Selective PPARα Modulator Pemafibrate and Sodium-Glucose Cotransporter 2 Inhibitor Tofogliflozin Combination Treatment Improved Histopathology in Experimental Mice Model of Non-Alcoholic Steatohepatitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Animal Treatment
2.2.1. Progression Prevention Study
2.2.2. Survival Study
2.3. Blood Parameter
2.4. Histology
2.5. RNA-Sequencing
2.6. Quantitative Real-Time PCR (qPCR)
2.7. LD Analysis
2.8. Statistical Analyses
3. Results
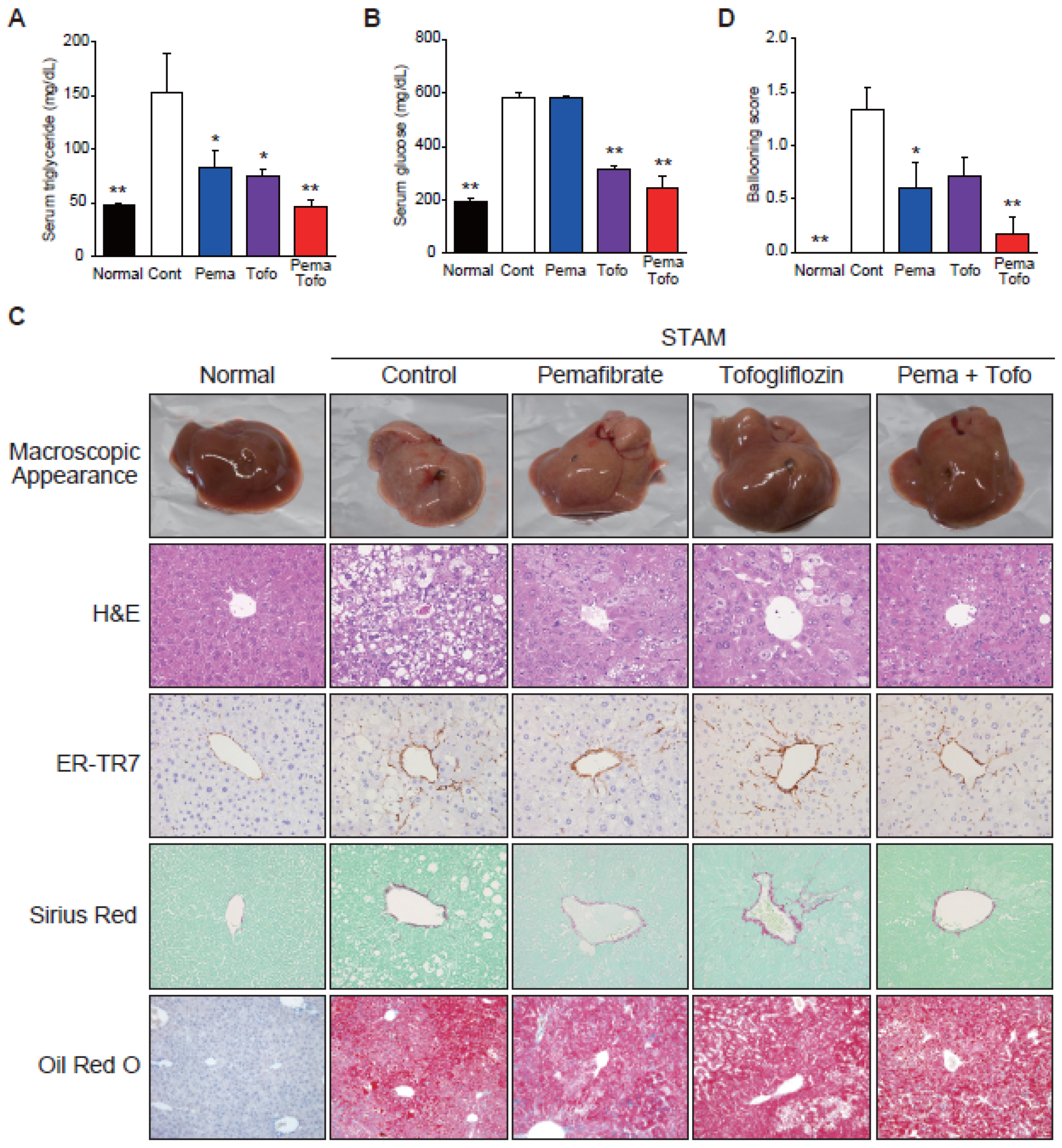
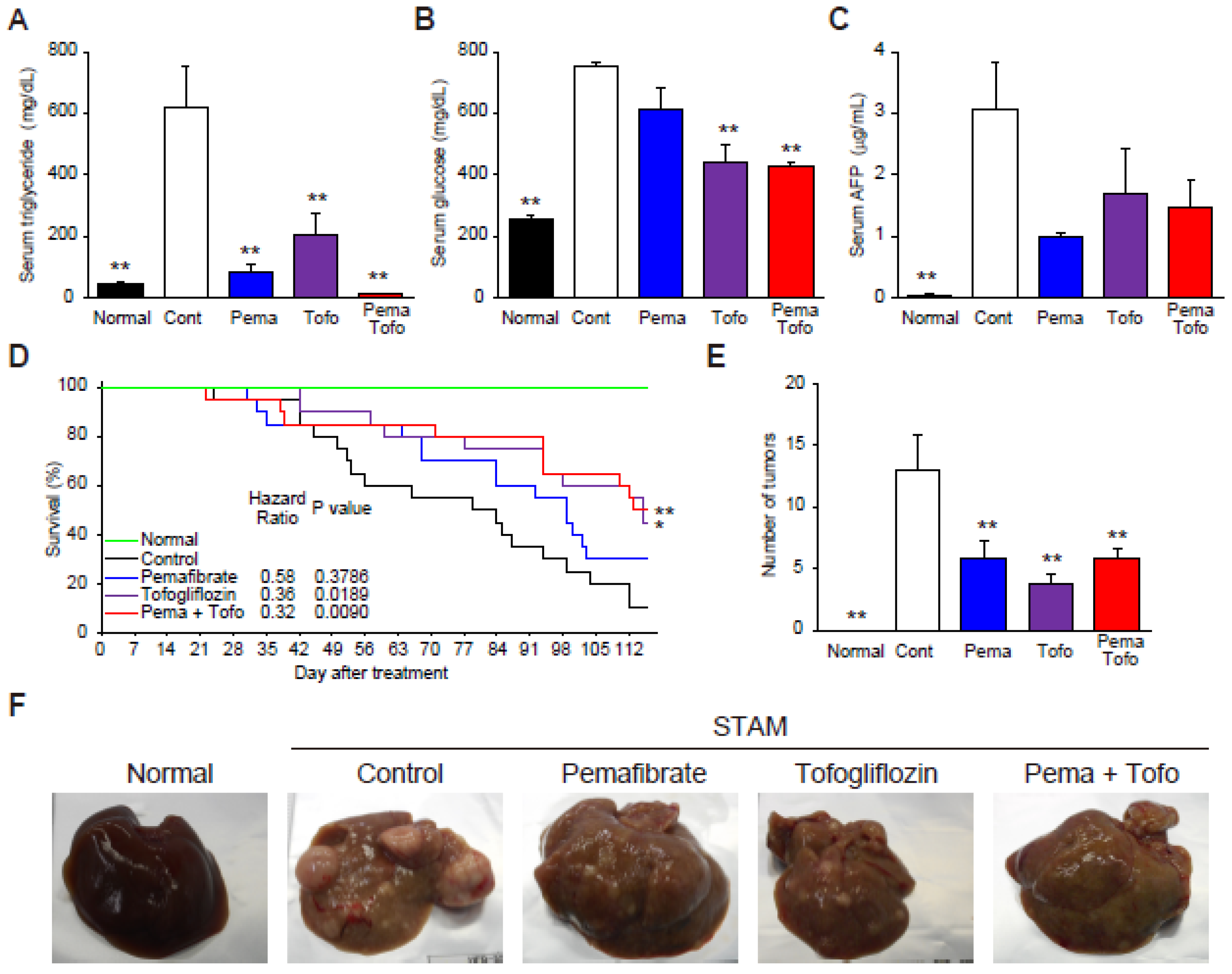
3.1. Pema and Tofo Combination Prevents Ballooning Degeneration of Hepatocytes
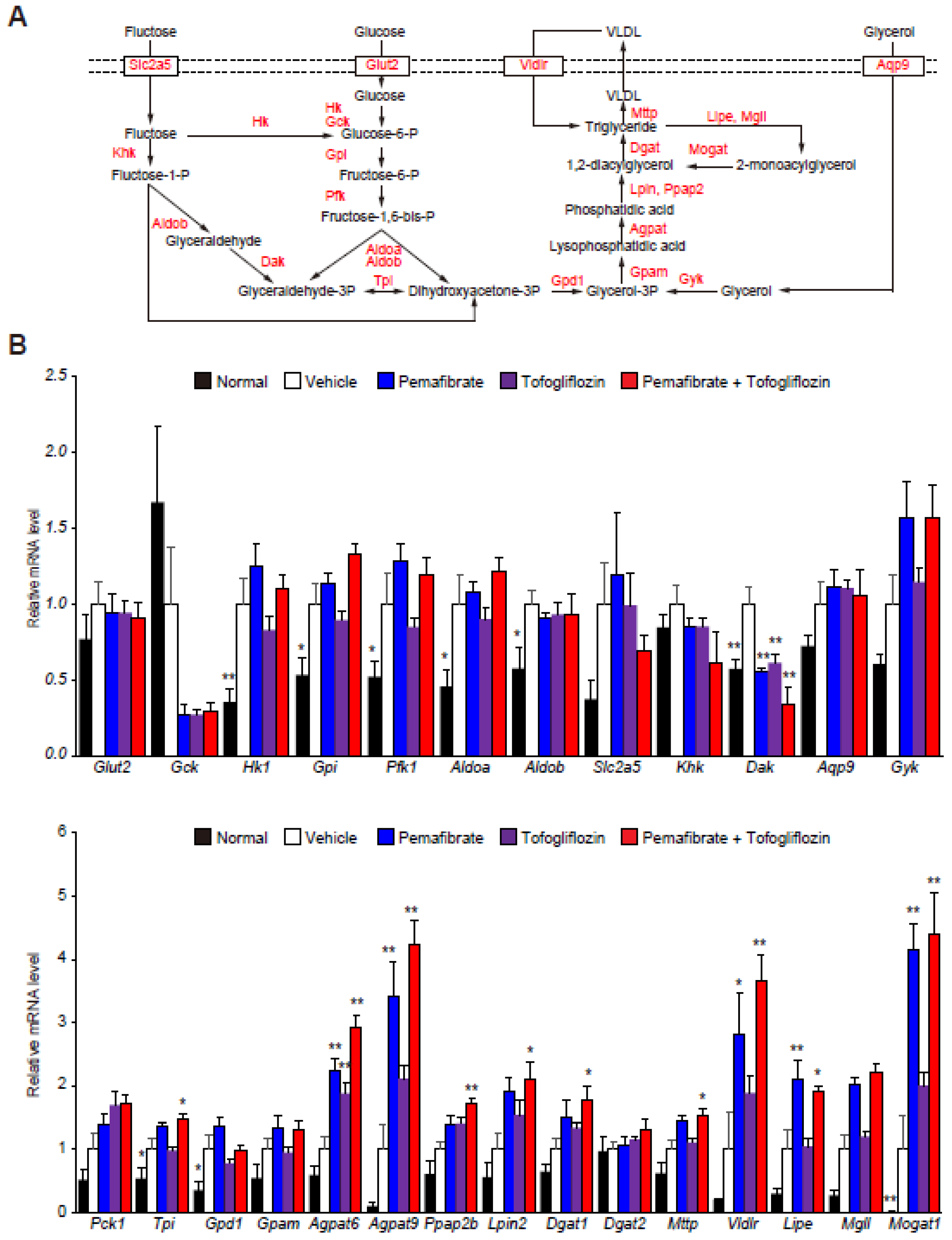
3.2. Pema and Tofo Combination Treatment Induces Lipolysis and Re-Esterification Cycles of TG in STAM Mouse Livers
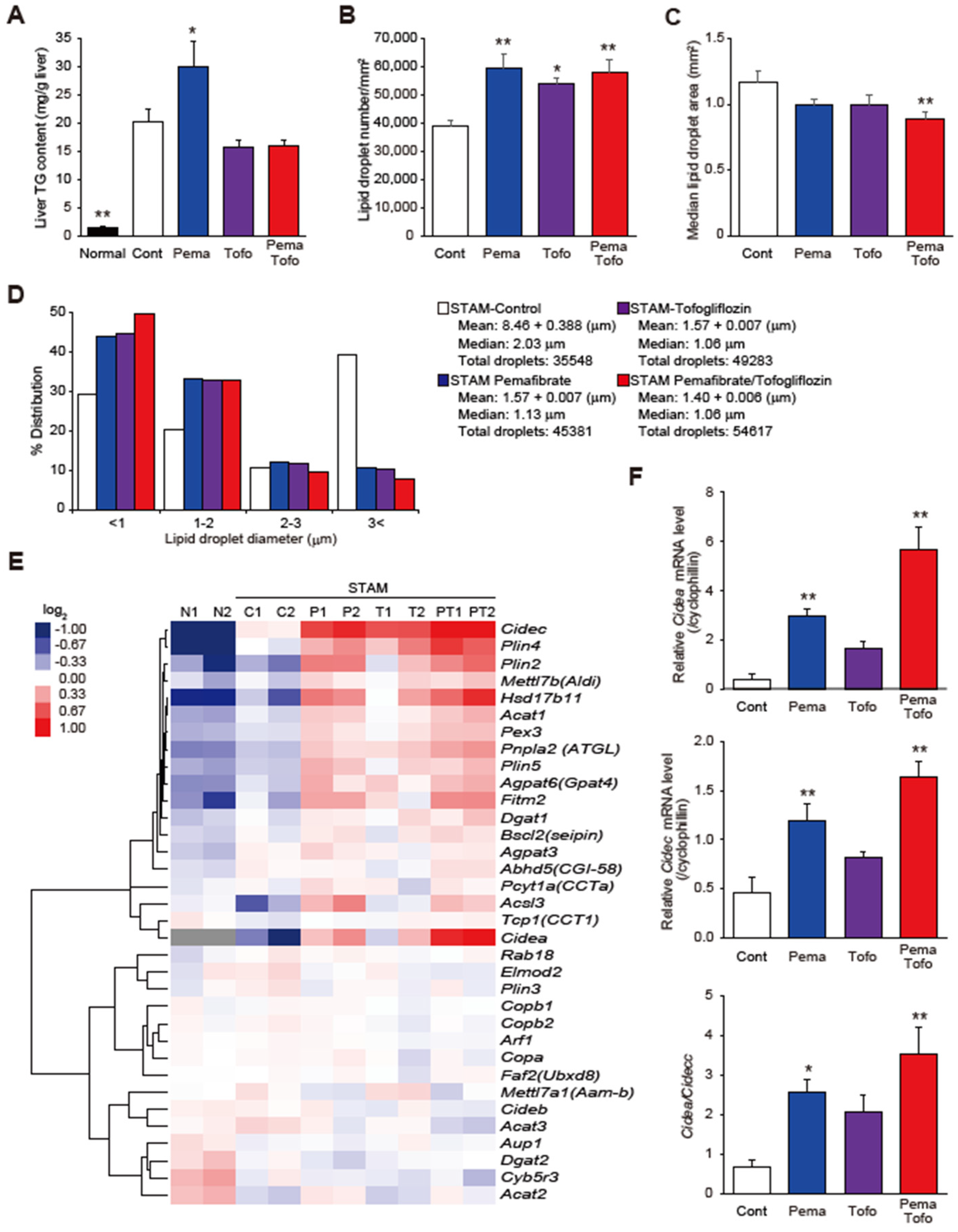
3.3. Pema and Tofo Combination Increases Small LDs in STAM Mouse Livers
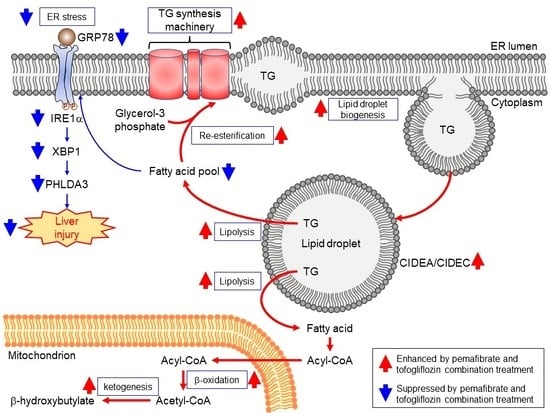
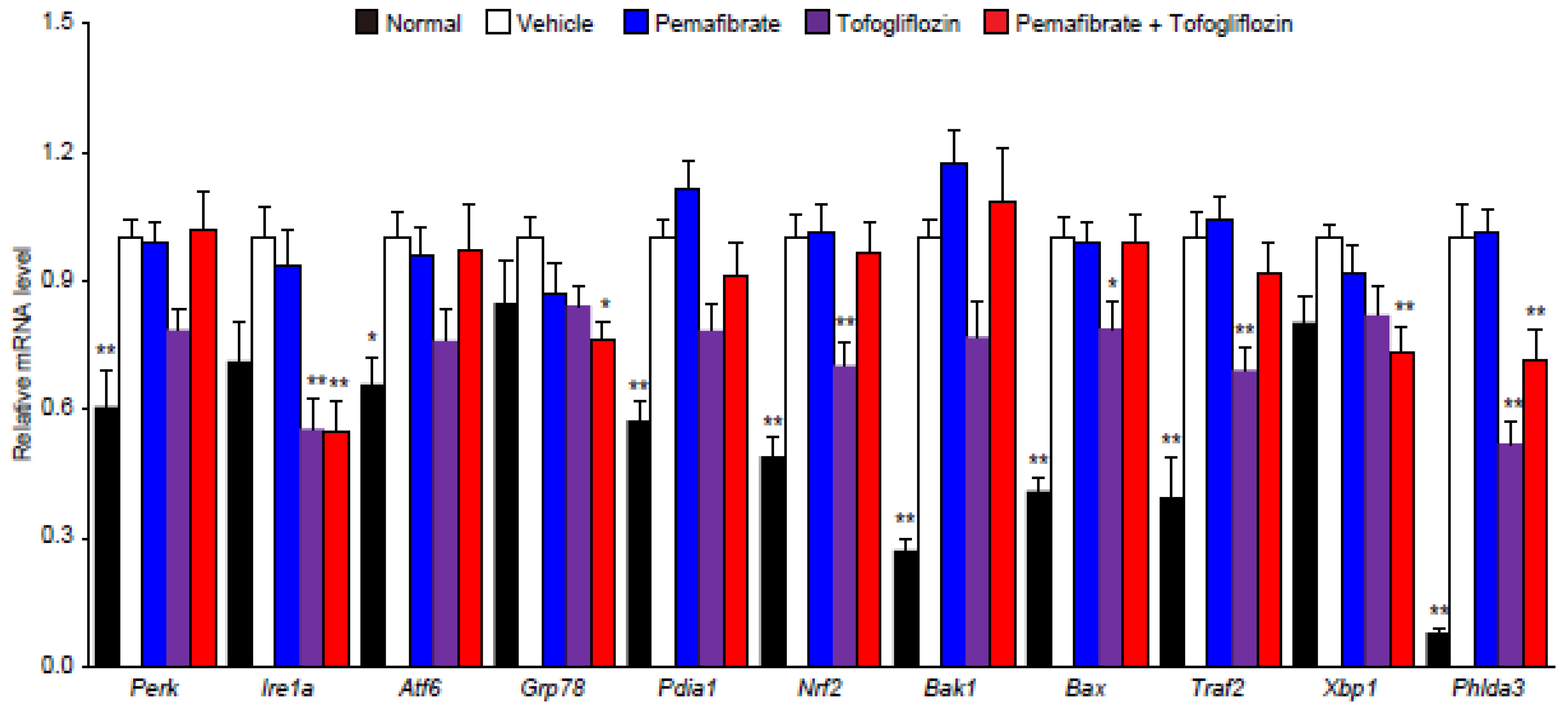
3.4. Pema and Tofo Combination Inhibits the IRE1α-XBP1-PHLDA3 Pathway
3.5. Pema and Tofo Combination Improves HCC-Related Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sheka, A.C.; Adeyi, O.; Thompson, J.; Hameed, B.; Crawford, P.A.; Ikramuddin, S. Nonalcoholic Steatohepatitis: A Review. JAMA 2020, 323, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Tilg, H.; Moschen, A.R.; Roden, M. NAFLD and diabetes mellitus. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Diehl, A.M. Nonalcoholic Steatohepatitis. Annu. Rev. Med. 2017, 68, 85–98. [Google Scholar] [CrossRef]
- Haas, J.T.; Francque, S.; Staels, B. Pathophysiology and Mechanisms of Nonalcoholic Fatty Liver Disease. Annu. Rev. Physiol. 2016, 78, 181–205. [Google Scholar] [CrossRef]
- Taylor, R.S.; Taylor, R.J.; Bayliss, S.; Hagström, H.; Nasr, P.; Schattenberg, J.M.; Ishigami, M.; Toyoda, H.; Wai-Sun Wong, V.; Peleg, N.; et al. Association Between Fibrosis Stage and Outcomes of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Gastroenterology 2020, 158, 1611–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walther, T.C.; Chung, J.; Farese, R.V., Jr. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 2017, 33, 491–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef]
- Scorletti, E.; Carr, R.M. A new perspective on NAFLD: Focusing on lipid droplets. J. Hepatol. 2021; in press. [Google Scholar] [CrossRef]
- Fu, S.; Watkins, S.M.; Hotamisligil, G.S. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012, 15, 623–634. [Google Scholar] [CrossRef] [Green Version]
- Colgan, S.M.; Hashimi, A.A.; Austin, R.C. Endoplasmic reticulum stress and lipid dysregulation. Expert Rev. Mol. Med. 2011, 13, e4. [Google Scholar] [CrossRef] [PubMed]
