Microtubule and Actin Cytoskeletal Dynamics in Male Meiotic Cells of Drosophila melanogaster
Abstract
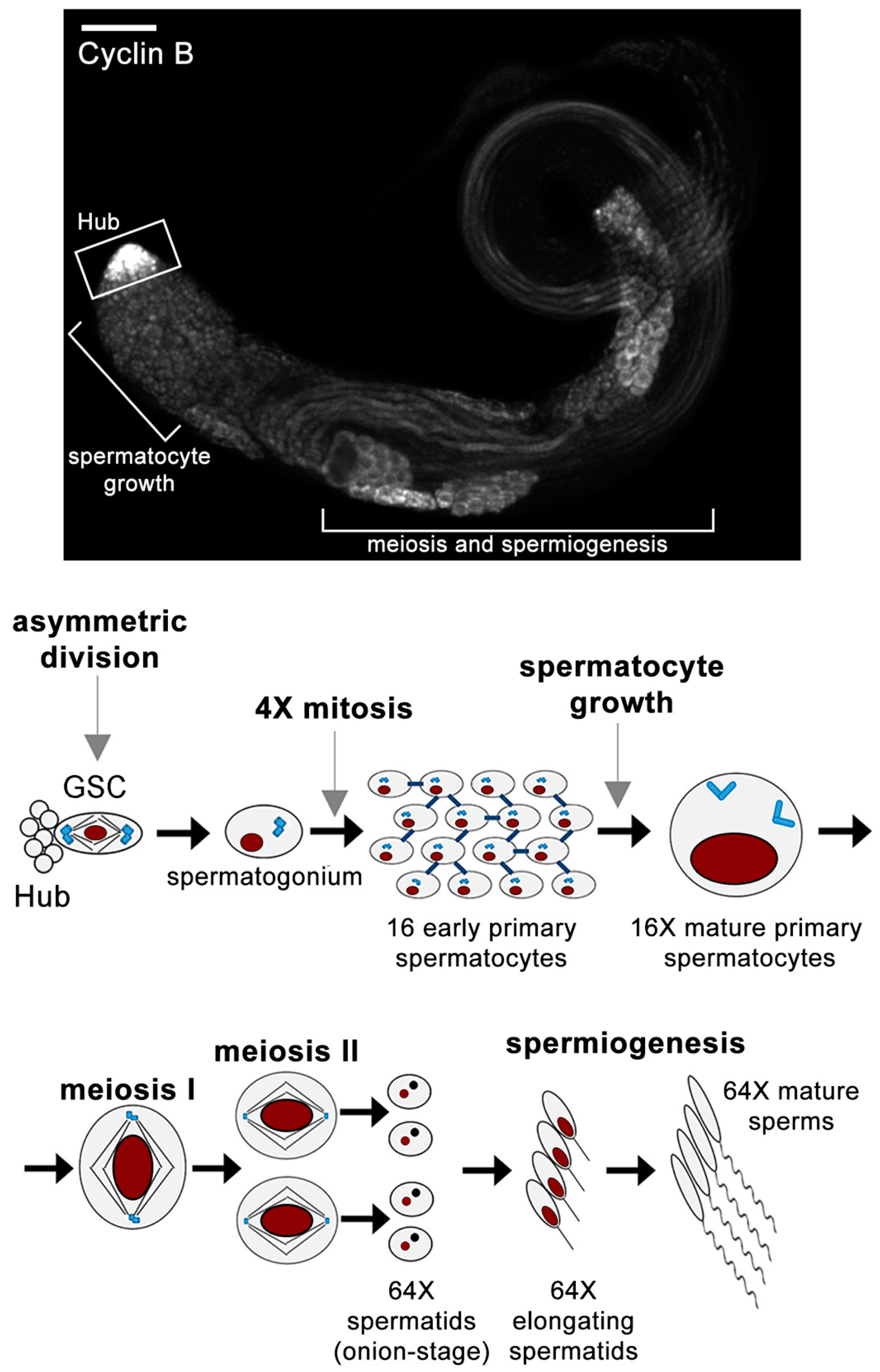
1. An Overview of Spermatogenesis in Drosophila melanogaster
2. Translational Control and Cytoskeletal Remodeling in Drosophila Spermatocytes
3. Centrioles and Centrosomes in Male Meiotic Cells
4. Centrosome Pericentriolar Material Assembly (PCM) in Male Meiotic Cells
5. The Centrosome Is Essential to Assemble a Functional Bipolar Spindle in Spermatocytes
6. Spindle Scaling in Primary Spermatocytes
7. The Advantages Offered by Drosophila Male Meiotic Cells to Explore Cytokinesis
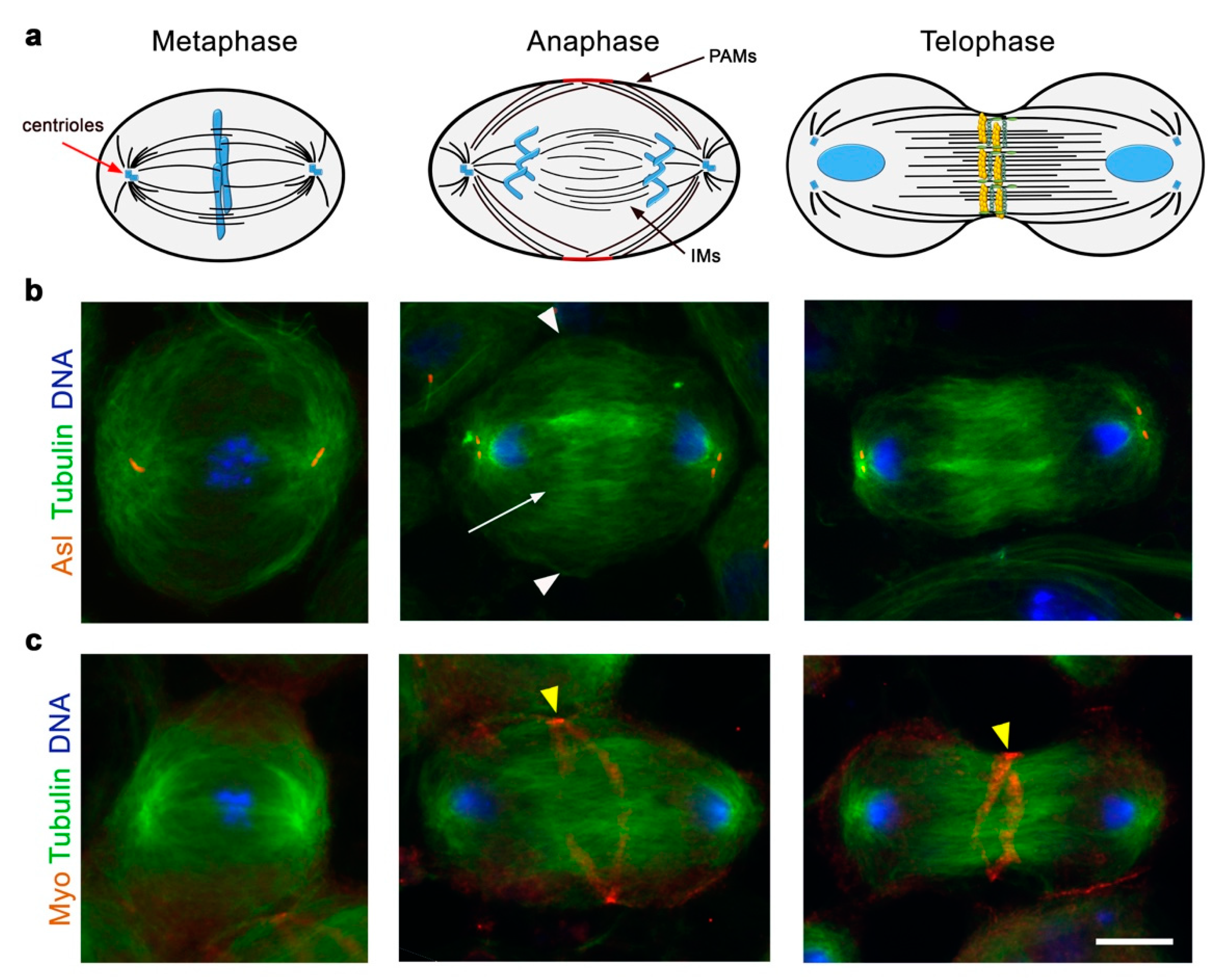
8. The Role of Spindle Microtubules in Cleavage Furrow Formation in Drosophila Spermatocytes
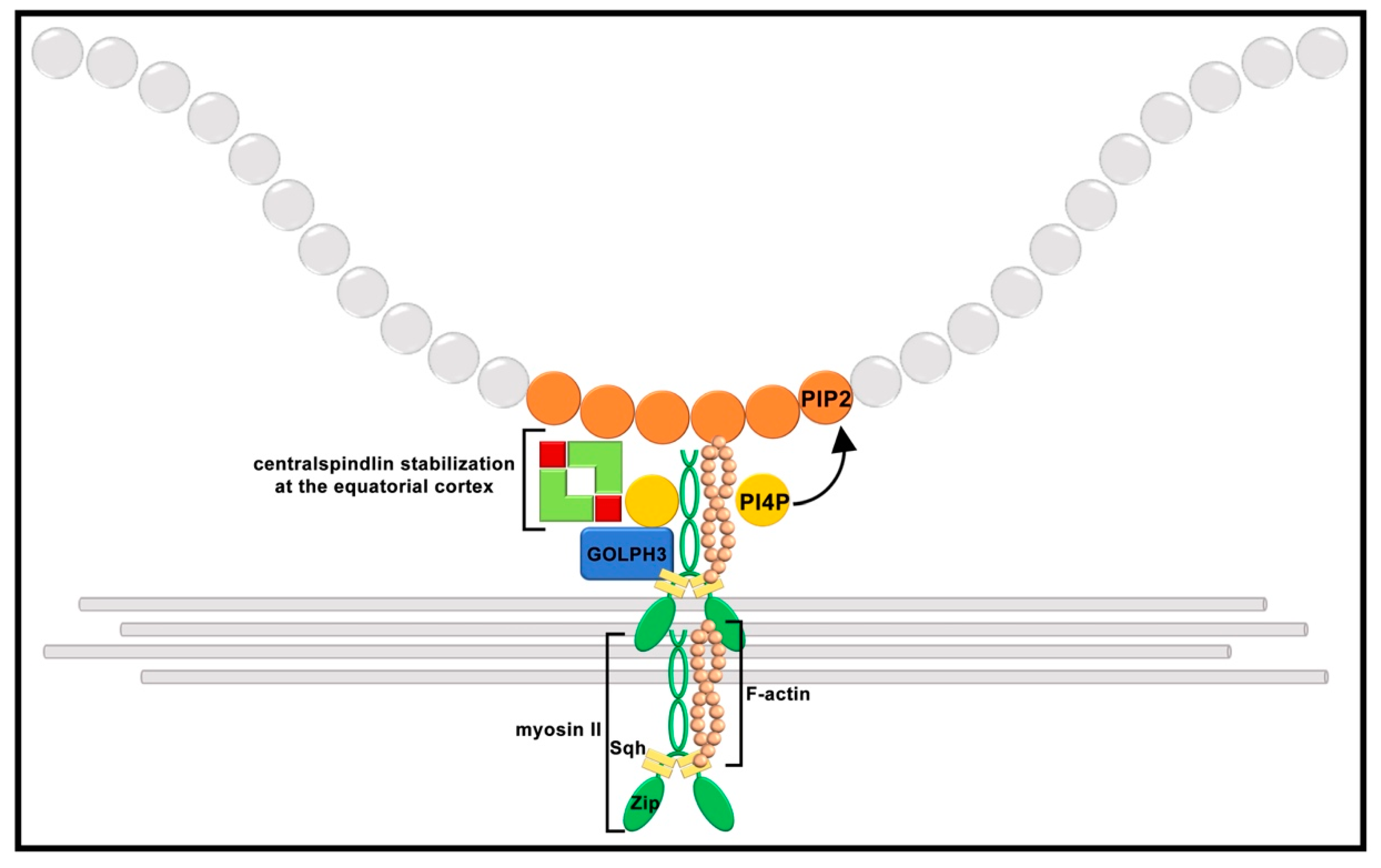
9. Scaffolding Proteins and Phosphoinositide Binding Proteins Contribute to Cleavage Furrow Ingression
10. Interconnectivity during Drosophila Spermatogenesis
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fuller, M.T. Spermatogenesis. In The Development of Drosophila melanogaster; Bate, M., Martinez-Arias, A., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1993; pp. 71–147. [Google Scholar]
- de Cuevas, M.; Matunis, E.L. The stem cell niche: Lessons from the Drosophila testis. Development 2011, 138, 2861–2869. [Google Scholar] [CrossRef]
- Fuller, M.T.; Spradling, A.C. Male and female Drosophila germline stem cells: Two versions of immortality. Science 2007, 316, 402–404. [Google Scholar] [CrossRef] [PubMed]
- Tokuyasu, K.T.; Peacock, W.J.; Hardy, R.W. Dynamics of spermiogenesis in Drosophila melanogaster. VII. Effects of segregation distorter (SD) chromosome. J. Ultrastruct. Res. 1977, 58, 96–107. [Google Scholar] [CrossRef]
- Lindsley, D.L.; Tokuyasu, K.T. Spermatogenesis. In Genetics and Biology of Drosophila; Ashburner, M., Wright, T.R.F., Eds.; Academic Press: New York, NY, USA, 1980; pp. 225–294. [Google Scholar]
- McKearin, D. The Drosophila fusome, organelle biogenesis and germ cell differentiation: If you build it. Bioessays 1997, 19, 147–152. [Google Scholar] [CrossRef]
- Giansanti, M.G.; Bonaccorsi, S.; Williams, B.; Williams, E.V.; Santolamazza, C.; Goldberg, M.L.; Gatti, M. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 1998, 12, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Lighthouse, D.V.; Buszczak, M.; Spradling, A.C. New components of the Drosophila fusome suggest it plays novel roles in signaling and transport. Dev. Biol. 2008, 317, 59–71. [Google Scholar] [CrossRef]
- Kaufman, R.S.; Price, K.L.; Mannix, K.M.; Ayers, K.M.; Hudson, A.M.; Cooley, L. Drosophila sperm development and intercellular cytoplasm sharing through ring canals do not require an intact fusome. Development 2020, 147, dev190140. [Google Scholar] [CrossRef] [PubMed]
- White-Cooper, H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction 2010, 139, 11–21. [Google Scholar] [CrossRef]
- Chandley, A.C.; Bateman, A.J. Timing of spermatogenesis in Drosophila melanogaster using tritiated thymidine. Nature 1962, 193, 299–300. [Google Scholar] [CrossRef]
- McKee, B.D.; Yan, R.; Tsai, J.H. Meiosis in male Drosophila. Spermatogenesis 2012, 2, 167–184. [Google Scholar] [CrossRef]
- Giansanti, M.G.; Sechi, S.; Frappaolo, A.; Belloni, G.; Piergentili, R. Cytokinesis in Drosophila male meiosis. Spermatogenesis 2012, 2, 185–196. [Google Scholar] [CrossRef][Green Version]
- Giansanti, M.G.; Fuller, M.T. What Drosophila spermatocytes tell us about the mechanisms underlying cytokinesis. Cytoskeleton 2012, 69, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Fabian, L.; Brill, J.A. Drosophila spermiogenesis: Big things come from little packages. Spermatogenesis 2012, 2, 197–212. [Google Scholar] [CrossRef]
- Demarco, R.S.; Eikenes, Å.H.; Haglund, K.; Jones, D.L. Investigating spermatogenesis in Drosophila melanogaster. Methods 2014, 68, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Alphey, L.; Jimenez, J.; White-Cooper, H.; Dawson, I.; Nurse, P.; Glover, D.M. twine, a cdc25 homolog that functions in the male and female germline of Drosophila. Cell 1992, 69, 977–988. [Google Scholar] [CrossRef]
- Courtot, C.; Fankhauser, C.; Simanis, V.; Lehner, C.F. The Drosophila cdc25 homolog twine is required for meiosis. Development 1992, 116, 405–416. [Google Scholar] [CrossRef] [PubMed]
- White-Cooper, H.; Alphey, L.; Glover, D.M. The cdc25 homologue twine is required for only some aspects of the entry into meiosis in Drosophila. J. Cell Sci. 1993, 106, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Sigrist, S.; Ried, G.; Lehner, C.F. Dmcdc2 kinase is required for both meiotic divisions during Drosophila spermatogenesis and is activated by the Twine/cdc25 phosphatase. Mech. Dev. 1995, 53, 247–260. [Google Scholar] [CrossRef]
- Lin, T.Y.; Viswanathan, S.; Wood, C.; Wilson, P.G.; Wolf, N.; Fuller, M.T. Coordinate developmental control of the meiotic cell cycle and spermatid differentiation in Drosophila males. Development 1996, 122, 1331–1341. [Google Scholar] [CrossRef]
- White-Cooper, H.; Schäfer, M.A.; Alphey, L.S.; Fuller, M.T. Transcriptional and post-transcriptional control mechanisms coordinate the onset of spermatid differentiation with meiosis I in Drosophila. Development 1998, 125, 125–134. [Google Scholar] [CrossRef]
- Baker, C.C.; Fuller, M.T. Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development 2007, 134, 2863–2869. [Google Scholar] [CrossRef]
- Baker, C.C.; Gim, B.S.; Fuller, M.T. Cell type-specific translational repression of Cyclin B during meiosis in males. Development 2015, 142, 3394–3402. [Google Scholar] [CrossRef] [PubMed]
- Franklin-Dumont, T.M.; Chatterjee, C.; Wasserman, S.A.; Dinardo, S. A novel eIF4G homolog, Off-schedule, couples translational control to meiosis and differentiation in Drosophila spermatocytes. Development 2007, 134, 2851–2861. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Lasko, P. Translational control in cellular and developmental processes. Nat. Rev. Genet. 2012, 13, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N.; Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 2009, 136, 731–745. [Google Scholar] [CrossRef]
- Hernández, G.; Han, H.; Gandin, V.; Fabian, L.; Ferreira, T.; Zuberek, J.; Sonenberg, N.; Brill, J.A.; Lasko, P. Eukaryotic initiation factor 4E-3 is essential for meiotic chromosome segregation, cytokinesis and male fertility in Drosophila. Development 2012, 139, 3211–3220. [Google Scholar] [CrossRef]
- Ghosh, S.; Lasko, P. Loss-of-function analysis reveals distinct requirements of the translation initiation factors eIF4E, eIF4E-3, eIF4G and eIF4G2 in Drosophila spermatogenesis. PLoS ONE 2015, 10, e0122519. [Google Scholar] [CrossRef]
- Sechi, S.; Frappaolo, A.; Karimpour-Ghahnavieh, A.; Gottardo, M.; Burla, R.; Di Francesco, L.; Szafer-Glusman, E.; Schininà, E.; Fuller, M.T.; Saggio, I.; et al. Drosophila Doublefault protein coordinates multiple events during male meiosis by controlling mRNA translation. Development 2019, 146, dev183053. [Google Scholar] [CrossRef]
- McDermott, S.M.; Meignin, C.; Rappsilber, J.; Davis, I. Drosophila Syncrip binds the gurken mRNA localisation signal and regulates localised transcripts during axis specification. Biol. Open 2012, 1, 488–497. [Google Scholar] [CrossRef]
