BAG Family Members as Mitophagy Regulators in Mammals
Abstract
:1. Introduction
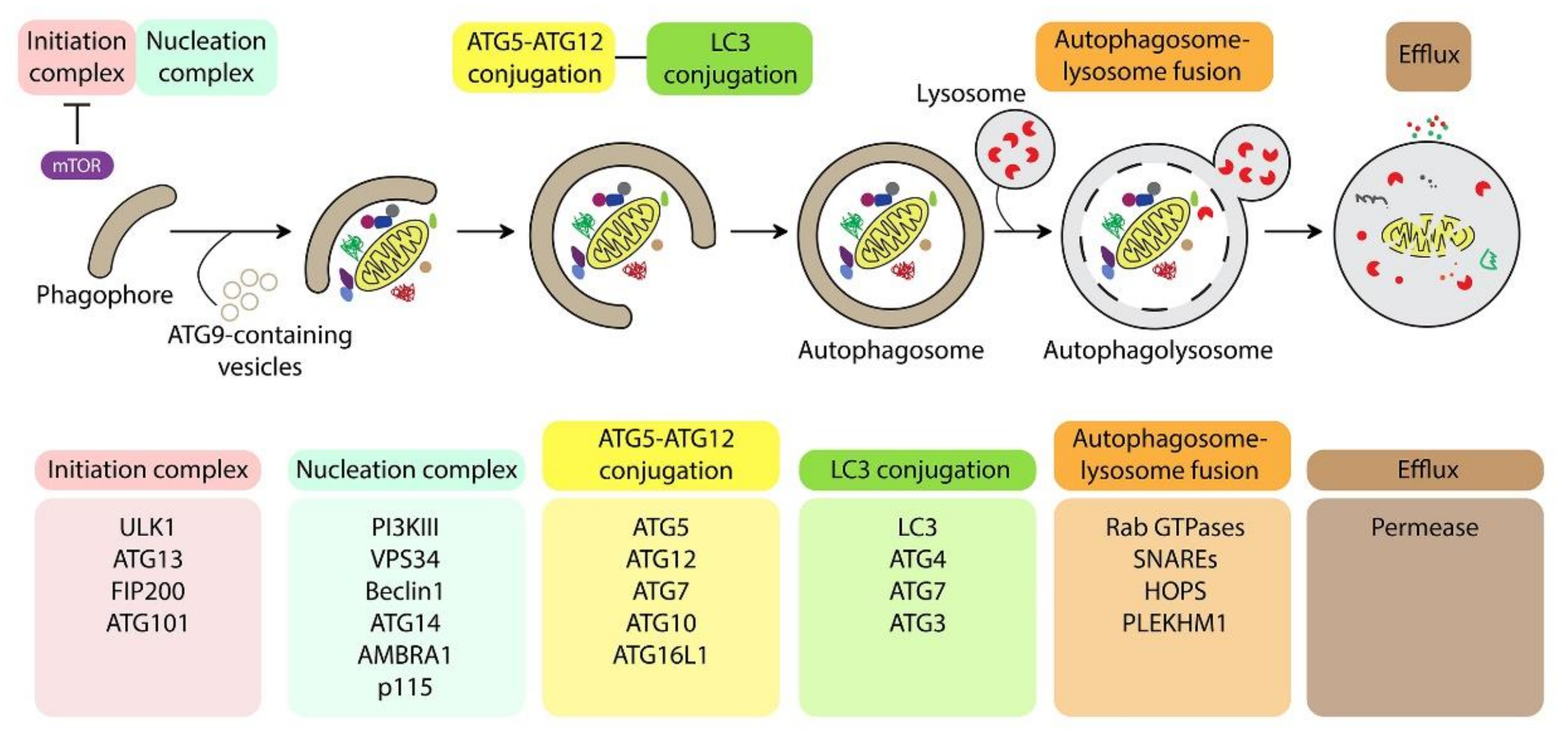
2. BAG Family Members in the Regulation of Autophagy and Selective Autophagy
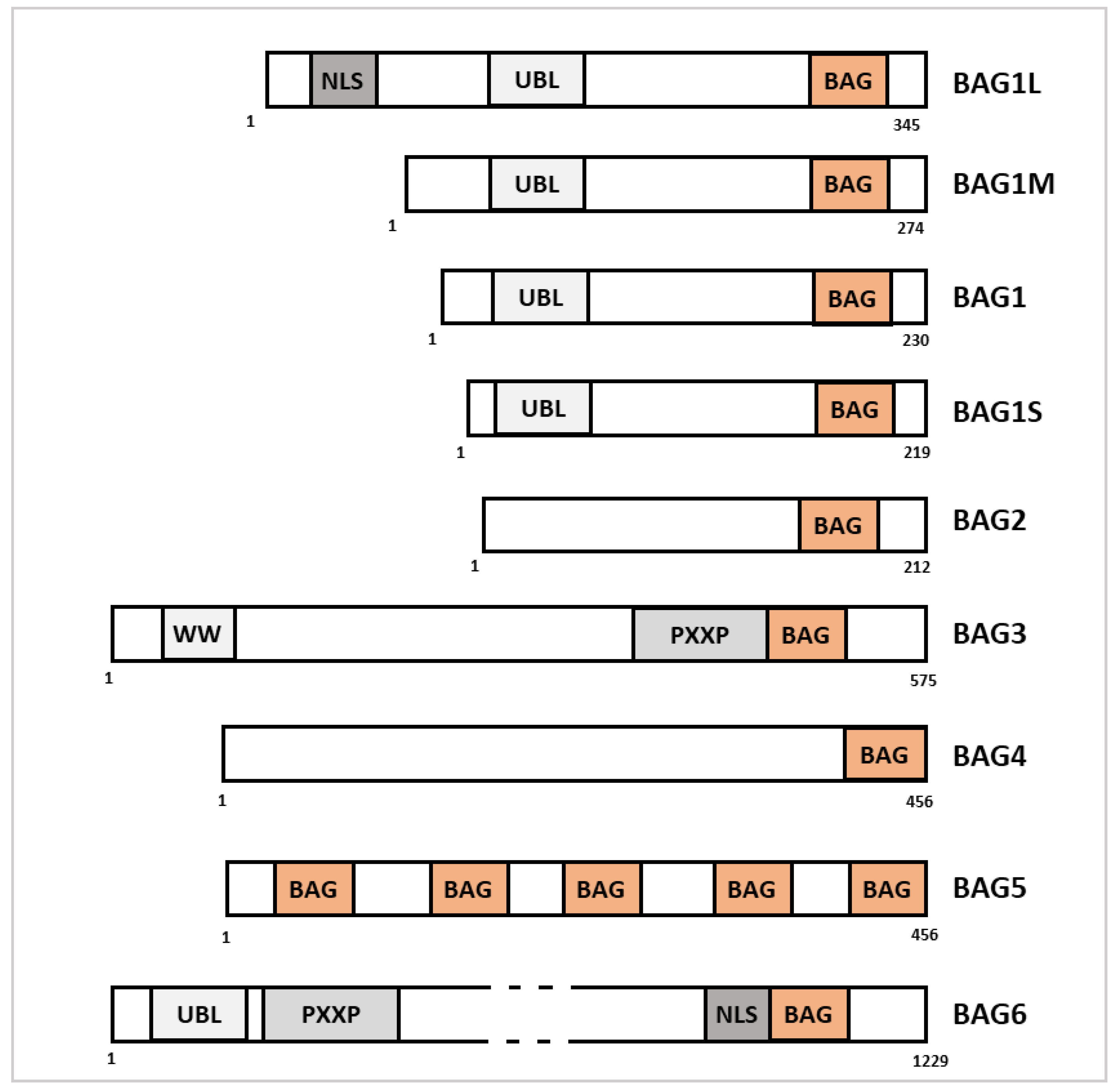
2.1. BAG1
2.2. BAG2
2.3. BAG3
2.4. BAG4
2.5. BAG5
2.6. BAG6
| Bag Family Member | Role in Autophagy |
|---|---|
| BAG1 | Stimulates autophagy during cardiac adaptation after ischemia/reperfusion injury [22]:
|
| BAG2 | Induction of reticulophagy after mycotuberculosis infection [25]:
|
| BAG3 | Promotion of chaperone-assisted selective autophagy [28] Stimulation of autophagy leading to drug resistance in colon and pancreatic cancer [37] |
| BAG5 | Stimulation of autophagy during sorafenib treatment in hepatocellular carcinoma leading to drug resistance [30] Promotion of aggrephagy through interaction with P62 in PD [40] |
| BAG6 | Modulation of autophagy in function of its intracellular localization [42,43]:
|
3. BAG Family Members in Mitophagy Regulation
3.1. BAG Family Members and the Regulation of Mitochondrial Morphology
3.2. BAG Family Members and the Regulation of the PINK/Parkin Signaling Pathway
3.2.1. PINK1
3.2.2. Parkin
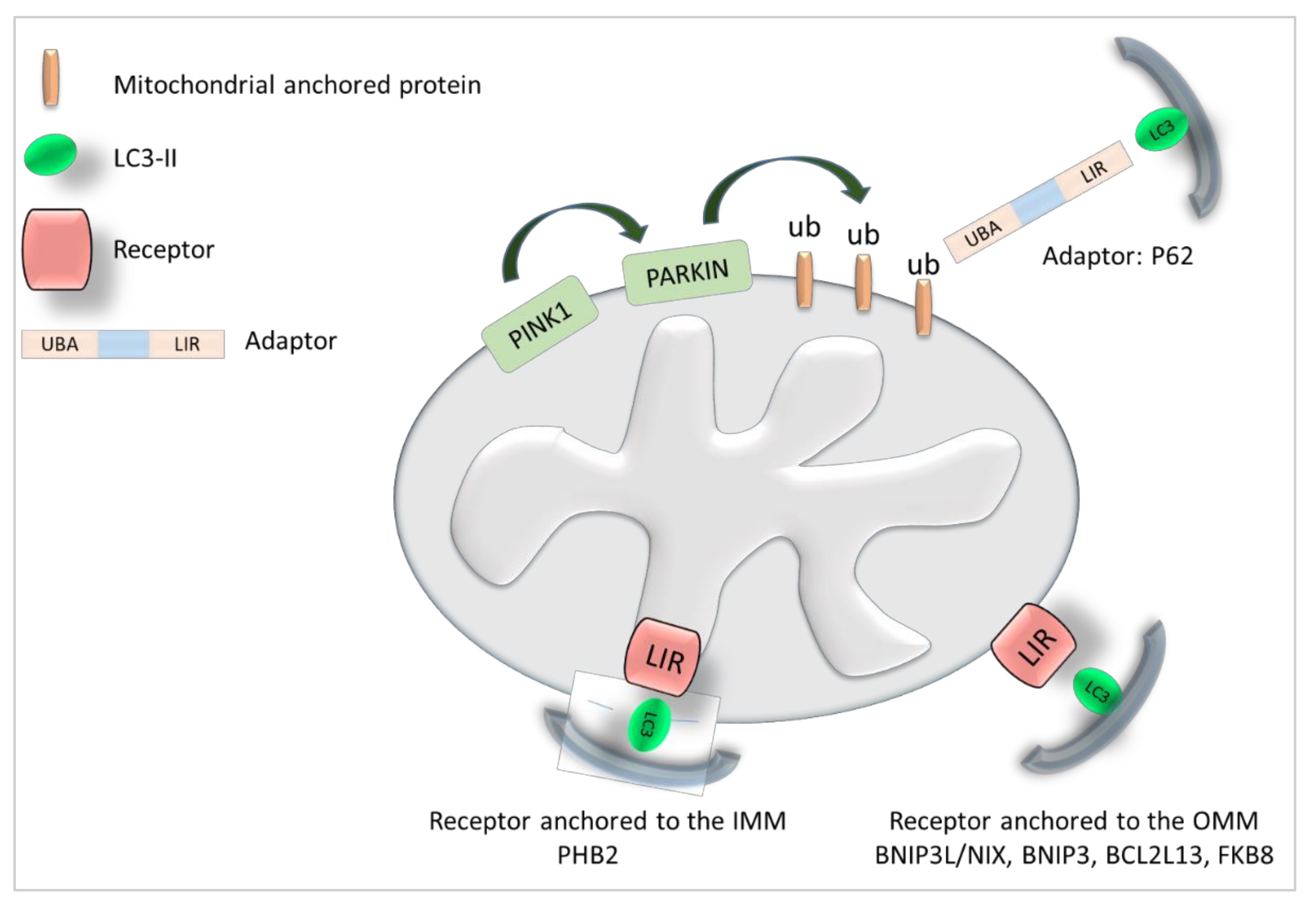
3.3. BAG Family Members as Mitophagy Receptors
| Bag Family Member | Role in Mitophagy |
|---|---|
| BAG2 | Stimulates mitophagy [52,53]: Interacts with PINK1 and inhibits its degradation. Promotes Parkin recruitment to the mitochondria and mitophagy. |
| BAG3 | Stimulates mitophagy: Recruited to mitochondria [59]. BAG3 silencing decreases Parkin expression and altered mitochondria are accumulated [59]. Interaction with the mitophagic receptor FUNDC1 [66], P62, BNIP3, NIX [67], Optoneurin [68]. The LIR motifs of BAG3 are conserved among species but their function is still unkown [65]. |