- Mashek, D.G. Hepatic lipid droplets: A balancing act between energy storage and metabolic dysfunction in NAFLD. Mol. Metab. 2021, 50, 101115. [Google Scholar] [CrossRef]
- Thiam, A.R.; Beller, M. The why, when and how of lipid droplet diversity. J. Cell Sci. 2017, 130, 315–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso, C.; Fernández-Ramos, D.; Varela-Rey, M.; Martínez-Arranz, I.; Navasa, N.; Van Liempd, S.M. Metabolomic identification of subtypes of nonalcoholic steatohepatitis. Gastroenterology 2017, 152, 1449–1461. [Google Scholar] [CrossRef] [Green Version]
- Chaurasia, B.; Tippetts, T.S.; Monibas, R.M.; Liu, J.; Li, Y.; Wang, L. Targeting a ceramide double bond improves insulin resistance and hepatic steatosis. Science 2019, 365, 386–392. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Abe, K.; Toma, T.; Nishikawa, M.; Ozawa, H.; Okuda, A.; Araki, T.; Oda, S.; Inoue, K.; Shibuya, K.; et al. Design and synthesis of highly potent and selective human peroxisome proliferator-activated receptor agonists. Bioorg. Med. Chem. Lett. 2007, 17, 4689–4693. [Google Scholar] [CrossRef]
- Fruchart, J.C. Peroxisome proliferator-activated receptor-α (PPARα): At the crossroads of obesity, diabetes and cardiovascular disease. Atherosclerosis 2009, 205, 1–8. [Google Scholar] [CrossRef]
- Fruchart, J.C. Selective peroxisome proliferator-activated α receptor modulators (SPPARM): The next generation of peroxisome proliferator-activated receptor α-agonists. Cardiovasc. Diabetol. 2013, 12, 82. [Google Scholar] [CrossRef] [Green Version]
- Fruchart, J.C. Pemafibrate (K-877), a novel selective peroxisome proliferator-activated receptor α modulator for management of atherogenic dyslipidaemia. Cardiovasc. Diabetol. 2017, 16, 124. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Han, S.I.; Murayama, Y.; Satoh, A.; Oikawa, F.; Ohno, H.; Osaki, Y.; Matsuzaka, T.; Sekiya, M.; Iwasaki, H.; et al. Selective peroxisome proliferator-activated receptor-α modulator K-877 efficiently activates the peroxisome proliferator-activated receptor-α pathway and improves lipid metabolism in mice. J. Diabetes Investig. 2017, 8, 446–452. [Google Scholar] [CrossRef] [Green Version]
- Raza-Iqbal, S.; Tanaka, T.; Anai, M.; Inagaki, T.; Matsumura, Y.; Ikeda, K.; Taguchi, A.; Gonzalez, F.J.; Sakai, J.; Kodama, T. Transcriptome Analysis of K-877 (a Novel Selective PPARα Modulator (SPPARMα))-Regulated Genes in Primary Human Hepatocytes and the Mouse Liver. J. Atheroscler. Thromb. 2015, 22, 754–772. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, Y.; Raza-Iqbal, S.; Tanaka, T.; Murakami, K.; Anai, M.; Osawa, T.; Matsumura, Y.; Sakai, J.; Kodama, T. Gene Expression Profiles Induced by a Novel Selective Peroxisome Proliferator-Activated Receptor α Modulator (SPPARMα) Pemafibrate. Int. J. Mol. Sci. 2019, 20, 5682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, Y.; Asahiyama, M.; Tanaka, T.; Yamamoto, S.; Murakami, K.; Kamiya, W.; Matsumura, Y.; Osawa, T.; Anai, M.; Fruchart, J.C.; et al. Pemafibrate, a selective PPARα modulator, prevents non-alcoholic steatohepatitis development without reducing the hepatic triglyceride content. Sci. Rep. 2020, 10, 7818. [Google Scholar] [CrossRef]
- Ferrannini, E.; Solini, A. SGLT2 inhibition in diabetes mellitus: Rationale and clinical prospects. Nat. Rev. Endocrinol. 2012, 8, 495–502. [Google Scholar] [CrossRef]
- Scheen, A.J. Pharmacodynamics, efficacy and safety of sodium-glucose co-transporter type 2 (SGLT2) inhibitors for the treatment of type 2 diabetes mellitus. Drugs 2015, 75, 33–59. [Google Scholar] [CrossRef]
- Fonseca-Correa, J.I.; Correa-Rotter, R. Sodium-Glucose Cotransporter 2 Inhibitors Mechanisms of Action: A Review. Front. Med. 2021, 8, 777861. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, N.; Tanaka, M.; Ochi, K.; Watanabe, A.; Ono, K.; Sawada, M.; Ogi, T.; Itoh, M.; Ito, A.; Shiraki, Y.; et al. The sodium-glucose cotransporter-2 inhibitor Tofogliflozin prevents the progression of nonalcoholic steatohepatitis-associated liver tumors in a novel murine model. Biomed. Pharmacother. 2021, 140, 111738. [Google Scholar] [CrossRef]
- Honda, Y.; Imajo, K.; Kato, T.; Kessoku, T.; Ogawa, Y.; Tomeno, W.; Kato, S.; Mawatari, H.; Fujita, K.; Yoneda, M.; et al. The Selective SGLT2 Inhibitor Ipragliflozin Has a Therapeutic Effect on Nonalcoholic Steatohepatitis in Mice. PLoS ONE 2016, 11, e0146337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ito, S.; Hosaka, T.; Yano, W.; Itou, T.; Yasumura, M.; Shimizu, Y.; Kobayashi, H.; Nakagawa, T.; Inoue, K.; Tanabe, S.; et al. Metabolic effects of Tofogliflozin are efficiently enhanced with appropriate dietary carbohydrate ratio and are distinct from carbohydrate restriction. Physiol. Rep. 2018, 6, e13642. [Google Scholar] [CrossRef]
- Obata, A.; Kubota, N.; Kubota, T.; Iwamoto, M.; Sato, H.; Sakurai, Y.; Takamoto, I.; Katsuyama, H.; Suzuki, Y.; Fukazawa, M.; et al. Tofogliflozin Improves Insulin Resistance in Skeletal Muscle and Accelerates Lipolysis in Adipose Tissue in Male Mice. Endocrinology 2016, 157, 1029–1042. [Google Scholar] [CrossRef]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Nonalcoholic Steatohepatitis Clinical Research Network. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Tanaka, T.; Yamamoto, J.; Iwasaki, S.; Asaba, H.; Hamura, H.; Ikeda, Y.; Watanabe, M.; Magoori, K.; Ioka, R.X.; Tachibana, K.; et al. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. USA 2003, 100, 15924–15929. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Tahara-Hanaoka, S.; Nabekura, T.; Ikeda, K.; Jiang, S.; Tsutsumi, S.; Inagaki, T.; Magoori, K.; Higurashi, T.; Takahashi, H.; et al. PPARβ/δ activation of CD300a controls intestinal immunity. Sci. Rep. 2014, 4, 5412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, F.J.; Shah, Y.M. PPARα: Mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology 2008, 246, 2–8. [Google Scholar] [CrossRef]
- Misra, P.; Reddy, J.K. Peroxisome proliferator-activated receptor-α activation and excess energy burning in hepatocarcinogenesis. Biochimie 2014, 98, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Sans, A.; Bonnafous, S.; Rousseau, D.; Patouraux, S.; Canivet, C.M.; Leclere, P.S.; Tran-Van-Nhieu, J.; Luci, C.; Bailly-Maitre, B.; Xu, X.; et al. The Differential Expression of Cide Family Members is Associated with Nafld Progression from Steatosis to Steatohepatitis. Sci. Rep. 2019, 9, 7501. [Google Scholar] [CrossRef]
- Gao, G.; Chen, F.J.; Zhou, L.; Su, L.; Xu, D.; Xu, L.; Li, P. Control of lipid droplet fusion and growth by CIDE family proteins. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, G.; Revelo, X.; Malhi, H. Pathogenesis of Nonalcoholic Steatohepatitis: An Overview. Hepatol. Commun. 2020, 4, 478–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, C.; Stewart, A.G.; Woodman, O.L.; Ritchie, R.H.; Qin, C.X. Non-Alcoholic Steatohepatitis: A Review of Its Mechanism, Models and Medical Treatments. Front. Pharmacol. 2020, 11, 603926. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.H.; Han, C.Y. Signaling Nodes Associated with Endoplasmic Reticulum Stress during NAFLD Progression. Biomolecules 2021, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Flessa, C.M.; Kyrou, I.; Nasiri-Ansari, N.; Kaltsas, G.; Papavassiliou, A.G.; Kassi, E.; Randeva, H.S. Endoplasmic Reticulum Stress and Autophagy in the Pathogenesis of Non-alcoholic Fatty Liver Disease (NAFLD): Current Evidence and Perspectives. Curr. Obes. Rep. 2021, 10, 134–161. [Google Scholar] [CrossRef] [PubMed]
- Malhi, H.; Kaufman, R.J. Endoplasmic reticulum stress in liver disease. J. Hepatol. 2011, 54, 795–809. [Google Scholar] [CrossRef] [Green Version]
- Han, C.Y.; Lim, S.W.; Koo, J.H.; Kim, W.; Kim, S.G. PHLDA3 overexpression in hepatocytes by endoplasmic reticulum stress via IRE1-Xbp1s pathway expedites liver injury. Gut 2016, 65, 1377–1388. [Google Scholar] [CrossRef] [Green Version]