- McDermott, S.M.; Yang, L.; Halstead, J.M.; Hamilton, R.S.; Meignin, C.; Davis, I. Drosophila Syncrip modulates the expression of mRNAs encoding key synaptic proteins required for morphology at the neuromuscular junction. RNA 2014, 20, 1593–1606. [Google Scholar] [CrossRef]
- Bannai, H.; Fukatsu, K.; Mizutani, A.; Natsume, T.; Iemura, S.; Ikegami, T.; Inoue, T.; Mikoshiba, K. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 2004, 279, 53427–53434. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Dohmae, N.; Hirokawa, N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron 2004, 43, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Elvira, G.; Wasiak, S.; Blandford, V.; Tong, X.K.; Serrano, A.; Fan, X.; del Rayo Sánchez-Carbente, M.; Servant, F.; Bell, A.W.; Boismenu, D.; et al. Characterization of an RNA granule from developing brain. Mol. Cell. Proteom. 2006, 5, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Gould-Somero, M.; Holland, L. The timing of RNA synthesis for spermiogenesis in organ cultures of Drosophila melanogaster testes. Wilhelm Roux Arch. Entwickl. Mech. Org. 1974, 174, 133–148. [Google Scholar] [CrossRef]
- Olivieri, G.; Olivieri, A. Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster. Mutat. Res. 1965, 2, 366–380. [Google Scholar] [CrossRef]
- White-Cooper, H.; Caporilli, S. Transcriptional and post-transcriptional regulation of Drosophila germline stem cells and their differentiating progeny. Adv. Exp. Med. Biol. 2013, 786, 47–61. [Google Scholar] [CrossRef]
- Aizer, A.; Brody, Y.; Ler, L.W.; Sonenberg, N.; Singer, R.H.; Shav-Tal, Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 2008, 19, 4154–4166. [Google Scholar] [CrossRef]
- Johnson, C.A.; Malicki, J.J. The Nuclear Arsenal of Cilia. Dev. Cell 2019, 49, 161–170. [Google Scholar] [CrossRef]
- Moser, J.J.; Fritzler, M.J.; Rattner, J.B. Repression of GW/P body components and the RNAi microprocessor impacts primary ciliogenesis in human astrocytes. BMC Cell Biol. 2011, 12, 37. [Google Scholar] [CrossRef]
- Youn, J.Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532.e11. [Google Scholar] [CrossRef]
- Iaconis, D.; Monti, M.; Renda, M.; van Koppen, A.; Tammaro, R.; Chiaravalli, M.; Cozzolino, F.; Pignata, P.; Crina, C.; Pucci, P.; et al. The centrosomal OFD1 protein interacts with the translation machinery and regulates the synthesis of specific targets. Sci. Rep. 2017, 7, 1224. [Google Scholar] [CrossRef] [PubMed]
- Fingerhut, J.M.; Yamashita, Y.M. mRNA localization mediates maturation of cytoplasmic cilia in Drosophila spermatogenesis. J. Cell Biol. 2020, 219, e202003084. [Google Scholar] [CrossRef]
- Lattao, R.; Kovács, L.; Glover, D.M. The Centrioles, Centrosomes, Basal Bodies, and Cilia of Drosophila melanogaster. Genetics 2017, 206, 33–53. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.L.; Pelletier, L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Tillery, M.M.L.; Blake-Hedges, C.; Zheng, Y.; Buchwalter, R.A.; Megraw, T.L. Centrosomal and Non-Centrosomal Microtubule-Organizing Centers (MTOCs) in Drosophila melanogaster. Cells 2018, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Jana, S.C.; Bettencourt-Dias, M.; Durand, B.; Megraw, T.L. Drosophila melanogaster as a model for basal body research. Cilia 2016, 5, 22. [Google Scholar] [CrossRef]
- Nigg, E.A.; Raff, J.W. Centrioles, centrosomes, and cilia in health and disease. Cell 2009, 139, 663–678. [Google Scholar] [CrossRef]
- Conduit, P.T.; Wainman, A.; Raff, J.W. Centrosome function and assembly in animal cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 611–624. [Google Scholar] [CrossRef]
- Tates, A.D. Cytodifferentiation during Spermatogenesis in Drosophila melanogaster: An Electron Microscope Study. Ph.D. Thesis, Rijksunivrsiteit Leiden, Leiden, The Netherlands, 1971. [Google Scholar]
- Delattre, M.; Canard, C.; Gönczy, P. Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 2006, 16, 1844–1849. [Google Scholar] [CrossRef]
- Pelletier, L.; O’Toole, E.; Schwager, A.; Hyman, A.A.; Müller-Reichert, T. Centriole assembly in Caenorhabditis elegans. Nature 2006, 444, 619–623. [Google Scholar] [CrossRef]
- Fu, J.; Glover, D.M. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012, 2, 120104. [Google Scholar] [CrossRef] [PubMed]
- Mennella, V.; Keszthelyi, B.; McDonald, K.L.; Chhun, B.; Kan, F.; Rogers, G.C.; Huang, B.; Agard, D.A. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 2012, 14, 1159–1168. [Google Scholar] [CrossRef]
- Dzhindzhev, N.S.; Tzolovsky, G.; Lipinszki, Z.; Schneider, S.; Lattao, R.; Fu, J.; Debski, J.; Dadlez, M.; Glover, D.M. Plk4 phosphorylates Ana2 to trigger Sas6 recruitment and procentriole formation. Curr. Biol. 2014, 24, 2526–2532. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Lipinszki, Z.; Rangone, H.; Min, M.; Mykura, C.; Chao-Chu, J.; Schneider, S.; Dzhindzhev, N.S.; Gottardo, M.; Riparbelli, M.G.; et al. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol. 2016, 18, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Cottee, M.A.; Muschalik, N.; Johnson, S.; Leveson, J.; Raff, J.W.; Lea, S.M. The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. Elife 2015, 4, e07236. [Google Scholar] [CrossRef]
- Cottee, M.A.; Muschalik, N.; Wong, Y.L.; Johnson, C.M.; Johnson, S.; Andreeva, A.; Oegema, K.; Lea, S.M.; Raff, J.W.; van Breugel, M. Crystal structures of the CPAP/STIL complex reveal its role in centriole assembly and human microcephaly. Elife 2013, 2, e01071. [Google Scholar] [CrossRef]
- Dammermann, A.; Maddox, P.S.; Desai, A.; Oegema, K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J. Cell Biol. 2008, 180, 771–785. [Google Scholar] [CrossRef]
- Hsu, W.B.; Hung, L.Y.; Tang, C.J.; Su, C.L.; Chang, Y.; Tang, T.K. Functional characterization of the microtubule-binding and -destabilizing domains of CPAP and d-SAS-4. Exp. Cell Res. 2008, 314, 2591–2602. [Google Scholar] [CrossRef]
- Tang, C.J.; Lin, S.Y.; Hsu, W.B.; Lin, Y.N.; Wu, C.T.; Lin, Y.C.; Chang, C.W.; Wu, K.S.; Tang, T.K. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J. 2011, 30, 790–804. [Google Scholar] [CrossRef]
- Vulprecht, J.; David, A.; Tibelius, A.; Castiel, A.; Konotop, G.; Liu, F.; Bestvater, F.; Raab, M.S.; Zentgraf, H.; Izraeli, S.; et al. STIL is required for centriole duplication in human cells. J. Cell Sci. 2012, 125, 1353–1362. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Y.; Duan, Q.; Jiang, N.; Huang, Y.; Darzynkiewicz, Z.; Dai, W. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev. Cell 2008, 14, 331–341. [Google Scholar] [CrossRef]
- Tsou, M.F.; Wang, W.J.; George, K.A.; Uryu, K.; Stearns, T.; Jallepalli, P.V. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell 2009, 17, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Riparbelli, M.G.; Gottardo, M.; Glover, D.M.; Callaini, G. Inhibition of Polo kinase by BI2536 affects centriole separation during Drosophila male meiosis. Cell Cycle 2014, 13, 2064–2072. [Google Scholar] [CrossRef] [PubMed]
- Varadarajan, R.; Ayeni, J.; Jin, Z.; Homola, E.; Campbell, S.D. Myt1 inhibition of Cyclin A/Cdk1 is essential for fusome integrity and premeiotic centriole engagement in Drosophila spermatocytes. Mol. Biol. Cell 2016, 27, 2051–2063. [Google Scholar] [CrossRef]
- Avidor-Reiss, T.; Gopalakrishnan, J. Building a centriole. Curr. Opin. Cell Biol. 2013, 25, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Arquint, C.; Nigg, E.A. The PLK4-STIL-SAS-6 module at the core of centriole duplication. Biochem. Soc. Trans. 2016, 44, 1253–1263. [Google Scholar] [CrossRef]