| BAG4 | Its role in mitophagy is unknown but BAG4 interacts with mitophagy regulators: Direct interaction with Parkin, inhibits its translocation to damaged mitochondria [62]. |
| BAG5 | Stimulates Mitophagy: Direct interaction with PINK1, increases its stability [55]. In aged bone marrow, the reduction of BAG5 destabilizes PINK1 and reduces mitophagy [56]. Inhibits mitophagy: Inhibits Parkin leading to dopaminergic neuron degeneration [60]. Direct interaction with Parkin and inhibition of its recruitment to the mitochondria leading to cell death after strong mitochondrial damages [61]. |
| BAG6 | Stimulates mitophagy: When localized in mitochondria, BAG6 promotes mitochondrial fission and PINK1/Parkin signaling [49]. Involved in the localization of mitochondria to the perinuclear region [51] New receptor for mitophagy [49]. Inhibits mitophagy in PD: Chronic MPP+ treatment increases the expression of BAG6 expression that interacts with PINK1 decreasing its stability [57]. |
4. Dual Role of BAG Family Members in the Regulation of Autophagy and Mitophagy: The Example of BAG6
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ATG | Autophagy-related proteins |
| BAG | BCL-2 associated athanogene |
| BCL-2 | B-cell lymphoma 2 |
| BCL2L13 | BCL2 Like 13 |
| BNIP3 | BCL2-adenovirus E1B 19 kDa protein-interacting protein 3 |
| BNIP3L | BCL2-adenovirus E1B 19 kDa protein-interacting protein 3-like |
| CASA | Chaperone-assisted selective autophagy |
| CCCP | Carbonyl cyanide m-chlorophenyl hydrazine |
| DRP1 | Dynamin-related protein 1 |
| ER | Endoplasmic reticulum |
| FKBP8 | FK506-binding protein 8 |