- Iynedjian, P.B. Molecular physiology of mammalian glucokinase. Cell Mol. Life Sci. 2009, 66, 27–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chitraju, C.; Mejhert, N.; Haas, J.T.; Diaz-Ramirez, L.G.; Grueter, C.A.; Imbriglio, J.E.; Pinto, S.; Koliwad, S.K.; Walther, T.C.; Farese, R.V., Jr. Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab. 2017, 26, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Becuwe, M.; Bond, L.M.; Pinto, A.F.M.; Boland, S.; Mejhert, N.; Elliott, S.D.; Cicconet, M.; Graham, M.M.; Liu, X.N.; Ilkayeva, O.; et al. FIT2 is an acyl-coenzyme A diphosphatase crucial for endoplasmic reticulum homeostasis. J. Cell Biol. 2020, 219, e202006111. [Google Scholar] [CrossRef] [PubMed]
- Hoyles, L.; Fernandez-Real, J.M.; Federici, M.; Serino, M.; Abbott, J.; Charpentier, J.; Heymes, C.; Luque, J.L.; Anthony, E.; Barton, R.H.; et al. Molecular phonemics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 2018, 24, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Mayneris-Perxachs, J.; Cardellini, M.; Hoyles, L.; Latorre, J.; Davato, F.; Moreno-Navarrete, J.M.; Arnoriaga-Rodríguez, M.; Serino, M.; Abbott, J.; Barton, R.H.; et al. Iron status influences non-alcoholic fatty liver disease in obesity through the gut microbiome. Microbiome 2021, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.Y.; Li, P.P.; Jin, F.S.; Yao, C.; Zhang, G.H.; Zang, T.; Ai, X. Ursolic acid induces ER stress response to activate ASK1-JNK signaling and induce apoptosis in human bladder cancer T24 cells. Cell Signal. 2013, 25, 206–213. [Google Scholar] [CrossRef]
- Challa, T.D.; Wueest, S.; Lucchini, F.C.; Dedual, M.; Modica, S.; Borsigova, M.; Wolfrum, C.; Blüher, M.; Konrad, D. Liver ASK1 protects from non-alcoholic fatty liver disease and fibrosis. EMBO Mol. Med. 2019, 11, e10124. [Google Scholar] [CrossRef]
- Harrison, S.A.; Wong, V.W.; Okanoue, T.; Bzowej, N.; Vuppalanchi, R.; Younes, Z.; Kohli, A.; Sarin, S.; Caldwell, S.H.; Alkhouri, N.; et al. Selonsertib for patients with bridging fibrosis or compensated cirrhosis due to NASH: Results from randomized phase III STELLAR trials. J. Hepatol. 2020, 73, 26–39. [Google Scholar] [CrossRef]
- Chen, Y.; Ohki, R. p53-PHLDA3-Akt Network: The Key Regulators of Neuroendocrine Tumorigenesis. Int. J. Mol. Sci. 2020, 21, 4098. [Google Scholar] [CrossRef] [PubMed]
- Roden, M. Mechanisms of Disease: Hepatic steatosis in type 2 diabetes—pathogenesis and clinical relevance. Nat. Clin. Pract. Endocrinol. Metab. 2006, 2, 335–348. [Google Scholar] [CrossRef] [PubMed]
- de Vries, M.; Westerink, J.; Kaasjager, K.H.; de Valk, H.W. Prevalence of Nonalcoholic Fatty Liver Disease (NAFLD) in Patients with Type 1 Diabetes Mellitus: A Systematic Review and Meta-Analysis. J. Clin. Endocrinol. Metab. 2020, 105, 3842–3853. [Google Scholar] [CrossRef] [PubMed]