- Bettencourt-Dias, M.; Rodrigues-Martins, A.; Carpenter, L.; Riparbelli, M.; Lehmann, L.; Gatt, M.K.; Carmo, N.; Balloux, F.; Callaini, G.; Glover, D.M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005, 15, 2199–2207. [Google Scholar] [CrossRef]
- Dzhindzhev, N.S.; Yu, Q.D.; Weiskopf, K.; Tzolovsky, G.; Cunha-Ferreira, I.; Riparbelli, M.; Rodrigues-Martins, A.; Bettencourt-Dias, M.; Callaini, G.; Glover, D.M. Asterless is a scaffold for the onset of centriole assembly. Nature 2010, 467, 714–718. [Google Scholar] [CrossRef]
- Galletta, B.J.; Fagerstrom, C.J.; Schoborg, T.A.; McLamarrah, T.A.; Ryniawec, J.M.; Buster, D.W.; Slep, K.C.; Rogers, G.C.; Rusan, N.M. A centrosome interactome provides insight into organelle assembly and reveals a non-duplication role for Plk4. Nat. Commun. 2016, 7, 12476. [Google Scholar] [CrossRef]
- Mottier-Pavie, V.; Megraw, T.L. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol. Biol. Cell 2009, 20, 2605–2614. [Google Scholar] [CrossRef]
- Dzhindzhev, N.S.; Tzolovsky, G.; Lipinszki, Z.; Abdelaziz, M.; Debski, J.; Dadlez, M.; Glover, D.M. Two-step phosphorylation of Ana2 by Plk4 is required for the sequential loading of Ana2 and Sas6 to initiate procentriole formation. Open Biol. 2017, 7, 170247. [Google Scholar] [CrossRef] [PubMed]
- McLamarrah, T.A.; Buster, D.W.; Galletta, B.J.; Boese, C.J.; Ryniawec, J.M.; Hollingsworth, N.A.; Byrnes, A.E.; Brownlee, C.W.; Slep, K.C.; Rusan, N.M.; et al. An ordered pattern of Ana2 phosphorylation by Plk4 is required for centriole assembly. J. Cell Biol. 2018, 217, 1217–1231. [Google Scholar] [CrossRef]
- Ohta, M.; Ashikawa, T.; Nozaki, Y.; Kozuka-Hata, H.; Goto, H.; Inagaki, M.; Oyama, M.; Kitagawa, D. Direct interaction of Plk4 with STIL ensures formation of a single procentriole per parental centriole. Nat. Commun. 2014, 5, 5267. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, M.; Erat, M.C.; Hachet, V.; Guichard, P.; Blank, I.D.; Flückiger, I.; Slater, L.; Lowe, E.D.; Hatzopoulos, G.N.; Steinmetz, M.O.; et al. Caenorhabditis elegans centriolar protein SAS-6 forms a spiral that is consistent with imparting a ninefold symmetry. Proc. Natl. Acad. Sci. USA 2013, 110, 11373–11378. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, D.; Vakonakis, I.; Olieric, N.; Hilbert, M.; Keller, D.; Olieric, V.; Bortfeld, M.; Erat, M.C.; Flückiger, I.; Gönczy, P.; et al. Structural basis of the 9-fold symmetry of centrioles. Cell 2011, 144, 364–375. [Google Scholar] [CrossRef]
- van Breuge, l.M.; Hirono, M.; Andreeva, A.; Yanagisawa, H.A.; Yamaguchi, S.; Nakazawa, Y.; Morgner, N.; Petrovich, M.; Ebong, I.O.; Robinson, C.V.; et al. Structures of SAS-6 suggest its organization in centrioles. Science 2011, 331, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Oegema, K.; Wiese, C.; Martin, O.C.; Milligan, R.A.; Iwamatsu, A.; Mitchison, T.J.; Zheng, Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 1999, 144, 721–733. [Google Scholar] [CrossRef]
- Gunawardane, R.N.; Martin, O.C.; Cao, K.; Zhang, L.; Dej, K.; Iwamatsu, A.; Zheng, Y. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J. Cell Biol. 2000, 151, 1513–1524. [Google Scholar] [CrossRef]
- Gunawardane, R.N.; Martin, O.C.; Zheng, Y. Characterization of a new gammaTuRC subunit with WD repeats. Mol. Biol. Cell 2003, 14, 1017–1026. [Google Scholar] [CrossRef]
- Sunkel, C.E.; Glover, D.M. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J. Cell Sci. 1988, 89, 25–38. [Google Scholar] [CrossRef]
- Haren, L.; Stearns, T.; Lüders, J. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE 2009, 4, e5976. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Rhee, K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol. 2011, 195, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Roque, H.; Saurya, S.; Pratt, M.B.; Johnson, E.; Raff, J.W. Drosophila PLP assembles pericentriolar clouds that promote centriole stability, cohesion and MT nucleation. PLoS Genet. 2018, 14, e1007198. [Google Scholar] [CrossRef] [PubMed]
- Dix, C.I.; Raff, J.W. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 2007, 17, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Giansanti, M.G.; Bucciarelli, E.; Bonaccorsi, S.; Gatti, M. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 2008, 18, 303–309. [Google Scholar] [CrossRef]
- Conduit, P.T.; Brunk, K.; Dobbelaere, J.; Dix, C.I.; Lucas, E.P.; Raff, J.W. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 2010, 20, 2178–2186. [Google Scholar] [CrossRef]
- Lucas, E.P.; Raff, J.W. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J. Cell Biol. 2007, 178, 725–732. [Google Scholar] [CrossRef]
- Megraw, T.L.; Li, K.; Kao, L.R.; Kaufman, T.C. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 1999, 126, 2829–2839. [Google Scholar] [CrossRef]
- Megraw, T.L.; Kao, L.R.; Kaufman, T.C. Zygotic development without functional mitotic centrosomes. Curr. Biol. 2001, 11, 116–120. [Google Scholar] [CrossRef]
- Vaizel-Ohayon, D.; Schejter, E.D. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 1999, 9, 889–898. [Google Scholar] [CrossRef]
- Conduit, P.T.; Richens, J.H.; Wainman, A.; Holder, J.; Vicente, C.C.; Pratt, M.B.; Dix, C.I.; Novak, Z.A.; Dobbie, I.M.; Schermelleh, L. A molecular mechanism of mitotic centrosome assembly in Drosophila. Elife 2014, 3, e03399. [Google Scholar] [CrossRef] [PubMed]
- Conduit, P.T.; Feng, Z.; Richens, J.H.; Baumbach, J.; Wainman, A.; Bakshi, S.D.; Dobbelaere, J.; Johnson, S.; Lea, S.M.; Raff, J.W. The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell 2014, 28, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Bonaccorsi, S.; Giansanti, M.G.; Gatti, M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J. Cell Biol. 1998, 142, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Basto, R.; Lau, J.; Vinogradova, T.; Gardiol, A.; Woods, C.G.; Khodjakov, A.; Raff, J.W. Flies without centrioles. Cell 2006, 125, 1375–1386. [Google Scholar] [CrossRef]
- Varmark, H.; Llamazares, S.; Rebollo, E.; Lange, B.; Reina, J.; Schwarz, H.; Gonzalez, C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 2007, 17, 1735–1745. [Google Scholar] [CrossRef]
- Gopalakrishnan, J.; Mennella, V.; Blachon, S.; Zhai, B.; Smith, A.H.; Megraw, T.L.; Nicastro, D.; Gygi, S.P.; Agard, D.A.; Avidor-Reiss, T. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2011, 2, 359. [Google Scholar] [CrossRef]
- Yamashita, Y.M.; Jones, D.L.; Fuller, M.T. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science 2003, 301, 1547–1550. [Google Scholar] [CrossRef]
- Yamashita, Y.M.; Mahowald, A.P.; Perlin, J.R.; Fuller, M.T. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 2007, 315, 518–521. [Google Scholar] [CrossRef]
- Bang, C.; Cheng, J. Dynamic interplay of spectrosome and centrosome organelles in asymmetric stem cell divisions. PLoS ONE 2015, 10, e0123294. [Google Scholar] [CrossRef]
- Cheng, J.; Türkel, N.; Hemati, N.; Fuller, M.T.; Hunt, A.J.; Yamashita, Y.M. Centrosome misorientation reduces stem cell division during ageing. Nature 2008, 456, 599–604. [Google Scholar] [CrossRef]
