The Landscape of Novel Expressed Chimeric RNAs in Rheumatoid Arthritis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of RNA-Seq Data
2.2. Identification of Chimeric RNAs from RNA-Seq Data
2.3. Differential Gene Expression Analysis
2.4. Annotation and Enrichment Analysis of the Parental Genes of Recurrent Chimeric RNAs in RA
2.5. Classification of Recurrent Chimeric RNAs into Coding and Non-Coding RNAs
3. Results
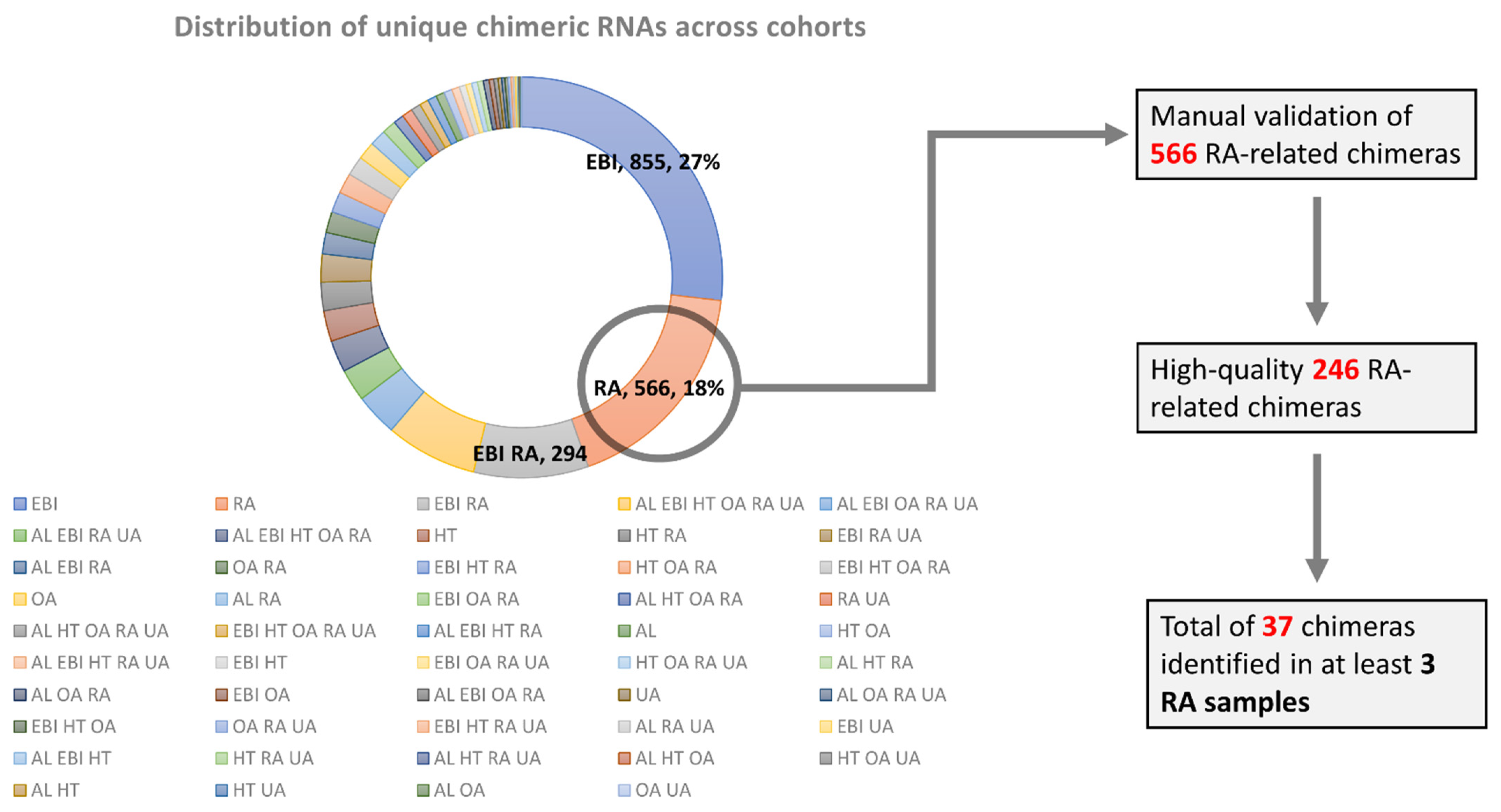
3.1. Identification of Chimeric RNAs across Normal and Arthritis Cohorts
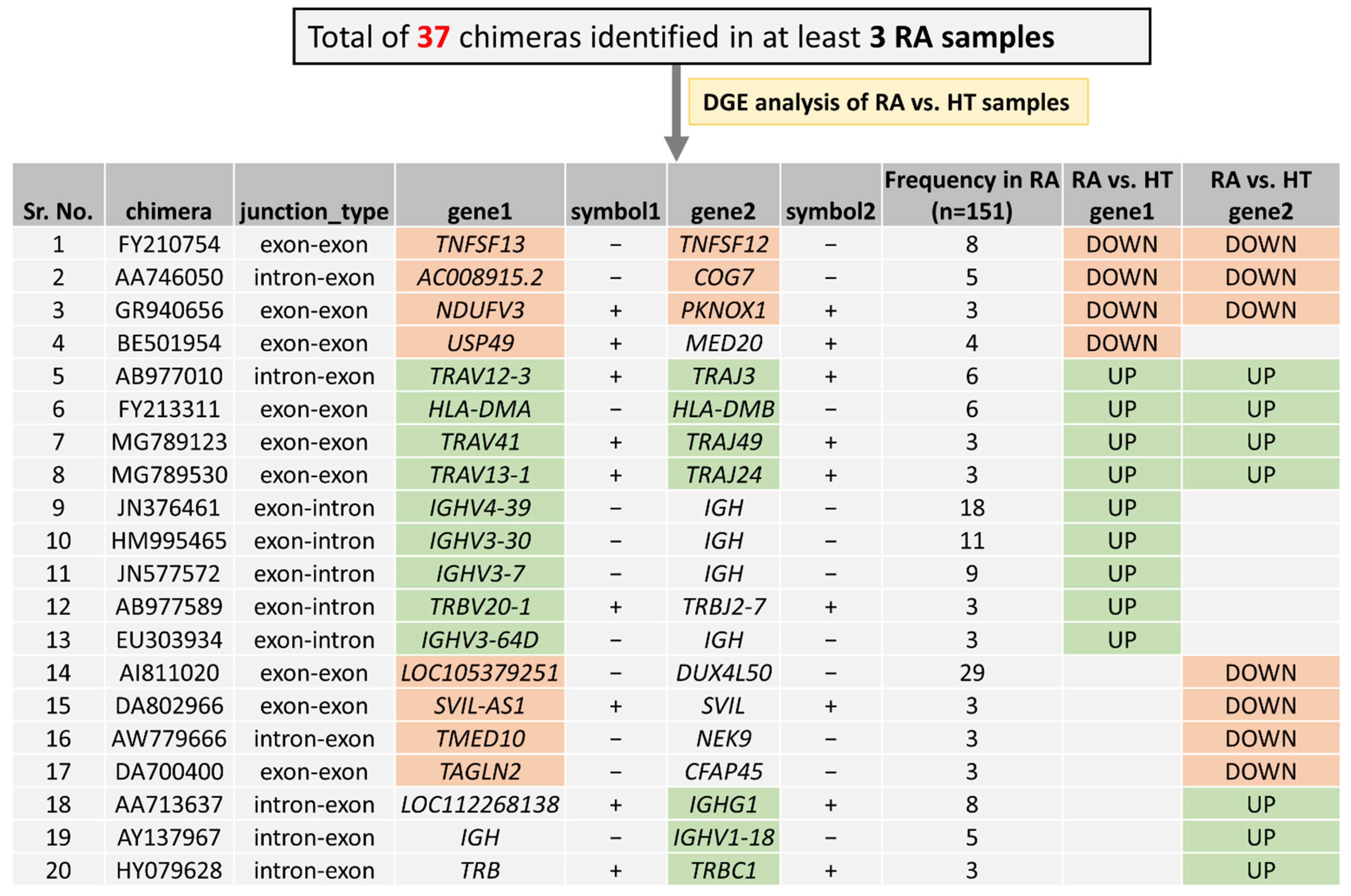
3.2. Expression Analysis of Recurrent Chimeric RNAs in RA Patients
3.3. Enrichment Analysis of the Parental Genes of RA-Specific Recurrent Chimeric RNAs
3.4. Differential Gene Expression Analysis of the Parental Genes of Recurrent Chimeric RNAs
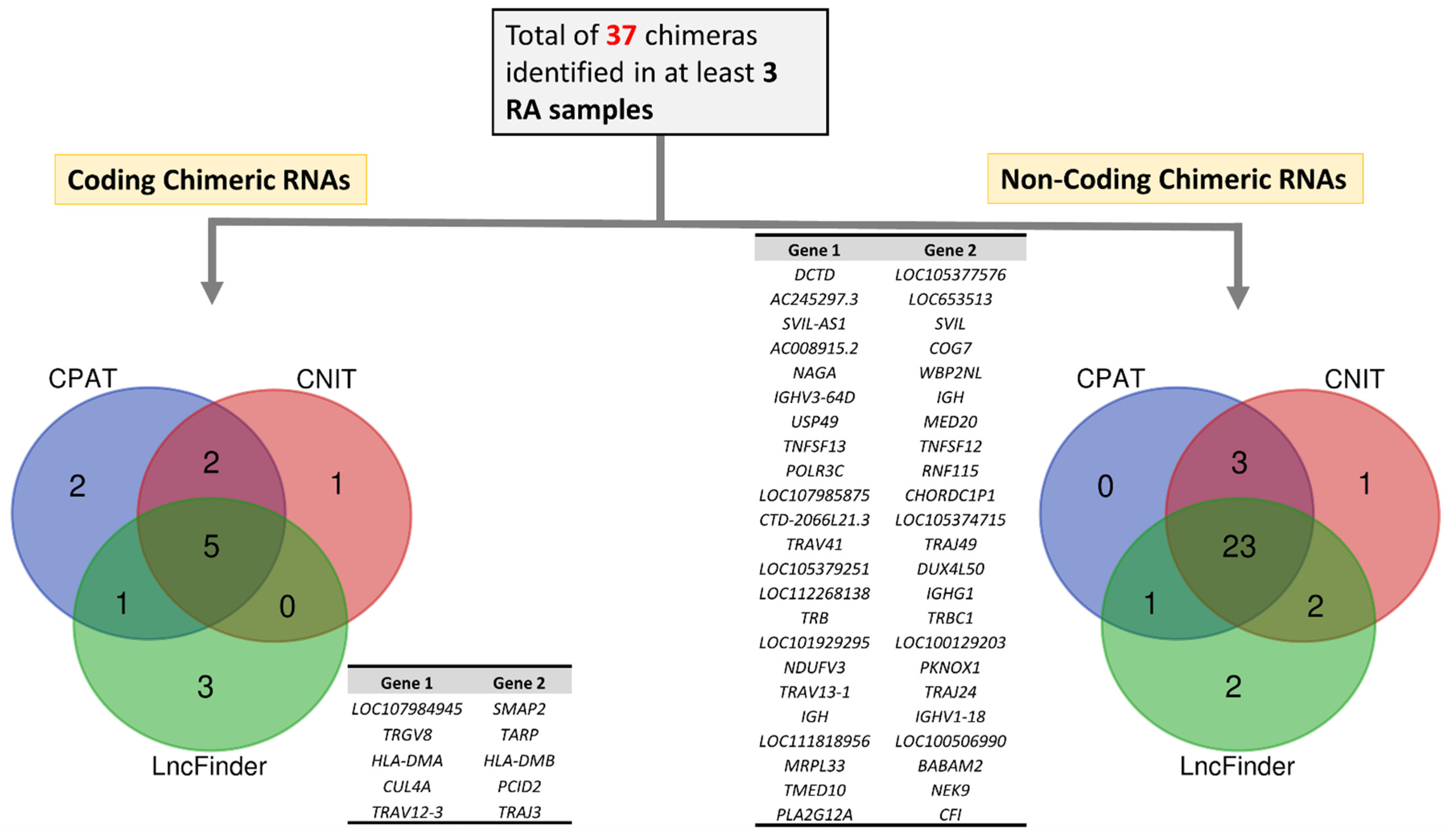
3.5. Functional Classification of RA-Specific Recurrent Chimeric RNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chwalenia, K.; Facemire, L.; Li, H. Chimeric RNAs in cancer and normal physiology. Wiley Interdiscip. Rev. RNA 2017, 8, e1427. [Google Scholar] [CrossRef] [PubMed]
- Mitelman, F.; Johansson, B.; Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 2007, 7, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Kannan, K.; Wang, L.; Wang, J.; Ittmann, M.M.; Li, W.; Yen, L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc. Natl. Acad. Sci. USA. 2011, 108, 9172–9177. [Google Scholar] [CrossRef] [Green Version]
- Asmann, Y.W.; Necela, B.M.; Kalari, K.R.; Hossain, A.; Baker, T.R.; Carr, J.M.; Davis, C.; Getz, J.E.; Hostetter, G.; Li, X.; et al. Detection of redundant fusion transcripts as biomarkers or disease-specific therapeutic targets in breast cancer. Cancer Res. 2012, 72, 1921–1928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, M.; Makinoshima, H.; Matsumoto, S.; Suzuki, A.; Mimaki, S.; Matsushima, K.; Yoh, K.; Goto, K.; Suzuki, Y.; Ishii, G.; et al. Identification of a lung adenocarcinoma cell line with CCDC6-RET fusion gene and the effect of RET inhibitors in vitro and in vivo. Cancer Sci. 2013, 104, 896–903. [Google Scholar] [CrossRef]
