Regulatory Role of Circadian Clocks on ABA Production and Signaling, Stomatal Responses, and Water-Use Efficiency under Water-Deficit Conditions
Abstract
:1. Introduction
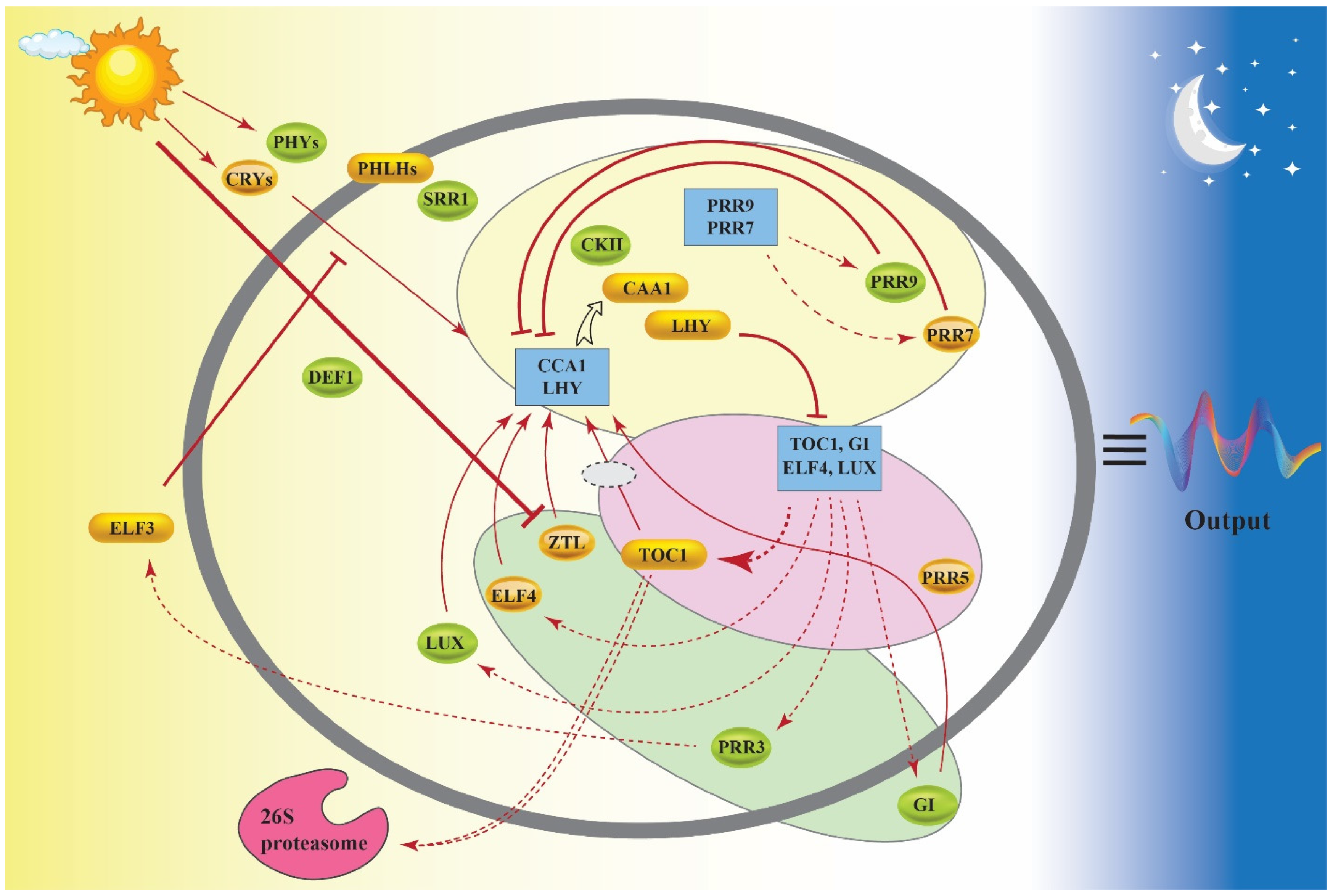
2. How Does the Circadian Clock Work?
3. Circadian Clock Plays a Central Role in Plant Fitness
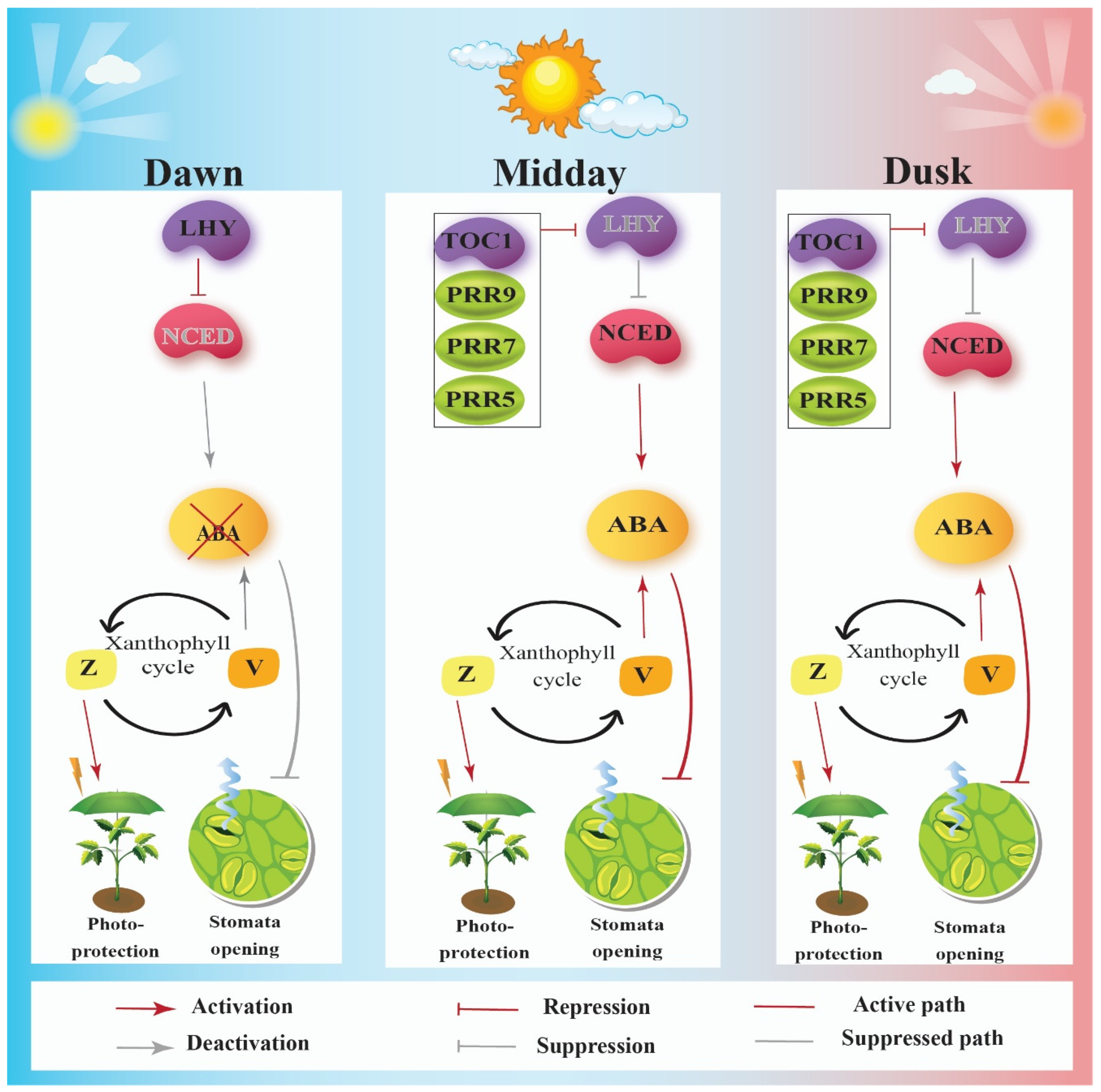
4. Circadian Clock Enhances Fitness by Controlling Photoprotection via the Xanthophyll Cycle
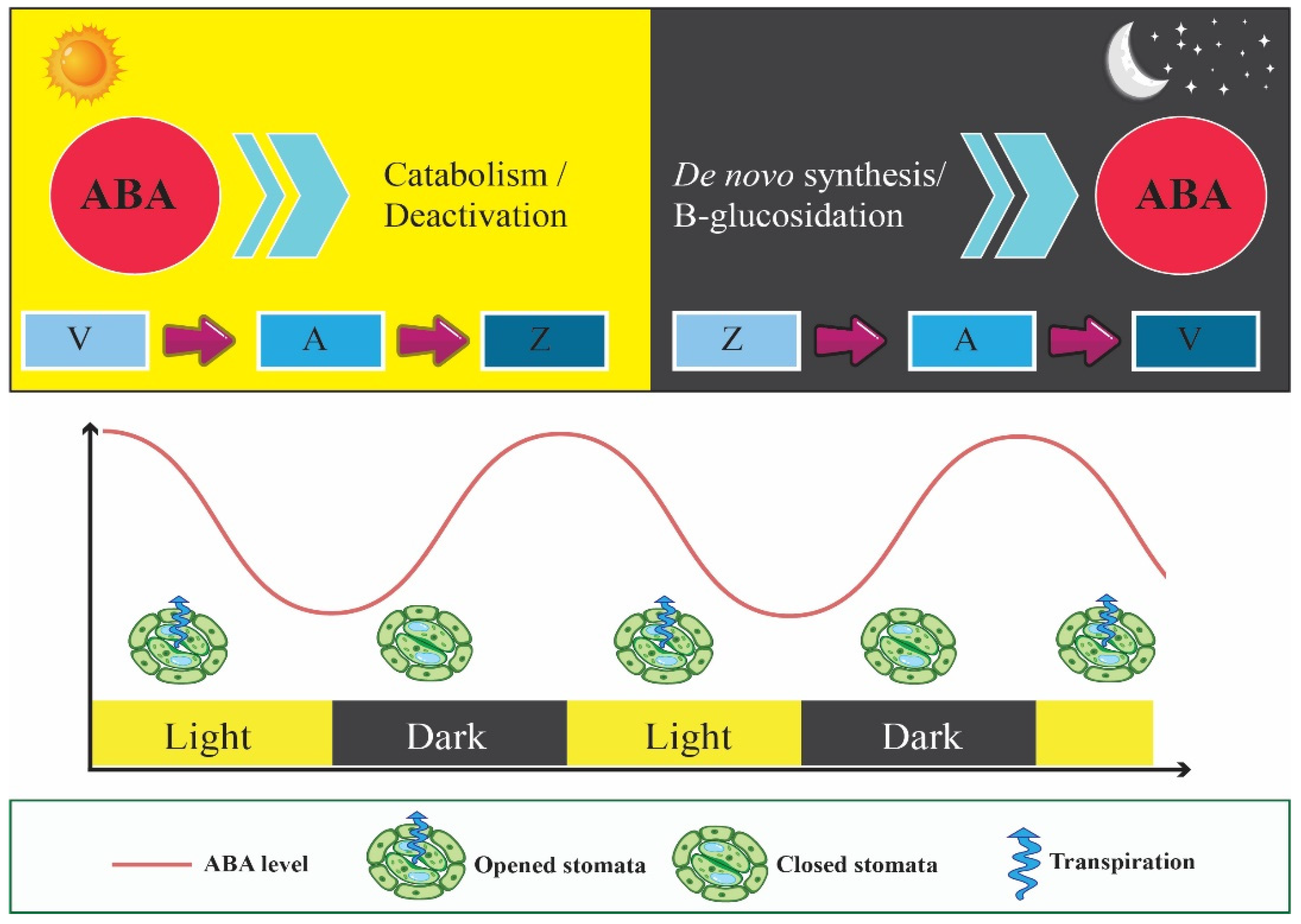
5. Circadian Clock Enhances Fitness by Assisting Adaptive Stomatal Movements
6. Circadian Clock Enhances Fitness by Controlling WUE through Affecting Stomatal Movements
7. Circadian Clock Enhances Fitness by Improving Plant Drought-Stress Responses
8. Circadian Clock Interacts with ABA Signaling Pathway
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rascher, U.; Nedbal, L. Dynamics of photosynthesis in fluctuating light. Curr. Opin. Plant Biol. 2006, 9, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Belbin, F.E.; Frank, A.; Webb, A.A.R. Interactions between circadian clocks and photosynthesis for the temporal and spatial coordination of metabolism. Front. Plant Sci. 2015, 6, 245. [Google Scholar] [CrossRef] [Green Version]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kévei, E.; Tóth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A.R. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [Green Version]
- Más, P.; Yanovsky, M.J. Time for circadian rhythms: Plants get synchronized. Curr. Opin. Plant Biol. 2009, 12, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Bell-Pedersen, D. Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryot. Cell 2006, 5, 1184–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, R.M.; Tingay, S.; Wang, Z.Y.; Tobin, E.M. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002, 129, 576–584. [Google Scholar] [CrossRef] [Green Version]
- Grundy, J.; Stoker, C.; Carré, I.A. Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 2015, 6, 648. [Google Scholar] [CrossRef]
- Haydon, M.J.; Mielczarek, O.; Robertson, F.C.; Hubbard, K.E.; Webb, A.A.R. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 2013, 502, 689–692. [Google Scholar] [CrossRef]
- Kondo, T.; Ishiura, M. The circadian clock of cyanobacteria. BioEssays 2000, 22, 10–15. [Google Scholar] [CrossRef]
- Sanchez, S.E.; Kay, S.A. The Plant Circadian Clock: From a Simple Timekeeper to a Complex Developmental Manager. Cold Spring Harb. Perspect. Biol. 2016, 8, a027748. [Google Scholar] [CrossRef]
- Shomali, A.; Aliniaeifard, S. Overview of Signal Transduction in Plants under Salt and Drought Stresses. In Salt and Drought Stress Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2020; pp. 231–258. [Google Scholar]
- Aliniaeifard, S.; van Meeteren, U. Can prolonged exposure to low VPD disturb the ABA signalling in stomatal guard cells? J. Exp. Bot. 2013, 64, 3551–3566. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought Stress Responses and Resistance in Plants: From Cellular Responses to Long-Distance Intercellular Communication. Front. Plant Sci. 2020, 11, 1407. [Google Scholar] [CrossRef] [PubMed]
- Todaka, D.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. ABA-responsive gene expression in response to drought stress: Cellular regulation and long-distance signaling. In Advances in Botanical Research; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; Volume 92, pp. 83–113. ISBN 9780081026205. [Google Scholar]
- Harmon, F.G.; Imaizumi, T.; Kay, S.A. The Plant Circadian Clock: Review of a Clockwork Arabidopsis. In Annual Plant Reviews Online; John Wiley & Sons, Ltd.: Chichester, UK, 2018; pp. 1–23. [Google Scholar]
- Yuan, L.; Yu, Y.; Liu, M.; Song, Y.; Li, H.; Sun, J.; Wang, Q.; Xie, Q.; Wang, L.; Xu, X. BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes. Plant Cell 2021, 33, 2602–2617. [Google Scholar] [CrossRef]