- Inaba, M.; Yuan, H.; Salzmann, V.; Fuller, M.T.; Yamashita, Y.M. E-cadherin is required for centrosome and spindle orientation in Drosophila male germline stem cells. PLoS ONE 2010, 5, e12473. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Bucciarelli, E.; Lattao, R.; Pellacani, C.; Mottier-Pavie, V.; Giansanti, M.G.; Somma, M.P.; Bonaccorsi, S. The relative roles of centrosomal and kinetochore-driven microtubules in Drosophila spindle formation. Exp. Cell Res. 2012, 318, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, E.; González, C. Visualizing the spindle checkpoint in Drosophila spermatocytes. EMBO Rep. 2000, 1, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Savoian, M.S.; Goldberg, M.L.; Rieder, C.L. The rate of poleward chromosome motion is attenuated in Drosophila zw10 and rod mutants. Nat. Cell Biol. 2000, 2, 948–952. [Google Scholar] [CrossRef]
- Sunkel, C.E.; Gomes, R.; Sampaio, P.; Perdigão, J.; González, C. Gamma-tubulin is required for the structure and function of the microtubule organizing centre in Drosophila neuroblasts. EMBO J. 1995, 14, 28–36. [Google Scholar] [CrossRef]
- Tavosanis, G.; Llamazares, S.; Goulielmos, G.; Gonzalez, C. Essential role for gamma-tubulin in the acentriolar female meiotic spindle of Drosophila. EMBO J. 1997, 16, 1809–1819. [Google Scholar] [CrossRef]
- Wilson, P.G.; Borisy, G.G. Maternally expressed gamma Tub37CD in Drosophila is differentially required for female meiosis and embryonic mitosis. Dev. Biol. 1998, 199, 273–290. [Google Scholar] [CrossRef]
- Sampaio, P.; Rebollo, E.; Varmark, H.; Sunkel, C.E.; González, C. Organized microtubule arrays in gamma-tubulin-depleted Drosophila spermatocytes. Curr. Biol. 2001, 11, 1788–1793. [Google Scholar] [CrossRef]
- Barbosa, V.; Gatt, M.; Rebollo, E.; Gonzalez, C.; Glover, D.M. Drosophila dd4 mutants reveal that gammaTuRC is required to maintain juxtaposed half spindles in spermatocytes. J. Cell Sci. 2003, 116, 929–941. [Google Scholar] [CrossRef]
- Bucciarelli, E.; Giansanti, M.G.; Bonaccorsi, S.; Gatti, M. Spindle assembly and cytokinesis in the absence of chromosomes during Drosophila male meiosis. J. Cell Biol. 2003, 160, 993–999. [Google Scholar] [CrossRef]
- Riparbelli, M.G.; Callaini, G.; Megraw, T.L. Assembly and persistence of primary cilia in dividing Drosophila spermatocytes. Dev. Cell. 2012, 23, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Rebollo, E.; Llamazares, S.; Reina, J.; Gonzalez, C. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2004, 2, E8. [Google Scholar] [CrossRef] [PubMed]
- Goshima, G.; Mayer, M.; Zhang, N.; Stuurman, N.; Vale, R.D. Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 2008, 181, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Kamasaki, T.; O’Toole, E.; Kita, S.; Osumi, M.; Usukura, J.; McIntosh, J.R.; Goshima, G. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J. Cell Biol. 2013, 202, 25–33. [Google Scholar] [CrossRef]
- Lawo, S.; Bashkurov, M.; Mullin, M.; Ferreria, M.G.; Kittler, R.; Habermann, B.; Tagliaferro, A.; Poser, I.; Hutchins, J.R.; Hegemann, B.; et al. AUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 2009, 19, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Petry, S.; Groen, A.C.; Ishihara, K.; Mitchison, T.J.; Vale, R.D. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 2013, 152, 768–777. [Google Scholar] [CrossRef]
- Song, J.G.; King, M.R.; Zhang, R.; Kadzik, R.S.; Thawani, A.; Petry, S. Mechanism of how augmin directly targets the gamma-tubulin ring complex to microtubules. J. Cell Biol. 2018, 217, 2417–2428. [Google Scholar] [CrossRef]
- Uehara, R.; Nozawa, R.S.; Tomioka, A.; Petry, S.; Vale, R.D.; Obuse, C.; Goshima, G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA 2009, 106, 6998–7003. [Google Scholar] [CrossRef]
- Wainman, A.; Buster, D.W.; Duncan, T.; Metz, J.; Ma, A.; Sharp, D.; Wakefield, J.G. A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev. 2009, 23, 1876–1881. [Google Scholar] [CrossRef]
- Bucciarelli, E.; Pellacani, C.; Naim, V.; Palena, A.; Gatti, M.; Somma, M.P. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and gamma-tubulin to promote kinetochore-driven MT formation. Curr. Biol. 2009, 19, 1839–1845. [Google Scholar] [CrossRef]
- Goshima, G.; Wollman, R.; Goodwin, S.S.; Zhang, N.; Scholey, J.M.; Vale, R.D.; Stuurman, N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 2007, 316, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Hayward, D.; Metz, J.; Pellacani, C.; Wakefield, J.G. Synergy between multiple microtubule-generating pathways confers robustness to centrosome-driven mitotic spindle formation. Dev. Cell 2014, 28, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Savoian, M.S.; Glover, D.M. Differing requirements for Augmin in male meiotic and mitotic spindle formation in Drosophila. Open Biol. 2014, 4, 140047. [Google Scholar] [CrossRef] [PubMed]
- Crowder, M.E.; Strzelecka, M.; Wilbur, J.D.; Good, M.C.; von Dassow, G.; Heald, R. A comparative analysis of spindle morphometrics across metazoans. Curr. Biol. 2015, 25, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Decker, M.; Jaensch, S.; Pozniakovsky, A.; Zinke, A.; O’Connell, K.F.; Zachariae, W.; Myers, E.; Hyman, A.A. Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 2011, 21, 1259–1267. [Google Scholar] [CrossRef] [PubMed]
- Manandhar, G.; Moreno, R.D.; Simerly, C.; Toshimori, K.; Schatten, G. Contractile apparatus of the normal and abortive cytokinetic cells during mouse male meiosis. J. Cell Sci. 2000, 113, 4275–4286. [Google Scholar] [CrossRef] [PubMed]
- Lattao, R.; Bonaccorsi, S.; Gatti, M. Giant meiotic spindles in males from Drosophila species with giant sperm tails. J. Cell Sci. 2012, 125, 584–588. [Google Scholar] [CrossRef]
- Pitnick, S.; Spicer, G.S.; Markow, T.A. How long is a giant sperm? Nature 1995, 375, 109. [Google Scholar] [CrossRef]
- Pitnick, S.; Markow, T.A.; Spicer, G.S. Delayed male maturity is a cost of producing large sperm in Drosophila. Proc. Natl. Acad. Sci. USA 1995, 92, 10614–10618. [Google Scholar] [CrossRef]
- Lüpold, S.; Manier, M.K.; Puniamoorthy, N.; Schoff, C.; Starmer, W.T.; Luepold, S.H.; Belote, J.M.; Pitnick, S. How sexual selection can drive the evolution of costly sperm ornamentation. Nature 2016, 533, 535–538. [Google Scholar] [CrossRef]
- Good, M.C.; Vahey, M.D.; Skandarajah, A.; Fletcher, D.A.; Heald, R. Cytoplasmic volume modulates spindle size during embryogenesis. Science 2013, 342, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Hazel, J.; Krutkramelis, K.; Mooney, P.; Tomschik, M.; Gerow, K.; Oakey, J.; Gatlin, J.C. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science 2013, 342, 853–856. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Li, Z.; Wang, S.; Barnes, P.M.; Liu, X.; Xu, H.; Jin, M.; Liu, A.P.; Yang, Q. A robust and tunable mitotic oscillator in artificial cells. Elife 2018, 7, e33549. [Google Scholar] [CrossRef]
- Arata, Y.; Takagi, H. Quantitative Studies for Cell-Division Cycle Control. Front. Physiol. 2019, 10, 1022. [Google Scholar] [CrossRef] [PubMed]
- Rieckhoff, E.M.; Berndt, F.; Elsner, M.; Golfier, S.; Decker, F.; Ishihara, K.; Brugués, J. Spindle Scaling Is Governed by Cell Boundary Regulation of Microtubule Nucleation. Curr. Biol. 2020, 30, 4973–4983.e10. [Google Scholar] [CrossRef]
- Lacroix, B.; Letort, G.; Pitayu, L.; Sallé, J.; Stefanutti, M.; Maton, G.; Ladouceur, A.M.; Canman, J.C.; Maddox, P.S.; Maddox, A.S.; et al. Microtubule Dynamics Scale with Cell Size to Set Spindle Length and Assembly Timing. Dev. Cell. 2018, 45, 496–511.e6. [Google Scholar] [CrossRef]