- Nome, T.; Thomassen, G.O.S.; Bruun, J.; Ahlquist, T.; Bakken, A.C.; Hoff, A.M.; Rognum, T.; Nesbakken, A.; Lorenz, S.; Sun, J.; et al. Common fusion transcripts identified in colorectal cancer cell lines by high-throughput RNA sequencing. Transl. Oncol. 2013, 6, 546-IN5. [Google Scholar] [CrossRef] [Green Version]
- Frenkel-Morgenstern, M.; Lacroix, V.; Ezkurdia, I.; Levin, Y.; Gabashvili, A.; Prilusky, J.; Del Pozo, A.; Tress, M.; Johnson, R.; Guigo, R.; et al. Chimeras taking shape: Potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012, 22, 1231–1242. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Heng, H.H.; Frenkel-Morgenstern, M. Emerging Role of Chimeric RNAs in Cell Plasticity and Adaptive Evolution of Cancer Cells. Cancers 2021, 13, 4328. [Google Scholar] [CrossRef]
- Zhu, Y.-C.; Wang, W.-X.; Zhang, Q.-X.; Xu, C.-W.; Zhuang, W.; Du, K.-Q.; Chen, G.; Lv, T.-F.; Song, Y. The KIF5B-RET Fusion Gene Mutation as a Novel Mechanism of Acquired EGFR Tyrosine Kinase Inhibitor Resistance in Lung Adenocarcinoma. Clin. Lung Cancer 2019, 20, e73–e76. [Google Scholar] [CrossRef]
- Ma, Y.; Miao, Y.; Peng, Z.; Sandgren, J.; De Ståhl, T.D.; Huss, M.; Lennartsson, L.; Liu, Y.; Nistér, M.; Nilsson, S.; et al. Identification of mutations, gene expression changes and fusion transcripts by whole transcriptome RNAseq in docetaxel resistant prostate cancer cells. Springerplus 2016, 5, 1861. [Google Scholar] [CrossRef] [Green Version]
- Christie, E.L.; Pattnaik, S.; Beach, J.; Copeland, A.; Rashoo, N.; Fereday, S.; Hendley, J.; Alsop, K.; Brady, S.L.; Lamb, G.; et al. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nat. Commun. 2019, 10, 1295. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Qin, F.; Li, H. Chimeric RNAs and their implications in cancer. Curr. Opin. Genet. Dev. 2018, 48, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Neckles, C.; Sundara Rajan, S.; Caplen, N.J. Fusion transcripts: Unexploited vulnerabilities in cancer? Wiley Interdiscip. Rev. RNA 2020, 11, e1562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frenkel-Morgenstern, M.; Gorohovski, A.; Tagore, S.; Sekar, V.; Vazquez, M.; Valencia, A. ChiPPI: A novel method for mapping chimeric protein-protein interactions uncovers selection principles of protein fusion events in cancer. Nucleic Acids Res. 2017, 45, 7094–7105. [Google Scholar] [CrossRef]
- Latysheva, N.S.; Babu, M.M. Molecular Signatures of Fusion Proteins in Cancer. ACS Pharmacol. Transl. Sci. 2019, 2, 122–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Li, X.; Li, H. Gene fusions and chimeric RNAs, and their implications in cancer. Genes Dis. 2019, 6, 385–390. [Google Scholar] [CrossRef]
- Qin, F.; Zhang, Y.; Liu, J.; Li, H. SLC45A3-ELK4 functions as a long non-coding chimeric RNA. Cancer Lett. 2017, 404, 53–61. [Google Scholar] [CrossRef]
- Mukherjee, S.; Detroja, R.; Balamurali, D.; Matveishina, E.; Medvedeva, Y.A.; Valencia, A.; Gorohovski, A.; Frenkel-Morgenstern, M. Computational analysis of sense-antisense chimeric transcripts reveals their potential regulatory features and the landscape of expression in human cells. NAR Genom. Bioinforma 2021, 3, lqab074. [Google Scholar] [CrossRef] [PubMed]
- Day, C.P.; Merlino, G.; Van Dyke, T. Preclinical Mouse Cancer Models: A Maze of Opportunities and Challenges. Cell 2015, 163, 39–53. [Google Scholar] [CrossRef] [Green Version]
- Frenkel-Morgenstern, M.; Valencia, A. Novel domain combinations in proteins encoded by chimeric transcripts. Bioinformatics 2012, 28, i67–i74. [Google Scholar] [CrossRef] [Green Version]