- Lai, X.; Bendix, C.; Yan, L.; Zhang, Y.; Schnable, J.C.; Harmon, F.G. Interspecific analysis of diurnal gene regulation in panicoid grasses identifies known and novel regulatory motifs. BMC Genom. 2020, 21, 428. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, L.; Yuan, L.; Song, Y.; Sun, J.; Jia, Q.; Xie, Q.; Xu, X. Molecular investigation of organ-autonomous expression of Arabidopsis circadian oscillators. Plant Cell Environ. 2020, 43, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.Y.; Chen, F.F.; Li, C.; Chen, C.; Lai, Y.C.; Chen, J.H.; Cui, Y.; Wu, K. The Arabidopsis LDL1/2-HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Res. 2018, 46, 10669–10681. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.Y.; Harmer, S.L. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014, 19, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Li, Y.; Sancar, A.; Oztas, O. The circadian clock shapes the Arabidopsis transcriptome by regulating alternative splicing and alternative polyadenylation. J. Biol. Chem. 2020, 295, 7608–7619. [Google Scholar] [CrossRef] [Green Version]
- McClung, C.R. Plant circadian rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef] [Green Version]
- Simon, N.M.L.; Graham, C.A.; Comben, N.E.; Hetherington, A.M.; Dodd, A.N. The circadian clock influences the long-term water use efficiency of Arabidopsis. Plant Physiol. 2020, 183, 317–330. [Google Scholar] [CrossRef] [Green Version]
- Uehara, T.N.; Mizutani, Y.; Kuwata, K.; Hirota, T.; Sato, A.; Mizoi, J.; Takao, S.; Matsuo, H.; Suzuki, T.; Ito, S.; et al. Casein kinase 1 family regulates PRR5 and TOC1 in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2019, 116, 11528–11536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Du, Z.; Huo, X.; Zhou, J.; Chen, Y.; Zhang, J.; Pan, A.; Wang, X.; Wang, F.; Zhang, J. Genome-wide analysis of PRR gene family uncovers their roles in circadian rhythmic changes and response to drought stress in Gossypium hirsutum L. PeerJ 2020, 8, e9936. [Google Scholar] [CrossRef] [PubMed]
- Pokhilko, A.; Fernández, A.P.; Edwards, K.D.; Southern, M.M.; Halliday, K.J.; Millar, A.J. The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 2012, 8, 574. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. The plant circadian oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, W.Y.; Fujiwara, S.; Suh, S.S.; Kim, J.; Kim, Y.; Han, L.; David, K.; Putterill, J.; Nam, H.G.; Somers, D.E. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 2007, 449, 356–360. [Google Scholar] [CrossRef]
- Yerushalmi, S.; Yakir, E.; Green, R.M. Circadian clocks and adaptation in Arabidopsis. Mol. Ecol. 2011, 20, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, P.; Zhang, Q.; Zhao, L.; Hao, X.; Liu, L.; Bu, C.; Pan, Y.; Zhang, D.; Song, Y. Dynamic physiological and transcriptome changes reveal a potential relationship between the circadian clock and salt stress response in Ulmus pumila. Mol. Genet. Genom. 2022, 297, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.-S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated Transcription of Key Pathways in Arabidopsis by the Circadian Clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari, C.; Proost, S.; Janowski, M.; Becker, J.; Nikoloski, Z.; Bhattacharya, D.; Price, D.; Tohge, T.; Bar-Even, A.; Fernie, A.; et al. Kingdom-wide comparison reveals the evolution of diurnal gene expression in Archaeplastida. Nat. Commun. 2019, 10, 737. [Google Scholar] [CrossRef]
- Monnier, A.; Liverani, S.; Bouvet, R.; Jesson, B.; Smith, J.Q.; Mosser, J.; Corellou, F.; Bouget, F.Y. Orchestrated transcription of biological processes in the marine picoeukaryote Ostreococcus exposed to light/dark cycles. BMC Genom. 2010, 11, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, T.P.; Mockler, T.C.; Breton, G.; McEntee, C.; Byer, A.; Trout, J.D.; Hazen, S.P.; Shen, R.; Priest, H.D.; Sullivan, C.M.; et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008, 4, e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano-Bueno, G.; Romero-Campero, F.J.; Lucas-Reina, E.; Romero, J.M.; Valverde, F. Evolution of photoperiod sensing in plants and algae. Curr. Opin. Plant Biol. 2017, 37, 10–17. [Google Scholar] [CrossRef]