- Farhadifar, R.; Yu, C.H.; Fabig, G.; Wu, H.Y.; Stein, D.B.; Rockman, M.; Müller-Reichert, T.; Shelley, M.J.; Needleman, D.J. Stoichiometric interactions explain spindle dynamics and scaling across 100 million years of nematode evolution. Elife 2020, 9, e55877. [Google Scholar] [CrossRef]
- Kemphues, K.J.; Kaufman, T.C.; Raff, R.A.; Raff, E.C. The testis-specific beta-tubulin subunit in Drosophila melanogaster has multiple functions in spermatogenesis. Cell 1982, 31, 655–670. [Google Scholar] [CrossRef]
- Hutchens, J.A.; Hoyle, H.D.; Turner, F.R.; Raff, E.C. Structurally similar Drosophila alpha-tubulins are functionally distinct in vivo. Mol. Biol. Cell 1997, 8, 481–500.e11. [Google Scholar] [CrossRef]
- Cenci, G.; Bonaccorsi, S.; Pisano, C.; Verni, F.; Gatti, M. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 1994, 107, 3521–3534. [Google Scholar] [CrossRef]
- Noguchi, T.; Koizumi, M.; Hayashi, S. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr. Biol. 2011, 21, 805–814. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, P.P.; Giansanti, M.G.; Petronczki, M. Cytokinesis in animal cells. Cold Spring Harb. Perspect. Biol. 2015, 7, a015834. [Google Scholar] [CrossRef] [PubMed]
- Giansanti, M.G.; Bonaccorsi, S.; Bucciarelli, E.; Gatti, M. Drosophila male meiosis as a model system for the study of cytokinesis in animal cells. Cell Struct. Funct. 2001, 26, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Church, K.; Lin, H.P. Meiosis in Drosophila melanogaster. II. The prometaphase-I kinetochore microtubule bundle and kinetochore orientation in males. J. Cell Biol. 1982, 93, 365–373. [Google Scholar] [CrossRef]
- Rappaport, R. Cytokinesis in Animal Cells; Cambridge University Press: Cambridge, UK, 1996. [Google Scholar]
- Glotzer, M. The 3Ms of central spindle assembly: Microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 2009, 10, 9–20. [Google Scholar] [CrossRef]
- Verma, V.; Mogilner, A.; Maresca, T.J. Classical and Emerging Regulatory Mechanisms of Cytokinesis in Animal Cells. Biology 2019, 8, 55. [Google Scholar] [CrossRef]
- Giansanti, M.G.; Bonaccorsi, S.; Gatti, M. The role of anillin in meiotic cytokinesis of Drosophila males. J. Cell Sci. 1999, 112, 2323–2334. [Google Scholar] [CrossRef]
- Giansanti, M.G.; Farkas, R.M.; Bonaccorsi, S.; Lindsley, D.L.; Wakimoto, B.T.; Fuller, M.T.; Gatti, M. Genetic dissection of meiotic cytokinesis in Drosophila males. Mol. Biol. Cell 2004, 15, 2509–2522. [Google Scholar] [CrossRef]
- Williams, B.C.; Riedy, M.F.; Williams, E.V.; Gatti, M.; Goldberg, M.L. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 1995, 129, 709–723. [Google Scholar] [CrossRef]
- Inoue, Y.H.; Savoian, M.S.; Suzuki, T.; Máthé, E.; Yamamoto, M.T.; Glover, D.M. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 2004, 166, 49–60. [Google Scholar] [CrossRef]
- Scholey, J.M.; Rogers, G.C.; Sharp, D.J. Mitosis, microtubules, and the matrix. J. Cell Biol. 2001, 154, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Vernì, F.; Somma, M.P.; Gunsalus, K.C.; Bonaccorsi, S.; Belloni, G.; Goldberg, M.L.; Gatti, M. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr. Biol. 2004, 14, 1569–1575. [Google Scholar] [CrossRef] [PubMed]
- Szafer-Glusman, E.; Fuller, M.T.; Giansanti, M.G. Role of Survivin in cytokinesis revealed by a separation-of-function allele. Mol. Biol. Cell 2011, 22, 3779–3790. [Google Scholar] [CrossRef] [PubMed]
- D’Avino, P.P.; Archambault, V.; Przewloka, M.R.; Zhang, W.; Lilley, K.S.; Laue, E.; Glover, D.M. Recruitment of Polo kinase to the spindle midzone during cytokinesis requires the Feo/Klp3A complex. PLoS ONE 2007, 2, e572. [Google Scholar] [CrossRef]
- Adams, R.R.; Tavares, A.A.; Salzberg, A.; Bellen, H.J.; Glover, D.M. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998, 12, 1483–1494. [Google Scholar] [CrossRef]
- D’Avino, P.P.; Savoian, M.S.; Capalbo, L.; Glover, D.M. RacGAP50C is sufficient to signal cleavage furrow formation during cytokinesis. J. Cell Sci. 2006, 119, 4402–4408. [Google Scholar] [CrossRef] [PubMed]
- Somers, W.G.; Saint, R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell. 2003, 4, 29–39. [Google Scholar] [CrossRef]
- Yüce, O.; Piekny, A.; Glotzer, M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 2005, 170, 571–582. [Google Scholar] [CrossRef]
- Zhao, W.M.; Fang, G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. USA 2005, 102, 13158–13163. [Google Scholar] [CrossRef]
- Piekny, A.; Werner, M.; Glotzer, M. Cytokinesis: Welcome to the Rho zone. Trends Cell Biol. 2005, 15, 651–658. [Google Scholar] [CrossRef]
- Kamijo, K.; Ohara, N.; Abe, M.; Uchimura, T.; Hosoya, H.; Lee, J.S.; Miki, T. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol. Biol. Cell 2006, 17, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yonemura, S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 2006, 119, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Guse, A.; Mishima, M.; Glotzer, M. Phosphorylation of ZEN-4/MKLP1 by aurora B regulates completion of cytokinesis. Curr. Biol. 2005, 15, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Neef, R.; Klein, U.R.; Kopajtich, R.; Barr, F.A. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr. Biol. 2006, 16, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Davies, T.; Joseph, N.; Mishima, M. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr. Biol. 2010, 20, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Brennan, I.M.; Peters, U.; Kapoor, T.M.; Straight, A.F. Polo-like kinase controls vertebrate spindle elongation and cytokinesis. PLoS ONE 2007, 2, e409. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, A.; Neef, R.; Eberspächer, U.; Eis, K.; Husemann, M.; Mumberg, D.; Prechtl, S.; Schulze, V.; Siemeister, G.; Wortmann, L.; et al. Use of the novel Plk1 inhibitor ZK-thiazolidinone to elucidate functions of Plk1 in early and late stages of mitosis. Mol. Biol. Cell 2007, 18, 4024–4036. [Google Scholar] [CrossRef]
- Burkard, M.E.; Maciejowski, J.; Rodriguez-Bravo, V.; Repka, M.; Lowery, D.M.; Clauser, K.R.; Zhang, C.; Shokat, K.M.; Carr, S.A.; Yaffe, M.B.; et al. Plk1 self-organization and priming phosphorylation of HsCYK-4 at the spindle midzone regulate the onset of division in human cells. PLoS Biol. 2009, 7, e1000111. [Google Scholar] [CrossRef]
- Wolfe, B.A.; Takaki, T.; Petronczki, M.; Glotzer, M. Polo-like kinase 1 directs assembly of the HsCyk-4 RhoGAP/Ect2 RhoGEF complex to initiate cleavage furrow formation. PLoS Biol. 2009, 7, e1000110. [Google Scholar] [CrossRef]
- Barr, F.A.; Silljé, H.H.; Nigg, E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004, 5, 429–440. [Google Scholar] [CrossRef]
- van der Waal, M.S.; Hengeveld, R.C.; van der Horst, A.; Lens, S.M. Cell division control by the Chromosomal Passenger Complex. Exp. Cell Res. 2012, 318, 1407–1420. [Google Scholar] [CrossRef] [PubMed]
- Llamazares, S.; Moreira, A.; Tavares, A.; Girdham, C.; Spruce, B.A.; Gonzalez, C.; Karess, R.E.; Glover, D.M.; Sunkel, C.E. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991, 5, 2153–2165. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Riparbelli, M.G.; Minestrini, G.; Tavares, A.M.; Adams, R.; Callaini, G.; Glover, D.M. Drosophila polo kinase is required for cytokinesis. J. Cell Biol. 1998, 143, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, S.; Amorim, I.; Sunkel, C.E. The POLO kinase is required at multiple stages during spermatogenesis in Drosophila melanogaster. Chromosoma 1998, 107, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.R.; Wheatley, S.P.; Gouldsworthy, A.M.; Kandels-Lewis, S.E.; Carmena, M.; Smythe, C.; Gerloff, D.L.; Earnshaw, W.C. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000, 10, 1075–1078. [Google Scholar] [CrossRef]