- Regnell, S.E.; Lernmark, A. Hepatic steatosis in type 1 diabetes. Rev. Diabet. Stud. 2011, 8, 454–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravikumar, B.; Carey, P.E.; Snaar, J.E.; Deelchand, D.K.; Cook, D.B.; Neely, R.D.; English, P.T.; Firbank, M.J.; Morris, P.G.; Taylor, R. Real-time assessment of postprandial fat storage in liver and skeletal muscle in health and type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 2005, 288, E789–E797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donnelly, K.L.; Smith, C.I.; Schwarzenberg, S.J.; Jessurun, J.; Boldt, M.D.; Parks, E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Investig. 2005, 115, 1343–1351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, S.; Shimomura, I. Contribution of adipose tissue and de novo lipogenesis to nonalcoholic fatty liver disease. J. Clin. Investig. 2005, 115, 1139–1142. [Google Scholar] [CrossRef]
- Grefhorst, A.; van de Peppel, I.P.; Larsen, L.E.; Jonker, J.W.; Holleboom, A.G. The Role of Lipophagy in the Development and Treatment of Non-Alcoholic Fatty Liver Disease. Front. Endocrinol. 2021, 11, 601627. [Google Scholar] [CrossRef] [PubMed]
- Soto-Gutierrez, A.; Gough, A.; Vernetti, L.A.; Taylor, D.L.; Monga, S.P. Pre-clinical and clinical investigations of metabolic zonation in liver diseases: The potential of microphysiology systems. Exp. Biol. Med. 2017, 242, 1605–1616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kietzmann, T. Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 2017, 11, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, R.P.; Porat-Shliom, N. Liver Zonation—Revisiting Old Questions With New Technologies. Front. Physiol. 2021, 12, 732929. [Google Scholar] [CrossRef] [PubMed]
- Steinman, J.B.; Salomao, M.A.; Pajvani, U.B. Zonation in NASH—A key paradigm for understanding pathophysiology and clinical outcomes. Liver Int. 2021, 41, 2534–2546. [Google Scholar] [CrossRef]
- Bedossa, P. Histological Assessment of NAFLD. Dig. Dis. Sci. 2016, 61, 1348–1355. [Google Scholar] [CrossRef] [PubMed]
- Brunt, E.M.; Kleiner, D.E.; Wilson, L.A.; Unalp, A.; Behling, C.E.; Lavine, J.E.; Neuschwander-Tetri, B.A. NASH Clinical Research NetworkA list of members of the Nonalcoholic Steatohepatitis Clinical Research Network can be found in the Appendix. Portal chronic inflammation in nonalcoholic fatty liver disease (NAFLD): A histologic marker of advanced NAFLD-Clinicopathologic correlations from the nonalcoholic steatohepatitis clinical research network. Hepatology 2009, 49, 809–820. [Google Scholar] [PubMed] [Green Version]
- Kumar, S.; Duan, Q.; Wu, R.; Harris, E.N.; Su, Q. Pathophysiological communication between hepatocytes and non-parenchymal cells in liver injury from NAFLD to liver fibrosis. Adv. Drug Deliv. Rev. 2021, 176, 113869. [Google Scholar] [CrossRef]
