+ Modulates the Proliferation and Differentiation of Adult Neural Stem/Progenitor Cells via Akt Signaling Pathway" Cells 11, no. 8: 1283. https://doi.org/10.3390/cells11081283
APA StyleHuang, X., Guo, H., Cheng, X., Zhang, J., Qu, W., Ding, Q., Sun, Q., Shu, Q., & Li, X. (2022). NAD+ Modulates the Proliferation and Differentiation of Adult Neural Stem/Progenitor Cells via Akt Signaling Pathway. Cells, 11(8), 1283. https://doi.org/10.3390/cells11081283