- Frenkel-Morgenstern, M.; Gorohovski, A.; Lacroix, V.; Rogers, M.; Ibanez, K.; Boullosa, C.; Leon, E.A.; Ben-Hur, A.; Valencia, A. ChiTaRS: A database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Res. 2013, 41, D142–D151. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Frenkel-Morgenstern, M. Evolutionary impact of chimeric RNAs on generating phenotypic plasticity in human cells. Trends Genet. 2021, 38, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Seldin, M.F.; Amos, C.I.; Ward, R.; Gregersen, P.K. The genetics revolution and the assault on rheumatoid arthritis. Arthritis Rheum. 1999, 42, 1071–1079. [Google Scholar] [CrossRef]
- Toonen, E.J.M.; Barrera, P.; Radstake, T.R.D.J.; Van Riel, P.L.C.M.; Scheffer, H.; Franke, B.; Coenen, M.J.H. Gene expression profiling in rheumatoid arthritis: Current concepts and future directions. Ann. Rheum. Dis. 2008, 67, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Van Der Pouw Kraan, T.C.T.M.; Wijbrandts, C.A.; Van Baarsen, L.G.M.; Voskuyl, A.E.; Rustenburg, F.; Baggen, J.M.; Ibrahim, S.M.; Fero, M.; Dijkmans, B.A.C.; Tak, P.P.; et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: Assignment of a type I interferon signature in a subpopulation of patients. Ann. Rheum. Dis. 2007, 66, 1008–1014. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, S.; Schrodi, S.J.; He, D. Molecular and Cellular Heterogeneity in Rheumatoid Arthritis: Mechanisms and Clinical Implications. Front. Immunol. 2021, 12, 790122. [Google Scholar] [CrossRef] [PubMed]
- Zendman, A.J.W.; van Venrooij, W.J.; Pruijn, G.J.M. Use and significance of anti-CCP autoantibodies in rheumatoid arthritis. Rheumatology 2006, 45, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Hueber, W.; Kidd, B.A.; Tomooka, B.H.; Lee, B.J.; Bruce, B.; Fries, J.F.; Sønderstrup, G.; Monach, P.; Drijfhout, J.W.; Van Venrooij, W.J.; et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005, 52, 2645–2655. [Google Scholar] [CrossRef]