| HOPS | Homotypic fusion and protein sorting |
| HSC70 | Heat shock cognate 71 kDa protein |
| HSP70 | Heat shock protein 70 family |
| IMM | Inner mitochondrial membrane |
| I/R | Ischemia/reperfusion |
| (LAMP2a) | Lysosome-associated membrane glycoprotein 2a |
| LC3 | Protein light chain 3 |
| LIR | LC3 interacting region |
| MFN2 | Mitofusin-2 |
| MPP+ | 1-methyl-4-phenylpyridinium |
| NEF | Nucleotide exchange factor |
| NIX | Nip3-like protein X |
| OMM | Outer mitochondrial membrane |
| PI3KIII | Phosphatidylinositol 3-kinase class III |
| PINK1 | PTEN-induced putative kinase 1 |
| PLEKHM1 | Pleckstrin homology domain-containing family member 1 |
| PD | Parkinson’s disease |
| PE | Phosphatidyethanolamine |
| PHB2 | Prohibitin 2 |
| ROS | Reactive oxygen species |
| SNARE | Soluble NSF (N-ethylmaleimide-sensitive factor) attachment protein receptor |
References
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, Regulation and Pathophysiological Implications of Autophagosome Maturation. Nat. Rev. Mol. Cell Biol. 2021, 22, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. The ATG Conjugation Systems in Autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef]
- Suzuki, H.; Osawa, T.; Fujioka, Y.; Noda, N.N. Structural Biology of the Core Autophagy Machinery. Curr. Opin. Struct. Biol. 2017, 43, 10–17. Available online: https://pubmed-ncbi-nlm-nih-gov.proxy.insermbiblio.inist.fr/27723509/ (accessed on 11 December 2021). [CrossRef] [PubMed] [Green Version]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (4th Edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Torres, J.L.; Hansen, M. Macroautophagy and Aging: The Impact of Cellular Recycling on Health and Longevity. Mol. Asp. Med. 2021, 82, 101020. [Google Scholar] [CrossRef]