- Huner, N.P.A.; Oquist, G.; Hurry, V.M.; Krol, M.; Falk, S.; Griffith, M. Photosynthesis, photoinhibition and low temperature acclimation in cold tolerant plants. Photosynth. Res. 1993, 37, 19–39. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.; Li, X.P.; Niyogi, K.K. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001, 125, 1558–1566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niyogi, K.K. Photoprotection revisited: Genetic and molecular approaches. Annu. Rev. Plant Biol. 1999, 50, 333–359. [Google Scholar] [CrossRef]
- Walters, R.G. Towards an understanding of photosynthetic acclimation. J. Exp. Bot. 2005, 56, 435–447. [Google Scholar] [CrossRef] [PubMed]
- Davison, P.A.; Hunter, C.N.; Horton, P. Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 2002, 418, 203–206. [Google Scholar] [CrossRef]
- Hager, A.; Holocher, K. Localization of the xanthophyll-cycle enzyme violaxanthin de-epoxidase within the thylakoid lumen and abolition of its mobility by a (light-dependent) pH decrease. Planta 1994, 192, 581–589. [Google Scholar] [CrossRef]
- Faure, S.; Turner, A.S.; Gruszka, D.; Christodoulou, V.; Davis, S.J.; Von Korff, M.; Laurie, D.A. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. USA 2012, 109, 8328–8333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, C.R. Beyond Arabidopsis: The circadian clock in non-model plant species. Semin. Cell Dev. Biol. 2013, 24, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Yarkhunova, Y.; Guadagno, C.R.; Rubin, M.J.; Davis, S.J.; Ewers, B.E.; Weinig, C. Circadian rhythms are associated with variation in photosystem II function and photoprotective mechanisms. Plant Cell Environ. 2018, 41, 2518–2529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litthauer, S.; Battle, M.W.; Lawson, T.; Jones, M.A. Phototropins maintain robust circadian oscillation of PSII operating efficiency under blue light. Plant J. 2015, 83, 1034–1045. [Google Scholar] [CrossRef] [Green Version]
- Galvez-Valdivieso, G.; Fryer, M.J.; Lawson, T.; Slattery, K.; Truman, W.; Smirnoff, N.; Asami, T.; Davies, W.J.; Jones, A.M.; Baker, N.R.; et al. The high light response in arabidopsis involves ABA signaling between vascular and bundle sheath cells W. Plant Cell 2009, 21, 2143–2162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, K.; Li, J.; Liu, L.; Han, Y.; Du, Y.; Zhang, J.; Sun, H.; Zhao, Q. Exogenous ABA induces drought tolerance in upland rice: The role of chloroplast and ABA biosynthesis-related gene expression on photosystem II during PEG stress. Acta Physiol. Plant. 2014, 36, 2219–2227. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Chory, J. The Arabidopsis Transcriptome Responds Specifically and Dynamically to High Light Stress. Cell Rep. 2019, 29, 4186–4199.e3. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, N.; Miller, G.; Salazar, C.; Mondal, H.A.; Shulaev, E.; Cortes, D.F.; Shuman, J.L.; Luo, X.; Shah, J.; Schlauch, K.; et al. Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 2013, 25, 3553–3569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamrani, Y.Y.; Matsuo, T.; Mittag, M.; Minagawa, J. Roc75 is an attenuator for the circadian clock that controls lhcsr3 expression. Plant Cell Physiol. 2018, 59, 2602–2607. [Google Scholar] [CrossRef] [PubMed]
- Peers, G.; Truong, T.B.; Ostendorf, E.; Busch, A.; Elrad, D.; Grossman, A.R.; Hippler, M.; Niyogi, K.K. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 2009, 462, 518–521. [Google Scholar] [CrossRef] [PubMed]
- van Meeteren, U.; Aliniaeifard, S. Stomata and postharvest physiology. Postharvest Ripening Physiol. Crop. 2016, 1, 157–216. [Google Scholar]
- Hetherington, A.M.; Woodward, F.I. The role of stomata in sensing and driving environmental change. Nature 2003, 424, 901–908. [Google Scholar] [CrossRef]