- Lipsky, P.E.; van der Heijde, D.M.F.M.; St. Clair, E.W.; Furst, D.E.; Breedveld, F.C.; Kalden, J.R.; Smolen, J.S.; Weisman, M.; Emery, P.; Feldmann, M.; et al. Infliximab and Methotrexate in the Treatment of Rheumatoid Arthritis. N. Engl. J. Med. 2000, 343, 1594–1602. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.C.W.; Szczepański, L.; Szechiński, J.; Filipowicz-Sosnowska, A.; Emery, P.; Close, D.R.; Stevens, R.M.; Shaw, T. Efficacy of B-Cell–Targeted Therapy with Rituximab in Patients with Rheumatoid Arthritis. N. Engl. J. Med. 2004, 350, 2572–2581. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Jiang, P.; Guo, S.; Schrodi, S.J.; He, D. Apoptosis, Autophagy, NETosis, Necroptosis, and Pyroptosis Mediated Programmed Cell Death as Targets for Innovative Therapy in Rheumatoid Arthritis. Front. Immunol. 2021, 12, 809806. [Google Scholar] [CrossRef] [PubMed]
- Athar, A.; Füllgrabe, A.; George, N.; Iqbal, H.; Huerta, L.; Ali, A.; Snow, C.; Fonseca, N.A.; Petryszak, R.; Papatheodorou, I.; et al. ArrayExpress update—From bulk to single-cell expression data. Nucleic Acids Res. 2019, 47, D711–D715. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 14 February 2022).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Detroja, R.; Gorohovski, A.; Giwa, O.; Baum, G.; Frenkel-Morgenstern, M. ChiTaH: A fast and accurate tool for identifying known human chimeric sequences from high-throughput sequencing data. NAR Genom. Bioinforma 2021, 3, lqab112. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Guo, J.C.; Fang, S.S.; Wu, Y.; Zhang, J.H.; Chen, Y.; Liu, J.; Wu, B.; Wu, J.R.; Li, E.M.; Xu, L.Y.; et al. CNIT: A fast and accurate web tool for identifying protein-coding and long non-coding transcripts based on intrinsic sequence composition. Nucleic Acids Res. 2019, 47, W516–W522. [Google Scholar] [CrossRef] [Green Version]
- Han, S.; Liang, Y.; Ma, Q.; Xu, Y.; Zhang, Y.; Du, W.; Wang, C.; Li, Y. LncFinder: An integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property. Brief. Bioinform. 2019, 20, 2009–2027. [Google Scholar] [CrossRef] [PubMed]