- Gadd, V.L.; Skoien, R.; Powell, E.E.; Fagan, K.J.; Winterford, C.; Horsfall, L.; Irvine, K.; Clouston, A.D. The portal inflammatory infiltrate and ductular reaction in human nonalcoholic fatty liver disease. Hepatology 2014, 59, 1393–1405. [Google Scholar] [CrossRef] [PubMed]
- Schwimmer, J.B.; Lavine, J.E.; Wilson, L.A.; Neuschwander-Tetri, B.A.; Xanthakos, S.A.; Kohli, R.; Barlow, S.E.; Vos, M.B.; Karpen, S.J.; Molleston, J.P.; et al. In children with non-alcoholic fatty liver disease, cysteamine bitartrate delayed release improves liver enzymes but does not reduce disease activity scores. Gastroenterology 2016, 151, 1141–1154. [Google Scholar] [CrossRef]
- Park, S.R.; Cho, C.-S.; Xi, J.; Kang, H.M.; Lee, J.H. Holistic characterization of single-hepatocyte transcriptome responses to high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E244–E258. [Google Scholar] [CrossRef] [PubMed]
- Brosch, M.; Kattler, K.; Herrmann, A.; von Schönfels, W.; Nordström, K.; Seehofer, D.; Damm, G.; Becker, T.; Zeissig, S.; Nehring, S.; et al. Epigenomic map of human liver reveals principles of zonated morphogenic and metabolic control. Nat. Commun. 2018, 9, 4150. [Google Scholar] [CrossRef]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Tóth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017, 542, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Kuang, H.; Ansari, S.; Liu, T.; Gong, J.; Wang, S.; Zhao, X.Y.; Ji, Y.; Li, C.; Guo, L.; et al. Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol. Cell. 2019, 75, 644–660. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, F.; Andersson, A.; Saarenpää, S.; Larsson, L.; Van Hul, N.; Kanatani, S.; Masek, J.; Ellis, E.; Barragan, A.; Mollbrink, A.; et al. Spatial Transcriptomics to define transcriptional patterns of zonation and structural components in the mouse liver. Nat. Commun. 2021, 12, 7046. [Google Scholar] [CrossRef] [PubMed]
| Normal | STAM | ||||
|---|---|---|---|---|---|
| Vehicle | Pemafibrate | Tofogliflozin | Pemafibrate/ Tofogliflozin | ||
| n | 6 | 6 | 5 | 7 | 6 |
| Body weight (g) | 23.49 ± 0.35 ** | 18.65 ± 0.45 | 18.15 ± 0.82 | 18.46 ± 0.49 | 18.25 ± 0.1 |
| Liver weight (g) | 1.31 ± 0.08 ** | 0.97 ± 0.03 | 1.43 ± 0.15 ** | 1.34 ± 0.03 ** | 1.65 ± 0.08 ** |
| TC (mg/dL) | 97.3 ± 0.7 ** | 171.3 ± 10.6 | 234.4 ± 12.1 ** | 194.6 ± 10.7 | 225.2 ± 10.7 ** |
| PL (mg/dL) | 207.7 ± 3.5 ** | 330.2 ± 12.9 | 380.4 ± 10.3 * | 323.6 ± 15.7 | 331.3 ± 13.4 |
| NEFA (mEq/L) | 0.77 ± 0.03 | 0.85 ± 0.10 | 0.71 ± 0.06 | 0.91 ± 0.07 | 0.75 ± 0.07 |
| β-hydroxybutylate (nmol) | 17.5 ± 2.5 ** | 140.2 ± 8.1 | 149.1 ± 0.7 | 152.8 ± 1.6 | 152.7 ± 2.9 |