- Giet, R.; Glover, D.M. Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 2001, 152, 669–682. [Google Scholar] [CrossRef]
- Vagnarelli, P.; Earnshaw, W.C. Chromosomal passengers: The four-dimensional regulation of mitotic events. Chromosoma 2004, 113, 211–222. [Google Scholar] [CrossRef]
- Vader, G.; Medema, R.H.; Lens, S.M. The chromosomal passenger complex: Guiding Aurora-B through mitosis. J. Cell Biol. 2006, 173, 833–837. [Google Scholar] [CrossRef]
- Gao, S.; Giansanti, M.G.; Buttrick, G.J.; Ramasubramanyan, S.; Auton, A.; Gatti, M.; Wakefield, J.G. Australin: A chromosomal passenger protein required specifically for Drosophila melanogaster male meiosis. J. Cell Biol. 2008, 180, 521–535. [Google Scholar] [CrossRef]
- Capalbo, L.; Montembault, E.; Takeda, T.; Bassi, Z.I.; Glover, D.M.; D’Avino, P.P. The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis. Open Biol. 2012, 2, 120070. [Google Scholar] [CrossRef]
- Roth, M.; Roubinet, C.; Iffländer, N.; Ferrand, A.; Cabernard, C. Asymmetrically dividing Drosophila neuroblasts utilize two spatially and temporally independent cytokinesis pathways. Nat. Commun. 2015, 6, 6551. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Gallaud, E.; Pascal, A.; Serre, L.; Arnal, I.; Richard-Parpaillon, L.; Savoian, M.S.; Giet, R. Peripheral astral microtubules ensure asymmetric furrow positioning in neural stem cells. Cell Rep. 2021, 37, 109895. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Lombardia, M.O.; Ogawa, H.; Earnshaw, W.C. Polo kinase regulates the localization and activity of the chromosomal passenger complex in meiosis and mitosis in Drosophila melanogaster. Open Biol. 2014, 4, 140162. [Google Scholar] [CrossRef] [PubMed]
- Ketchum, A.S.; Stewart, C.T.; Stewart, M.; Kiehart, D.P. Complete sequence of the Drosophila nonmuscle myosin heavy-chain transcript: Conserved sequences in the myosin tail and differential splicing in the 5′ untranslated sequence. Proc. Natl. Acad. Sci. USA 1990, 87, 6316–6320. [Google Scholar] [CrossRef] [PubMed]
- Kiehart, D.P.; Lutz, M.S.; Chan, D.; Ketchum, A.S.; Laymon, R.A.; Nguyen, B.; Goldstein, L.S. Identification of the gene for fly non-muscle myosin heavy chain: Drosophila myosin heavy chains are encoded by a gene family. EMBO J. 1989, 8, 913–922. [Google Scholar] [CrossRef]
- Mansfield, S.G.; al-Shirawi, D.Y.; Ketchum, A.S.; Newbern, E.C.; Kiehart, D.P. Molecular organization and alternative splicing in zipper, the gene that encodes the Drosophila non-muscle myosin II heavy chain. J. Mol. Biol. 1996, 255, 98–109. [Google Scholar] [CrossRef]
- Young, P.E.; Richman, A.M.; Ketchum, A.S.; Kiehart, D.P. Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 1993, 7, 29–41. [Google Scholar] [CrossRef]
- Shutova, M.S.; Svitkina, T.M. Mammalian nonmuscle myosin II comes in three flavors. Biochem. Biophys. Res. Commun. 2018, 506, 394–402. [Google Scholar] [CrossRef]
- Murakami, N.; Kotula, L.; Hwang, Y.W. Two distinct mechanisms for regulation of nonmuscle myosin assembly via the heavy chain: Phosphorylation for MIIB and mts 1 binding for MIIA. Biochemistry 2000, 39, 11441–11451. [Google Scholar] [CrossRef]
- Ronen, D.; Ravid, S. Myosin II tailpiece determines its paracrystal structure, filament assembly properties, and cellular localization. J. Biol. Chem. 2009, 284, 24948–24957. [Google Scholar] [CrossRef]
- Vicente-Manzanares, M.; Ma, X.; Adelstein, R.S.; Horwitz, A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Sirajuddin, M.; Farkasovsky, M.; Hauer, F.; Kühlmann, D.; Macara, I.G.; Weyand, M.; Stark, H.; Wittinghofer, A. Structural insight into filament formation by mammalian septins. Nature 2007, 449, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Bridges, A.A.; Gladfelter, A.S. Septin Form and Function at the Cell Cortex. J. Biol. Chem. 2015, 290, 17173–17180. [Google Scholar] [CrossRef] [PubMed]
- Spiliotis, E.T. Spatial effects - site-specific regulation of actin and microtubule organization by septin GTPases. J. Cell Sci. 2018, 131, jcs207555. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, D.C.; Macedo, J.N.; Guimarães, S.L.; Barroso da Silva, F.L.; Cassago, A.; Garratt, R.C.; Portugal, R.V.; Araujo, A.P.U. A revised order of subunits in mammalian septin complexes. Cytoskeleton 2019, 76, 457–466. [Google Scholar] [CrossRef]
- Soroor, F.; Kim, M.S.; Palander, O.; Balachandran, Y.; Collins, R.F.; Benlekbir, S.; Rubinstein, J.L.; Trimble, W.S. Revised subunit order of mammalian septin complexes explains their in vitro polymerization properties. Mol. Biol. Cell 2021, 32, 289–300. [Google Scholar] [CrossRef]
- Field, C.M.; al-Awar, O.; Rosenblatt, J.; Wong, M.L.; Alberts, B.; Mitchison, T.J. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 1996, 133, 605–616. [Google Scholar] [CrossRef]
- Hime, G.R.; Brill, J.A.; Fuller, M.T. Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 1996, 109, 2779–2788. [Google Scholar] [CrossRef]
- Sechi, S.; Colotti, G.; Belloni, G.; Mattei, V.; Frappaolo, A.; Raffa, G.D.; Fuller, M.T.; Giansanti, M.G. GOLPH3 is essential for contractile ring formation and Rab11 localization to the cleavage site during cytokinesis in Drosophila melanogaster. PLoS Genet. 2014, 10, e1004305. [Google Scholar] [CrossRef]
- Zhang, J.; Kong, C.; Xie, H.; McPherson, P.S.; Grinstein, S.; Trimble, W.S. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr. Biol. 1999, 9, 1458–1467. [Google Scholar] [CrossRef]
- Tanaka-Takiguchi, Y.; Kinoshita, M.; Takiguchi, K. Septin-mediated uniform bracing of phospholipid membranes. Curr. Biol. 2009, 19, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Bertin, A.; McMurray, M.A.; Thai, L.; Garcia, G., 3rd; Votin, V.; Grob, P.; Allyn, T.; Thorner, J.; Nogales, E. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J. Mol. Biol. 2010, 404, 711–731. [Google Scholar] [CrossRef] [PubMed]
- Field, C.M.; Alberts, B.M. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 1995, 131, 165–178. [Google Scholar] [CrossRef]
- Field, C.M.; Coughlin, M.; Doberstein, S.; Marty, T.; Sullivan, W. Characterization of anillin mutants reveals essential roles in septin localization and plasma membrane integrity. Development 2005, 132, 2849–2860. [Google Scholar] [CrossRef]
- Jananji, S.; Risi, C.; Lindamulage, I.K.S.; Picard, L.P.; Van Sciver, R.; Laflamme, G.; Albaghjati, A.; Hickson, G.R.X.; Kwok, B.H.; Galkin, V.E. Multimodal and Polymorphic Interactions between Anillin and Actin: Their Implications for Cytokinesis. J. Mol. Biol. 2017, 429, 715–731. [Google Scholar] [CrossRef]
- Straight, A.F.; Field, C.M.; Mitchison, T.J. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol. Biol. Cell 2005, 16, 193–201. [Google Scholar] [CrossRef]
- Kinoshita, M.; Field, C.M.; Coughlin, M.L.; Straight, A.F.; Mitchison, T.J. Self- and actin-templated assembly of Mammalian septins. Dev. Cell 2002, 3, 791–802. [Google Scholar] [CrossRef]
- Hickson, G.R.; O’Farrell, P.H. Rho-dependent control of anillin behavior during cytokinesis. J. Cell Biol. 2008, 180, 285–294. [Google Scholar] [CrossRef]
- D’Avino, P.P. How to scaffold the contractile ring for a safe cytokinesis - lessons from Anillin-related proteins. J. Cell Sci. 2009, 122, 1071–1079. [Google Scholar] [CrossRef]