- Taniue, K.; Akimitsu, N. Fusion genes and RNAs in cancer development. Non-Coding RNA 2021, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Firestein, G.S. Evolving concepts of rheumatoid arthritis. Nature 2003, 423, 356–361. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. Mechanism of Disease The Pathogenesis of Rheumatoid Arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef] [Green Version]
- Majithia, V.; Geraci, S.A. Rheumatoid Arthritis: Diagnosis and Management. Am. J. Med. 2007, 120, 936–939. [Google Scholar] [CrossRef]


| Description | No. of Samples | Average Raw Paired-End Reads | Average Trimmed Paired-End Reads |
|---|---|---|---|
| Rheumatoid Arthritis | 151 | 86,568,972 | 86,432,629 |
| Healthy | 28 | 84,069,634 | 83,923,697 |
| Osteoarthritis | 22 | 86,614,846 | 86,458,277 |
| Arthralgia | 10 | 90,722,882 | 90,617,905 |
| Undifferentiated Arthritis | 6 | 87,407,948 | 87,278,313 |
| Description | No. of Samples | No. of Chimeric RNAs |
|---|---|---|
| Rheumatoid Arthritis | 151 | 2102 |
| Healthy | 28 | 833 |
| Osteoarthritis | 22 | 856 |
| Arthralgia | 10 | 783 |
| Undifferentiated Arthritis | 6 | 671 |
| Healthy Human Tissues From EBI | 199 | 2066 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Detroja, R.; Mukherjee, S.; Frenkel-Morgenstern, M. The Landscape of Novel Expressed Chimeric RNAs in Rheumatoid Arthritis. Cells 2022, 11, 1092. https://doi.org/10.3390/cells11071092
Detroja R, Mukherjee S, Frenkel-Morgenstern M. The Landscape of Novel Expressed Chimeric RNAs in Rheumatoid Arthritis. Cells. 2022; 11(7):1092. https://doi.org/10.3390/cells11071092
Chicago/Turabian StyleDetroja, Rajesh, Sumit Mukherjee, and Milana Frenkel-Morgenstern. 2022. "The Landscape of Novel Expressed Chimeric RNAs in Rheumatoid Arthritis" Cells 11, no. 7: 1092. https://doi.org/10.3390/cells11071092
APA StyleDetroja, R., Mukherjee, S., & Frenkel-Morgenstern, M. (2022). The Landscape of Novel Expressed Chimeric RNAs in Rheumatoid Arthritis. Cells, 11(7), 1092. https://doi.org/10.3390/cells11071092