- Lei, Y.; Klionsky, D.J. The Emerging Roles of Autophagy in Human Diseases. Biomedicines 2021, 9, 1651. [Google Scholar] [CrossRef]
- Kettern, N.; Dreiseidler, M.; Tawo, R.; Höhfeld, J. Chaperone-Assisted Degradation: Multiple Paths to Destruction. Biol. Chem. 2010, 391, 481–489. [Google Scholar] [CrossRef]
- Birgisdottir, Å.B.; Lamark, T.; Johansen, T. The LIR Motif—Crucial for Selective Autophagy. J. Cell Sci. 2013, 126, 3237–3247. [Google Scholar] [CrossRef] [Green Version]
- Morishita, H.; Mizushima, N. Diverse Cellular Roles of Autophagy. Annu. Rev. Cell Dev. Biol. 2019, 35, 453–475. [Google Scholar] [CrossRef]
- Novak, I.; Kirkin, V.; McEwan, D.G.; Zhang, J.; Wild, P.; Rozenknop, A.; Rogov, V.; Löhr, F.; Popovic, D.; Occhipinti, A.; et al. Nix Is a Selective Autophagy Receptor for Mitochondrial Clearance. EMBO Rep. 2010, 11, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Hanna, R.A.; Quinsay, M.N.; Orogo, A.M.; Giang, K.; Rikka, S.; Gustafsson, Å.B. Microtubule-Associated Protein 1 Light Chain 3 (LC3) Interacts with Bnip3 Protein to Selectively Remove Endoplasmic Reticulum and Mitochondria via Autophagy. J. Biol. Chem. 2012, 287, 19094–19104. [Google Scholar] [CrossRef] [Green Version]
- Murakawa, T.; Yamaguchi, O.; Hashimoto, A.; Hikoso, S.; Takeda, T.; Oka, T.; Yasui, H.; Ueda, H.; Akazawa, Y.; Nakayama, H.; et al. Bcl-2-like Protein 13 Is a Mammalian Atg32 Homologue That Mediates Mitophagy and Mitochondrial Fragmentation. Nat. Commun. 2015, 6, 7527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Chiang, W.-C.; Sumpter, R.; Mishra, P.; Levine, B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell 2017, 168, 224–238.