- Piekny, A.J.; Maddox, A.S. The myriad roles of Anillin during cytokinesis. Semin. Cell Dev. Biol. 2010, 21, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Guan, R.; Lee, I.J.; Liu, Y.; Chen, M.; Wang, J.; Wu, J.Q.; Chen, Z. Mechanistic insights into the anchorage of the contractile ring by anillin and Mid1. Dev. Cell 2015, 33, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Budnar, S.; Husain, K.B.; Gomez, G.A.; Naghibosadat, M.; Varma, A.; Verma, S.; Hamilton, N.A.; Morris, R.G.; Yap, A.S. Anillin Promotes Cell Contractility by Cyclic Resetting of RhoA Residence Kinetics. Dev. Cell 2019, 49, 894–906.e12. [Google Scholar] [CrossRef]
- D’Avino, P.P.; Takeda, T.; Capalbo, L.; Zhang, W.; Lilley, K.S.; Laue, E.D.; Glover, D.M. Interaction between Anillin and RacGAP50C connects the actomyosin contractile ring with spindle microtubules at the cell division site. J. Cell Sci. 2008, 121, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Gregory, S.L.; Ebrahimi, S.; Milverton, J.; Jones, W.M.; Bejsovec, A.; Saint, R. Cell division requires a direct link between microtubule-bound RacGAP and Anillin in the contractile ring. Curr. Biol. 2008, 18, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gai, M.; Camera, P.; Dema, A.; Bianchi, F.; Berto, G.; Scarpa, E.; Germena, G.; Di Cunto, F. Citron kinase controls abscission through RhoA and anillin. Mol. Biol. Cell 2011, 22, 3768–3778. [Google Scholar] [CrossRef]
- Frenette, P.; Haines, E.; Loloyan, M.; Kinal, M.; Pakarian, P.; Piekny, A. An anillin-Ect2 complex stabilizes central spindle microtubules at the cortex during cytokinesis. PLoS ONE 2012, 7, e34888. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fairn, G.D.; Ceccarelli, D.F.; Sicheri, F.; Wilde, A. Cleavage furrow organization requires PIP(2)-mediated recruitment of anillin. Curr. Biol. 2012, 22, 64–69. [Google Scholar] [CrossRef]
- Frémont, S.; Echard, A. Membrane Traffic in the Late Steps of Cytokinesis. Curr. Biol. 2018, 28, R458–R470. [Google Scholar] [CrossRef]
- Farkas, R.M.; Giansanti, M.G.; Gatti, M.; Fuller, M.T. The Drosophila Cog5 homologue is required for cytokinesis, cell elongation, and assembly of specialized Golgi architecture during spermatogenesis. Mol. Biol. Cell 2003, 14, 190–200. [Google Scholar] [CrossRef]
- Belloni, G.; Sechi, S.; Riparbelli, M.G.; Fuller, M.T.; Callaini, G.; Giansanti, M.G. Mutations in Cog7 affect Golgi structure, meiotic cytokinesis and sperm development during Drosophila spermatogenesis. J. Cell Sci. 2012, 125, 5441–5452. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Brill, J.A.; Hsien, J.; McBride, R.; Boulianne, G.L.; Trimble, W.S. Syntaxin 5 is required for cytokinesis and spermatid differentiation in Drosophila. Dev. Biol. 2002, 251, 294–306. [Google Scholar] [CrossRef]
- Robinett, C.C.; Giansanti, M.G.; Gatti, M.; Fuller, M.T. TRAPPII is required for cleavage furrow ingression and localization of Rab11 in dividing male meiotic cells of Drosophila. J. Cell Sci. 2009, 122, 4526–4534. [Google Scholar] [CrossRef] [PubMed]
- Giansanti, M.G.; Belloni, G.; Gatti, M. Rab11 is required for membrane trafficking and actomyosin ring constriction in meiotic cytokinesis of Drosophila males. Mol. Biol. Cell 2007, 18, 5034–5047. [Google Scholar] [CrossRef]
- Dyer, N.; Rebollo, E.; Domínguez, P.; Elkhatib, N.; Chavrier, P.; Daviet, L.; González, C.; González-Gaitán, M. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development 2007, 134, 4437–4447. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, D.; Yamaguchi, M.; Mori, H.; Inoue, Y.H. COPI-mediated membrane trafficking is required for cytokinesis in Drosophila male meiotic divisions. J. Cell Sci. 2012, 125, 3649–3660. [Google Scholar] [CrossRef]
- Giansanti, M.G.; Vanderleest, T.E.; Jewett, C.E.; Sechi, S.; Frappaolo, A.; Fabian, L.; Robinett, C.C.; Brill, J.A.; Loerke, D.; Fuller, M.T.; et al. Exocyst-Dependent Membrane Addition Is Required for Anaphase Cell Elongation and Cytokinesis in Drosophila. PLoS Genet 2015, 11, e1005632. [Google Scholar] [CrossRef]
- Sechi, S.; Frappaolo, A.; Fraschini, R.; Capalbo, L.; Gottardo, M.; Belloni, G.; Glover, D.M.; Wainman, A.; Giansanti, M.G. Rab1 interacts with GOLPH3 and controls Golgi structure and contractile ring constriction during cytokinesis in Drosophila melanogaster. Open Biol. 2017, 7, 160257. [Google Scholar] [CrossRef]
- Frappaolo, A.; Sechi, S.; Kumagai, T.; Robinson, S.; Fraschini, R.; Karimpour-Ghahnavieh, A.; Belloni, G.; Piergentili, R.; Tiemeyer, K.H.; Tiemeyer, M.; et al. COG7 deficiency in Drosophila generates multifaceted developmental, behavioral and protein glycosylation phenotypes. J. Cell Sci. 2017, 130, 3637–3649. [Google Scholar] [CrossRef]
- Wong, R.; Hadjiyanni, I.; Wei, H.C.; Polevoy, G.; McBride, R.; Sem, K.P.; Brill, J.A. PIP2 hydrolysis and calcium release are required for cytokinesis in Drosophila spermatocytes. Curr. Biol. 2005, 15, 1401–1406. [Google Scholar] [CrossRef]
- Echard, A. Phosphoinositides and cytokinesis: The “PIP” of the iceberg. Cytoskeleton 2012, 69, 893–912. [Google Scholar] [CrossRef] [PubMed]
- Brill, J.A.; Wong, R.; Wilde, A. Phosphoinositide function in cytokinesis. Curr. Biol. 2011, 21, R930–R934. [Google Scholar] [CrossRef]
- Yin, H.L.; Janmey, P.A. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 2003, 65, 761–789. [Google Scholar] [CrossRef] [PubMed]
- Lekomtsev, S.; Su, K.C.; Pye, V.E.; Blight, K.; Sundaramoorthy, S.; Takaki, T.; Collinson, L.M.; Cherepanov, P.; Divecha, N.; Petronczki, M. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature 2012, 492, 276–279. [Google Scholar] [CrossRef] [PubMed]
- Sechi, S.; Frappaolo, A.; Belloni, G.; Giansanti, M.G. The roles of the oncoprotein GOLPH3 in contractile ring assembly and membrane trafficking during cytokinesis. Biochem. Soc. Trans. 2015, 43, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Sechi, S.; Frappaolo, A.; Karimpour-Ghahnavieh, A.; Fraschini, R.; Giansanti, M.G. A novel coordinated function of Myosin II with GOLPH3 controls centralspindlin localization during cytokinesis in Drosophila. J. Cell Sci. 2020, 133, jcs252965. [Google Scholar] [CrossRef] [PubMed]
- Sechi, S.; Karimpour-Ghahnavieh, A.; Frappaolo, A.; Di Francesco, L.; Piergentili, R.; Schininà, E.; D’Avino, P.P.; Giansanti, M.G. Identification of GOLPH3 Partners in Drosophila Unveils Potential Novel Roles in Tumorigenesis and Neural Disorders. Cells 2021, 10, 2336. [Google Scholar] [CrossRef] [PubMed]
- Vale, R.D.; Spudich, J.A.; Griffis, E.R. Dynamics of myosin, microtubules, and Kinesin-6 at the cortex during cytokinesis in Drosophila S2 cells. J. Cell Biol. 2009, 186, 727–738. [Google Scholar] [CrossRef]
- Eda, M.; Yonemura, S.; Kato, T.; Watanabe, N.; Ishizaki, T.; Madaule, P.; Narumiya, S. Rho-dependent transfer of Citron-kinase to the cleavage furrow of dividing cells. J. Cell Sci. 2001, 114, 3273–3284. [Google Scholar] [CrossRef]
- Ishizaki, T.; Maekawa, M.; Fujisawa, K.; Okawa, K.; Iwamatsu, A.; Fujita, A.; Watanabe, N.; Saito, Y.; Kakizuka, A.; Morii, N.; et al. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996, 15, 1885–1893. [Google Scholar] [CrossRef]
- Matsumura, F.; Yamakita, Y.; Yamashiro, S. Myosin light chain kinases and phosphatase in mitosis and cytokinesis. Arch. Biochem. Biophys. 2011, 510, 76–82. [Google Scholar] [CrossRef]