- Hubbard, K.E.; Webb, A.A.R. Circadian Rhythms in Stomata: Physiological and Molecular Aspects. In Rhythms in Plants; Springer International Publishing: Cham, Switzerland, 2015; pp. 231–255. ISBN 9783319205175. [Google Scholar]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carré, I.A. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018, 220, 893–907. [Google Scholar] [CrossRef] [PubMed]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokhilko, A.; Mas, P.; Millar, A.J. Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst. Biol. 2013, 7, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, M.J.; Pereira, J.S.; Chaves, M.M.; Rodrigues, M.L.; Pacheco, C.A. ABA xylem concentrations determine maximum daily leaf conductance of field-grown Vitis vinifera L. plants. Plant Cell Environ. 1995, 18, 511–521. [Google Scholar] [CrossRef]
- Snaith, P.J.; Mansfield, T.A. The circadian rhythm of stomatal opening: Evidence for the involvement of potassium and chloride fluxes. J. Exp. Bot. 1986, 37, 188–199. [Google Scholar] [CrossRef]
- Zhang, C.; Xie, Q.; Anderson, R.G.; Ng, G.; Seitz, N.C.; Peterson, T.; McClung, C.R.; McDowell, J.M.; Kong, D.; Kwak, J.M.; et al. Crosstalk between the Circadian Clock and Innate Immunity in Arabidopsis. PLoS Pathog. 2013, 9, e1003370. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Han, X.; Yang, J.; Jiang, Y.; Hu, Y. The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell 2021, 33, 3022–3041. [Google Scholar] [CrossRef]
- Lee, H.G.; Mas, P.; Seo, P.J. MYB96 shapes the circadian gating of ABA signaling in Arabidopsis. Sci. Rep. 2016, 6, 17754. [Google Scholar] [CrossRef]
- Webb, A.A.R.; Satake, A. Understanding Circadian Regulation of Carbohydrate Metabolism in Arabidopsis Using Mathematical Models. Plant Cell Physiol. 2015, 56, 586–593. [Google Scholar] [CrossRef] [Green Version]
- Göring, H.; Koshuchowa, S.; Deckert, C. Influence of Gibberellic Acid on Stomatal Movement. Biochem. Physiol. Pflanz. 1990, 186, 367–374. [Google Scholar] [CrossRef]
- Tallman, G. Are diurnal patterns of stomatal movement the result of alternating metabolism of endogenous guard cell ABA and accumulation of ABA delivered to the apoplast around guard cells by transpiration? J. Exp. Bot. 2004, 55, 1963–1976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassidim, M.; Dakhiya, Y.; Turjeman, A.; Hussien, D.; Shor, E.; Anidjar, A.; Goldberg, K.; Green, R.M. CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and the Circadian Control of Stomatal Aperture. Plant Physiol. 2017, 175, 1864–1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, F.I.; Arteaga, C.; Alam, M.S.; Alam, I.; Resco de Dios, V. Drivers of nocturnal stomatal conductance in C3 and C4 plants. Sci. Total Environ. 2022, 814, 151952. [Google Scholar] [CrossRef]
- Nakamichi, N.; Takao, S.; Kudo, T.; Kiba, T.; Wang, Y.; Kinoshita, T.; Sakakibara, H. Improvement of Arabidopsis Biomass and Cold, Drought and Salinity Stress Tolerance by Modified Circadian Clock-Associated PSEUDO-RESPONSE REGULATORs. Plant Cell Physiol. 2016, 57, 1085–1097. [Google Scholar] [CrossRef] [Green Version]
- Panchal, S.; Melotto, M. Stomate-based defense and environmental cues. Plant Signal. Behav. 2017, 12, e1362517. [Google Scholar] [CrossRef]
- Aliniaeifard, S.; Shomali, A.; Seifikalhor, M.; Lastochkina, O. Calcium Signaling in Plants under Drought. In Salt and Drought Stress Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2020; pp. 259–298. [Google Scholar]
- Zhang, S.; Wu, Q.R.; Liu, L.L.; Zhang, H.M.; Gao, J.W.; Pei, Z.M. Osmotic stress alters circadian cytosolic Ca2+ oscillations and OSCA1 is required in circadian gated stress adaptation. Plant Signal. Behav. 2020, 15, 1836883. [Google Scholar] [CrossRef]
- Somers, D.E.; Webb, A.A.R.; Pearson, M.; Kay, S.A. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 1998, 125, 485–494. [Google Scholar] [CrossRef]
- Dodd, A.N.; Parkinson, K.; Webb, A.A.R. Independent circadian regulation of assimilation and stomatal conductance in the ztl-1 mutant of Arabidopsis. New Phytol. 2004, 162, 63–70. [Google Scholar] [CrossRef]