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhujabal, Z.; Birgisdottir, Å.B.; Sjøttem, E.; Brenne, H.B.; Øvervatn, A.; Habisov, S.; Kirkin, V.; Lamark, T.; Johansen, T. FKBP8 Recruits LC3A to Mediate Parkin-independent Mitophagy. EMBO Rep. 2017, 18, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Montava-Garriga, L.; Ganley, I.G. Outstanding Questions in Mitophagy: What We Do and Do Not Know. J. Mol. Biol. 2019, 432, 206–230. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskaite, A.; Kondapalli, C.; Gourlay, R.; Campbell, D.G.; Ritorto, M.S.; Hofmann, K.; Alessi, D.R.; Knebel, A.; Trost, M.; Muqit, M.M.K. Parkin Is Activated by PINK1-Dependent Phosphorylation of Ubiquitin at Ser65. Biochem. J. 2014, 460, 127–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of Selective Autophagy: The P62/SQSTM1 Paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar] [CrossRef]
- Altinok, S.; Sanchez-Hodge, R.; Stewart, M.; Smith, K.; Schisler, J.C. With or without You: Co-Chaperones Mediate Health and Disease by Modifying Chaperone Function and Protein Triage. Cells 2021, 10, 3121. [Google Scholar] [CrossRef]
- Sondermann, H.; Scheufler, C.; Schneider, C.; Hohfeld, J.; Hartl, F.U.; Moarefi, I. Structure of a Bag/Hsc70 Complex: Convergent Functional Evolution of Hsp70 Nucleotide Exchange Factors. Science 2001, 291, 1553–1557. [Google Scholar] [CrossRef]
- Takayama, S.; Sato, T.; Krajewski, S.; Kochel, K.; Irie, S.; Millan, J.A.; Reed, J.C. Cloning and Functional Analysis of BAG-1: A Novel Bcl-2-Binding Protein with Anti-Cell Death Activity. Cell 1995, 80, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Chernenko, G.; Hao, Y.; Ding, Z.; Pater, M.M.; Pater, A.; Tang, S.