- Yamashiro, S.; Yamakita, Y.; Totsukawa, G.; Goto, H.; Kaibuchi, K.; Ito, M.; Hartshorne, D.J.; Matsumura, F. Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Dev. Cell 2008, 14, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Uehara, R.; Goshima, G.; Mabuchi, I.; Vale, R.D.; Spudich, J.A.; Griffis, E.R. Determinants of myosin II cortical localization during cytokinesis. Curr. Biol. 2010, 20, 1080–1085. [Google Scholar] [CrossRef] [PubMed]
- Beach, J.R.; Egelhoff, T.T. Myosin II recruitment during cytokinesis independent of centralspindlin-mediated phosphorylation. J. Biol. Chem. 2009, 284, 27377–27383. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Shu, S.; Billington, N.; Williamson, C.D.; Yu, S.; Brzeska, H.; Donaldson, J.G.; Sellers, J.R.; Korn, E.D. Mammalian non muscle Myosin II binds to anionic phospholipids with concomitant dissociation of the regulatory light chain. J. Biol. Chem. 2016, 291, 24828–24837. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.N.; Cooley, L. Stable intercellular bridges in development: The cytoskeleton lining the tunnel. Trends Cell Biol. 1996, 6, 474–479. [Google Scholar] [CrossRef]
- Yamashita, Y.M. Subcellular Specialization and Organelle Behavior in Germ Cells. Genetics 2018, 208, 19–51. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, M.P.; Iwamori, T.; Buchold, G.M.; Matzuk, M.M. Germ cell intercellular bridges. Cold Spring Harb. Perspect. Biol. 2011, 3, a005850. [Google Scholar] [CrossRef]
- Lu, K.; Jensen, L.; Lei, L.; Yamashita, Y.M. Stay Connected: A Germ Cell Strategy. Trends Genet. 2017, 33, 971–978. [Google Scholar] [CrossRef]
- Haglund, K.; Nezis, I.P.; Stenmark, H. Structure and functions of stable intercellular bridges formed by incomplete cytokinesis during development. Commun. Integr. Biol. 2011, 4, 1–9. [Google Scholar] [CrossRef]
- Eikenes, Å.H.; Brech, A.; Stenmark, H.; Haglund, K. Spatiotemporal control of Cindr at ring canals during incomplete cytokinesis in the Drosophila male germline. Dev. Biol. 2013, 377, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, C.; Kitazawa, D.; Ando, I.; Hayashi, D.; Inoue, Y.H. Orbit/CLASP is required for germline cyst formation through its developmental control of fusomes and ring canals in Drosophila males. PLoS ONE 2013, 8, e58220. [Google Scholar] [CrossRef]
- Montembault, E.; Zhang, W.; Przewloka, M.R.; Archambault, V.; Sevin, E.W.; Laue, E.D.; Glover, D.M.; D’Avino, P.P. Nessun Dorma, a novel centralspindlin partner, is required for cytokinesis in Drosophila spermatocytes. J. Cell Biol. 2010, 191, 1351–1365. [Google Scholar] [CrossRef]
- Braun, R.E.; Behringer, R.R.; Peschon, J.J.; Brinster, R.L.; Palmiter, R.D. Genetically haploid spermatids are phenotypically diploid. Nature 1989, 337, 373–376. [Google Scholar] [CrossRef] [PubMed]
- Morales, C.R.; Lefrancois, S.; Chennathukuzhi, V.; El-Alfy, M.; Wu, X.; Yang, J.; Gerton, G.L.; Hecht, N.B. A TB-RBP and Ter ATPase complex accompanies specific mRNAs from nuclei through the nuclear pores and into intercellular bridges in mouse male germ cells. Dev. Biol. 2002, 246, 480–494. [Google Scholar] [CrossRef]
- Lu, K.L.; Yamashita, Y.M. Germ cell connectivity enhances cell death in response to DNA damage in the Drosophila testis. Elife 2017, 6, e27960. [Google Scholar] [CrossRef]
- Yue, L.; Spradling, A.C. hu-li tai shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes Dev. 1992, 6, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Yue, L.; Spradling, A.C. The Drosophila fusome, a germline-specific organelle, contains membrane skeletal proteins and functions in cyst formation. Development 1994, 120, 947–956. [Google Scholar] [CrossRef] [PubMed]
- de Cuevas, M.; Lee, J.K.; Spradling, A.C. alpha-spectrin is required for germline cell division and differentiation in the Drosophila ovary. Development 1996, 122, 3959–3968. [Google Scholar] [CrossRef]
- Yan, D.; Neumüller, R.A.; Buckner, M.; Ayers, K.; Li, H.; Hu, Y.; Yang-Zhou, D.; Pan, L.; Wang, X.; Kelley, C.; et al. A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell 2014, 28, 459–473. [Google Scholar] [CrossRef]
- Wilson, P.G. Centrosome inheritance in the male germ line of Drosophila requires hu-li tai-shao function. Cell Biol. Int. 2005, 29, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef]
- Mirzoyan, Z.; Sollazzo, M.; Allocca, M.; Valenza, A.M.; Grifoni, D.; Bellosta, P. Drosophila melanogaster: A Model Organism to Study Cancer. Front. Genet. 2019, 10, 51. [Google Scholar] [CrossRef]
- Brandt, A.; Vilcinskas, A. The fruit fly Drosophila melanogaster as a model for aging research. Adv. Biochem. Eng. Biotechnol. 2013, 135, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Frappaolo, A.; Sechi, S.; Kumagai, T.; Karimpour-Ghahnavieh, A.; Tiemeyer, M.; Giansanti, M.G. Modeling Congenital Disorders of N-Linked Glycoprotein Glycosylation in Drosophila melanogaster. Front. Genet. 2018, 9, 436. [Google Scholar] [CrossRef] [PubMed]
- Godinho, S.A.; Pellman, D. Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130467. [Google Scholar] [CrossRef]
- Dewhurst, S.M.; McGranahan, N.; Burrell, R.A.; Rowan, A.J.; Grönroos, E.; Endesfelder, D.; Joshi, T.; Mouradov, D.; Gibbs, P.; Ward, R.L.; et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 2014, 4, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Galofré, C.; Asensio, E.; Ubach, M.; Torres, I.M.; Quintanilla, I.; Castells, A.; Camps, J. Centrosome reduction in newly-generated tetraploid cancer cells obtained by separase depletion. Sci. Rep. 2020, 10, 9152. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.L.; Kabbarah, O.; Liang, M.C.; Ivanova, E.; Anagnostou, V.; Wu, J.; Dhakal, S.; Wu, M.; Chen, S.; Feinberg, T.; et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 2009, 459, 1085–1090. [Google Scholar] [CrossRef]
- Sechi, S.; Frappaolo, A.; Karimpour-Ghahnavieh, A.; Piergentili, R.; Giansanti, M.G. Oncogenic roles of GOLPH3 in the Physiopathology of cancer. Int. J. Mol. Sci. 2020, 21, 933. [Google Scholar] [CrossRef]
- Naydenov, N.; Koblinski, J.; Ivanov, A. Anillin is an emerging regulator of tumorigenesis, acting as a cortical cytoskeletal scaffold and a nuclear modulator of cancer cell differentiation. Cell Mol. Life Sci. 2021, 78, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Q.; Cheng, Y.; Gao, M.; Kuang, L.; Wang, C. Myosin. heavy chain 10 (MYH10) gene silencing reduces cell migration and invasion in the glioma cell lines U251, T98G, and SHG44 by inhibiting the Wnt/β-catenin pathway. Med. Sci. 2018, 24, 9110. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frappaolo, A.; Piergentili, R.; Giansanti, M.G. Microtubule and Actin Cytoskeletal Dynamics in Male Meiotic Cells of Drosophila melanogaster. Cells 2022, 11, 695. https://doi.org/10.3390/cells11040695
Frappaolo A, Piergentili R, Giansanti MG. Microtubule and Actin Cytoskeletal Dynamics in Male Meiotic Cells of Drosophila melanogaster. Cells. 2022; 11(4):695. https://doi.org/10.3390/cells11040695
Chicago/Turabian StyleFrappaolo, Anna, Roberto Piergentili, and Maria Grazia Giansanti. 2022. "Microtubule and Actin Cytoskeletal Dynamics in Male Meiotic Cells of Drosophila melanogaster" Cells 11, no. 4: 695. https://doi.org/10.3390/cells11040695
APA StyleFrappaolo, A., Piergentili, R., & Giansanti, M. G. (2022). Microtubule and Actin Cytoskeletal Dynamics in Male Meiotic Cells of Drosophila melanogaster. Cells, 11(4), 695. https://doi.org/10.3390/cells11040695