| FGF21 (pg/mL) | 156.1 ± 55.4 * | 1579.6 ± 458.8 | 2449.0 ± 500.1 | 1430.4 ± 212.9 | 2759.6 ± 143.5 * |
| CRN (mg/dL) | 0.108 ± 0.004 | 0.122 ± 0.005 | 0.100 ± 0.011 | 0.114 ± 0.006 | 0.093 ± 0.013 |
| AST (U/L) | 128.0 ± 16.4 * | 181.8 ± 9.6 | 192.2 ± 17.3 | 189.6 ± 15.4 | 192.7 ± 10.6 |
| ALT (U/L) | 38.3 ± 7.9 | 71.2 ± 7.6 | 76.6 ± 13.2 | 62.9 ± 5.5 | 84.8 ± 12.3 |
| Oil Red O score | 0.3 ± 0.2 ** | 2.8 ± 0.3 | 3.2 ± 0.2 | 2.6 ± 0.2 | 3.2 ± 0.2 |
| ER-TR-7 (% area) | 1.667 ± 0.037 ** | 3.718 ± 0.451 | 3.074 ± 0.205 | 3.347 ± 0.319 | 3.096 ± 0.143 |
| Sirius Red (% area) | 0.257 ± 0.019 ** | 1.160 ± 0.205 | 1.096 ± 0.146 | 1.301 ± 0.127 | 1.062 ± 0.136 |
| Steatosis | 0.00 ** | 2.2 ± 0.3 | 2.2 ± 0.2 | 1.7 ± 0.2 | 2.0 ± 0.0 |
| Inflammation | 0.00 ** | 1.17 ± 0.17 | 1.20 ± 0.20 | 1.29 ± 0.18 | 1.50 ± 0.22 |
| NAS | 0.00 ** | 4.67 ± 0.56 | 4.00 ± 0.32 | 3.71 ± 0.36 | 3.67 ± 0.33 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murakami, K.; Sasaki, Y.; Asahiyama, M.; Yano, W.; Takizawa, T.; Kamiya, W.; Matsumura, Y.; Anai, M.; Osawa, T.; Fruchart, J.-C.; et al. Selective PPARα Modulator Pemafibrate and Sodium-Glucose Cotransporter 2 Inhibitor Tofogliflozin Combination Treatment Improved Histopathology in Experimental Mice Model of Non-Alcoholic Steatohepatitis. Cells 2022, 11, 720. https://doi.org/10.3390/cells11040720
Murakami K, Sasaki Y, Asahiyama M, Yano W, Takizawa T, Kamiya W, Matsumura Y, Anai M, Osawa T, Fruchart J-C, et al. Selective PPARα Modulator Pemafibrate and Sodium-Glucose Cotransporter 2 Inhibitor Tofogliflozin Combination Treatment Improved Histopathology in Experimental Mice Model of Non-Alcoholic Steatohepatitis. Cells. 2022; 11(4):720. https://doi.org/10.3390/cells11040720
Chicago/Turabian StyleMurakami, Kentaro, Yusuke Sasaki, Masato Asahiyama, Wataru Yano, Toshiaki Takizawa, Wakana Kamiya, Yoshihiro Matsumura, Motonobu Anai, Tsuyoshi Osawa, Jean-Charles Fruchart, and et al. 2022. "Selective PPARα Modulator Pemafibrate and Sodium-Glucose Cotransporter 2 Inhibitor Tofogliflozin Combination Treatment Improved Histopathology in Experimental Mice Model of Non-Alcoholic Steatohepatitis" Cells 11, no. 4: 720. https://doi.org/10.3390/cells11040720
APA StyleMurakami, K., Sasaki, Y., Asahiyama, M., Yano, W., Takizawa, T., Kamiya, W., Matsumura, Y., Anai, M., Osawa, T., Fruchart, J.-C., Fruchart-Najib, J., Aburatani, H., Sakai, J., Kodama, T., & Tanaka, T. (2022). Selective PPARα Modulator Pemafibrate and Sodium-Glucose Cotransporter 2 Inhibitor Tofogliflozin Combination Treatment Improved Histopathology in Experimental Mice Model of Non-Alcoholic Steatohepatitis. Cells, 11(4), 720. https://doi.org/10.3390/cells11040720