- Graf, A.; Schlereth, A.; Stitt, M.; Smith, A.M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 2010, 107, 9458–9463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resco de Dios, V.; Gessler, A.; Ferrio, J.P.; Alday, J.G.; Bahn, M.; del Castillo, J.; Devidal, S.; García-Muñoz, S.; Kayler, Z.; Landais, D.; et al. Circadian rhythms have significant effects on leaf-to-canopy scale gas exchange under field conditions. Gigascience 2016, 5, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, M.M.; Costa, J.M.; Zarrouk, O.; Pinheiro, C.; Lopes, C.M.; Pereira, J.S. Controlling stomatal aperture in semi-arid regions—The dilemma of saving water or being cool? Plant Sci. 2016, 251, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Resco de Dios, V. Circadian regulation and diurnal variation in gas exchange. Plant Physiol. 2017, 175, 3–4. [Google Scholar] [CrossRef] [Green Version]
- Hauser, F.; Waadt, R.; Schroeder, J.I. Evolution of Abscisic Acid Synthesis and Signaling Mechanisms. Curr. Biol. 2011, 21, R346–R355. [Google Scholar] [CrossRef] [Green Version]
- Kimura, Y.; Aoki, S.; Ando, E.; Kitatsuji, A.; Watanabe, A.; Ohnishi, M.; Takahashi, K.; Inoue, S.; Nakamichi, N.; Tamada, Y.; et al. A flowering integrator, SOC1, affects stomatal opening in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 640–649. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, T.; Ono, N.; Hayashi, Y.; Morimoto, S.; Nakamura, S.; Soda, M.; Kato, Y.; Ohnishi, M.; Nakano, T.; Inoue, S.; et al. FLOWERING LOCUS T Regulates Stomatal Opening. Curr. Biol. 2011, 21, 1232–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ando, E.; Ohnishi, M.; Wang, Y.; Matsushita, T.; Watanabe, A.; Hayashi, Y.; Fujii, M.; Ma, J.F.; Inoue, S.; Kinoshita, T. TWIN SISTER OF FT, GIGANTEA, and CONSTANS Have a Positive But Indirect Effect on Blue Light-Induced Stomatal Opening in Arabidopsis. Plant Physiol. 2013, 162, 1529–1538. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.L. ELF3 Encodes a Circadian Clock-Regulated Nuclear Protein That Functions in an Arabidopsis PHYB Signal Transduction Pathway. Plant Cell 2001, 13, 1293–1304. [Google Scholar]
- Chen, C.; Xiao, Y.-G.; Li, X.; Ni, M. Light-Regulated Stomatal Aperture in Arabidopsis. Mol. Plant 2012, 5, 566–572. [Google Scholar] [CrossRef] [Green Version]
- Cominelli, E.; Galbiati, M.; Vavasseur, A.; Conti, L.; Sala, T.; Vuylsteke, M.; Leonhardt, N.; Dellaporta, S.L.; Tonelli, C. A Guard-Cell-Specific MYB Transcription Factor Regulates Stomatal Movements and Plant Drought Tolerance. Curr. Biol. 2005, 15, 1196–1200. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.-K.; Dubos, C.; Dodd, I.C.; Holroyd, G.H.; Hetherington, A.M.; Campbell, M.M. AtMYB61, an R2R3-MYB Transcription Factor Controlling Stomatal Aperture in Arabidopsis thaliana. Curr. Biol. 2005, 15, 1201–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Romero, J.L.; Inostroza-Blancheteau, C.; Orellana, D.; Aquea, F.; Reyes-Díaz, M.; Gil, P.M.; Matte, J.P.; Arce-Johnson, P. Stomata regulation by tissue-specific expression of the Citrus sinensis MYB61 transcription factor improves water-use efficiency in Arabidopsis. Plant Physiol. Biochem. 2018, 130, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Kie bowicz-Matuk, A.; Rey, P.; Rorat, T. Interplay between circadian rhythm, time of the day and osmotic stress constraints in the regulation of the expression of a Solanum Double B-box gene. Ann. Bot. 2014, 113, 831–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcolino-Gomes, J.; Rodrigues, F.A.; Fuganti-Pagliarini, R.; Bendix, C.; Nakayama, T.J.; Celaya, B.; Molinari, H.B.C.; de Oliveira, M.C.N.; Harmon, F.G.; Nepomuceno, A. Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS ONE 2014, 9, e86402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkins, O.; Bräutigam, K.; Campbell, M.M. Time of day shapes Arabidopsis drought transcriptomes. Plant J. 2010, 63, 715–727. [Google Scholar] [CrossRef]
- Li, J.; Liu, Y.-H.; Zhang, Y.; Chen, C.; Yu, X.; Yu, S.-W. Drought stress modulates diurnal oscillations of circadian clock and drought-responsive genes in Oryza sativa L. Yi Chuan = Hereditas 2017, 39, 837–846. [Google Scholar]
- Mizuno, T.; Yamashino, T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: Insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008, 49, 481–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Song, Y.; Zhang, H.; Zhang, D. Genome-Wide Analysis of Gene Expression in Response to Drought Stress in Populus simonii. Plant Mol. Biol. Rep. 2013, 31, 946–962. [Google Scholar] [CrossRef]
- Syed, N.H.; Prince, S.J.; Mutava, R.N.; Patil, G.; Li, S.; Chen, W.; Babu, V.; Joshi, T.; Khan, S.; Nguyen, H.T. Core clock, SUB1, and ABAR genes mediate flooding and drought responses via alternative splicing in soybean. J. Exp. Bot. 2015, 66, 7129–7149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valim, H.F.; McGale, E.; Yon, F.; Halitschke, R.; Fragoso, V.; Schuman, M.C.; Baldwin, I.T. The Clock Gene TOC1 in Shoots, Not Roots, Determines Fitness of Nicotiana attenuata under Drought. Plant Physiol. 2019, 181, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Kim, W.-Y.; Cha, J.-Y.; Park, H.J.; Shin, G.; Park, J.; Lim, C.J.; Chun, H.J.; Li, N.; Kim, D.H.; et al. The GIGANTEA-ENHANCED EM LEVEL Complex Enhances Drought Tolerance via Regulation of Abscisic Acid Synthesis. Plant Physiol. 2020, 184, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kudo, T.; Makita, N.; Kiba, T.; Kinoshita, T.; Sakakibara, H. Flowering time control in rice by introducing Arabidopsis clock-associated PSEUDO-RESPONSE REGULATOR 5. Biosci. Biotechnol. Biochem. 2020, 84, 970–979. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, A.; Shin, J.; Davis, S.J. Abiotic stress and the plant circadian clock. Plant Signal. Behav. 2011, 6, 223–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsui, A.; Ishida, J.; Morosawa, T.; Mochizuki, Y.; Kaminuma, E.; Endo, T.A.; Okamoto, M.; Nambara, E.; Nakajima, M.; Kawashima, M.; et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008, 49, 1135–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, F.C.; Skeffington, A.W.; Gardner, M.J.; Webb, A.A.R. Interactions between circadian and hormonal signalling in plants. Plant Mol. Biol. 2009, 69, 419–427. [Google Scholar] [CrossRef]
- Pizzio, G.A. Abscisic Acid Machinery Is under Circadian Clock Regulation at Multiple Levels. Stresses 2022, 2, 65–78. [Google Scholar] [CrossRef]
- Caldeira, C.F.; Jeanguenin, L.; Chaumont, F.; Tardieu, F. Circadian rhythms of hydraulic conductance and growth are enhanced by drought and improve plant performance. Nat. Commun. 2014, 5, 5365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, M.; Ding, G.; Cai, Q. The transcriptomic responses of Pinus massoniana to drought stress. Forests 2018, 9, 326. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, Y.; Abe, H.; Takase, T.; Kobayashi, M.; Kiyosue, T. Overexpression of LOV KELCH PROTEIN 2 confers dehydration tolerance and is associated with enhanced expression of dehydration-inducible genes in Arabidopsis thaliana. Plant Cell Rep. 2015, 34, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Villarreal, A.; Shin, J.; Bujdoso, N.; Obata, T.; Neumann, U.; Du, S.X.; Ding, Z.; Davis, A.M.; Shindo, T.; Schmelzer, E.; et al. TIME for COFFEE is an essential component in the maintenance of metabolic homeostasis in Arabidopsis thaliana. Plant J. 2013, 76, 188–200. [Google Scholar] [PubMed]
- Fornara, F.; De Montaigu, A.; Sánchez-Villarreal, A.; Takahashi, Y.; Ver Loren Van Themaat, E.; Huettel, B.; Davis, S.J.; Coupland, G. The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 2015, 81, 695–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubois, M.; Claeys, H.; Van den Broeck, L.; Inzé, D. Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant Cell Environ. 2017, 40, 180–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, D.X.; Manchester, L.C.; Hardeland, R.; Lopez-Burillo, S.; Mayo, J.C.; Sainz, R.M.; Reiter, R.J. Melatonin: A hormone, a tissue factor, an autocoid, a paracoid, and an antioxidant vitamin. J. Pineal Res. 2003, 34, 75–78. [Google Scholar] [CrossRef]
- Ruggiero, B.; Koiwa, H.; Manabe, Y.; Quist, T.M.; Inan, G.; Saccardo, F.; Joly, R.J.; Hasegawa, P.M.; Bressan, R.A.; Maggio, A. Uncoupling the effects of abscisic acid on plant growth and water relations. Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in arabidopsis. Plant Physiol. 2004, 136, 3134–3147. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Yang, X.; Weston, D.J.; Chen, J.G. Abscisic Acid Receptors: Past, Present and Future. J. Integr. Plant Biol. 2011, 53, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.F. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.K. Osmotic stress signaling via protein kinases. Cell. Mol. Life Sci. 2012, 69, 3165–3173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khianpho, O.; Lontom, W.; Theerakulpisut, P. Expression of Circadian Clock-related Genes in Flag Leaves of KDML 105 Rice Cultivar under Osmotic Stress Conditions. In Proceedings of the National and International Graduate Research Conference (BMP24), Khon Kaen, Thailand, 15 January 2016; pp. 579–586. Available online: https://gsbooks.gs.kku.ac.th/59/ingrc2016/pdf/BMP24.pdf (accessed on 15 September 2021).
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.-H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 Are Transcriptional Repressors in the Arabidopsis Circadian Clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalmani, A.; Jing, X.Q.; Shi, Y.; Muhammad, I.; Zhou, M.R.; Wei, X.Y.; Chen, Q.Q.; Li, W.Q.; Liu, W.T.; Chen, K.M. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genom. 2019, 20, 27. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Yuan, L.; Sun, Q.; Zhang, S.; Wang, X. A B-box zinc finger protein, MdBBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Plant Mol. Biol. 2019, 99, 437–447. [Google Scholar] [CrossRef]
- Talar, U.; Kieøbowicz-Matuk, A.; Czarnecka, J.; Rorat, T. Genome-wide survey of B-box proteins in potato (Solanum tuberosum)-Identification, characterization and expression patterns during diurnal cycle, etiolation and deetiolation. PLoS ONE 2017, 12, e0177471. [Google Scholar] [CrossRef] [Green Version]
- Min, J.H.; Chung, J.S.; Lee, K.H.; Kim, C.S. The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 313–324. [Google Scholar] [CrossRef]
- Kurup, S.; Jones, H.D.; Holdsworth, M.J. Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 2000, 21, 143–155. [Google Scholar] [CrossRef]
- Xu, D.; Li, J.; Gangappa, S.N.; Hettiarachchi, C.; Lin, F.; Andersson, M.X.; Jiang, Y.; Deng, X.W.; Holm, M. Convergence of Light and ABA Signaling on the ABI5 Promoter. PLoS Genet. 2014, 10, e1004197. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, X.; Aiwaili, P.; Mu, X.; Zhao, M.; Zhao, J.; Cheng, L.; Ma, C.; Gao, J.; Hong, B. A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 2020, 103, 1783–1795. [Google Scholar] [CrossRef]
- Wang, Y.; Zhai, Z.; Sun, Y.; Feng, C.; Peng, X.; Zhang, X.; Xiao, Y.; Zhou, X.; Wang, W.; Jiao, J.; et al. Genome-wide identification of the b-box genes that respond to multiple ripening related signals in sweet cherry fruit. Int. J. Mol. Sci. 2021, 22, 1622. [Google Scholar] [CrossRef]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; Li, H.; Ouyang, B. Genomic organization, phylogenetic and expression analysis of the B-Box gene family in tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The Rice B-Box Zinc Finger Gene Family: Genomic Identification, Characterization, Expression Profiling and Diurnal Analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Jiang, Y.; Li, J.; Lin, F.; Holm, M.; Deng, X.W. BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc. Natl. Acad. Sci. USA 2016, 113, 7655–7660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, T.; Ito, S.; Nakamichi, N.; Niwa, Y.; Murakami, M.; Yamashino, T.; Mizuno, T. The common function of a novel subfamily of B-box zinc finger proteins with reference to circadian-associated events in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2008, 72, 1539–1549. [Google Scholar] [CrossRef] [Green Version]
- Piechulla, B.; Merforth, N.; Rudolph, B. Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol. Biol. 1998, 38, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Kiełbowicz-Matuk, A.; Czarnecka, J.; Banachowicz, E.; Rey, P.; Rorat, T. Solanum tuberosum ZPR1 encodes a light-regulated nuclear DNA-binding protein adjusting the circadian expression of StBBX24 to light cycle. Plant Cell Environ. 2017, 40, 424–440. [Google Scholar] [CrossRef]
- Wei, H.; Wang, P.; Chen, J.; Li, C.; Wang, Y.; Yuan, Y.; Fang, J.; Leng, X. Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol. 2020, 20, 72. [Google Scholar] [CrossRef]
- Cao, Y.; Meng, D.; Han, Y.; Chen, T.; Jiao, C.; Chen, Y.; Jin, Q.; Cai, Y. Comparative analysis of B-BOX genes and their expression pattern analysis under various treatments in Dendrobium officinale. BMC Plant Biol. 2019, 19, 245. [Google Scholar] [CrossRef]
- Lyu, G.; Li, D.; Li, S. Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana. Plant Signal. Behav. 2020, 15, 1782647. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the Apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 2018, 293, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Zhang, Y.; Deng, Y.; Chen, G.; Yu, Y.; Wei, Q. Genomic identification and expression analysis of the BBX transcription factor gene family in Petunia hybrida. Mol. Biol. Rep. 2020, 47, 6027–6041. [Google Scholar] [CrossRef]
- Jin, H.; Xing, M.; Cai, C.; Li, S. B-box Proteins in Arachis duranensis: Genome-wide characterization and expression profiles analysis. Agronomy 2020, 10, 23. [Google Scholar] [CrossRef] [Green Version]

| Species | Gene/Mutant | Drought-Response Findings | Ref. |
|---|---|---|---|
| Populus simonii | LHY | Decrease in expression | [94] |
| Soybean | TOC1 LUX, ELF4 | Down-regulation of evening-specific components of the clock | [90] |
| Soybean | PRR7 TOC1 | Expression deviated from normal | [95] |
| Soybean | PRR3 | Drought-specific splicing pattern was observed | [95] |
| Rice | OsCCA1/LHY | Increased transcription at 4 a.m. | [118] |
| Rice | OsPRR1/TOC1 | Disruption of expression | [118] |
| Gossypium hirsutum | PRR gene family | GhPRR5a, b, d, GhPRR3a, and GhPRR3c may be involved in drought response | [25] |
| Arabidopsis thaliana | DREB1/CBF | [98] | |
| Rice | idr1-1 | Increase in ROS production and ROS-scavenging enzyme | [119] |
| Species | BBX Genes with ABRE and Circadian Motif | Ref. |
|---|---|---|
| Tomato | BBX3, BBX5, BBX7, BBX17, BBX18, BBX19, BBX23, BBX26, BBX28 | [128] |
| Grapevine | BBX3, BBX5, BBX6, BBX11, BBX12, BBX13, BBX21 | [135] |
| Dendrobium officinale | BBX1, BBX4, BBX13 | [136] |
| Phalaenopsis equestris | BBX03, BBX04, BBX05, BBX06, BBX07, BBX11, BBX13, BBX14, BBX16 | [136] |
| Arabidopsis thaliana | BBX2, BBX14, BBX17, BBX21 | [137] |
| Apple | BBX2, BBX3, BBX4, BBX5, BBX6, BBX8, BBX9, BBX10, BBX11, BBX19, BBX21, BBX22, BBX23, BBX25, BBX32, BBX35, BBX41, BBX43, BBX44, BBX48 | [138] |
| Petunia | BBX4, BBX5, BBX6, BBX9, BBX10, BBX12, BBX22, BBX28 | [139] |
| Arachis duranensis | BBX1, BBX11, BBX13, BBX15, BBX16, BBX22 | [140] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yari Kamrani, Y.; Shomali, A.; Aliniaeifard, S.; Lastochkina, O.; Moosavi-Nezhad, M.; Hajinajaf, N.; Talar, U. Regulatory Role of Circadian Clocks on ABA Production and Signaling, Stomatal Responses, and Water-Use Efficiency under Water-Deficit Conditions. Cells 2022, 11, 1154. https://doi.org/10.3390/cells11071154
Yari Kamrani Y, Shomali A, Aliniaeifard S, Lastochkina O, Moosavi-Nezhad M, Hajinajaf N, Talar U. Regulatory Role of Circadian Clocks on ABA Production and Signaling, Stomatal Responses, and Water-Use Efficiency under Water-Deficit Conditions. Cells. 2022; 11(7):1154. https://doi.org/10.3390/cells11071154
Chicago/Turabian StyleYari Kamrani, Yousef, Aida Shomali, Sasan Aliniaeifard, Oksana Lastochkina, Moein Moosavi-Nezhad, Nima Hajinajaf, and Urszula Talar. 2022. "Regulatory Role of Circadian Clocks on ABA Production and Signaling, Stomatal Responses, and Water-Use Efficiency under Water-Deficit Conditions" Cells 11, no. 7: 1154. https://doi.org/10.3390/cells11071154
APA StyleYari Kamrani, Y., Shomali, A., Aliniaeifard, S., Lastochkina, O., Moosavi-Nezhad, M., Hajinajaf, N., & Talar, U. (2022). Regulatory Role of Circadian Clocks on ABA Production and Signaling, Stomatal Responses, and Water-Use Efficiency under Water-Deficit Conditions. Cells, 11(7), 1154. https://doi.org/10.3390/cells11071154