-C. Human BAG-1/RAP46 Protein Is Generated as Four Isoforms by Alternative Translation Initiation and Overexpressed in Cancer Cells. Oncogene 1998, 17, 981–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurusamy, N.; Lekli, I.; Gorbunov, N.V.; Gherghiceanu, M.; Popescu, L.M.; Das, D.K. Cardioprotection by Adaptation to Ischaemia Augments Autophagy in Association with BAG-1 Protein. J. Cell Mol. Med. 2009, 13, 373–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turk, M.; Tatli, O.; Alkan, H.F.; Ozfiliz Kilbas, P.; Alkurt, G.; Dinler Doganay, G. Co-Chaperone Bag-1 Plays a Role in the Autophagy-Dependent Cell Survival through Beclin 1 Interaction. Molecules 2021, 26, 854. [Google Scholar] [CrossRef]
- Qin, L.; Guo, J.; Zheng, Q.; Zhang, H. BAG2 Structure, Function and Involvement in Disease. Cell Mol. Biol. Lett. 2016, 21, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, S.; Wang, F.; Bao, C.; Han, J.; Guo, Y.; Liu, F.; Zhang, Y. BAG2 Ameliorates Endoplasmic Reticulum Stress-Induced Cell Apoptosis in Mycobacterium Tuberculosis-Infected Macrophages through Selective Autophagy. Autophagy 2020, 16, 1453–1467. [Google Scholar] [CrossRef]
- Yang, K.-M.; Bae, E.; Ahn, S.G.; Pang, K.; Park, Y.; Park, J.; Lee, J.; Ooshima, A.; Park, B.; Kim, J.; et al. Co-Chaperone BAG2 Determines the Pro-Oncogenic Role of Cathepsin B in Triple-Negative Breast Cancer Cells. Cell Rep. 2017, 21, 2952–2964. [Google Scholar] [CrossRef] [Green Version]
- Behl, C. Breaking BAG: The Co-Chaperone BAG3 in Health and Disease. Trends Pharm. Sci. 2016, 37, 672–688. [Google Scholar] [CrossRef]
- Klimek, C.; Kathage, B.; Wördehoff, J.; Höhfeld, J. BAG3-Mediated Proteostasis at a Glance. J. Cell Sci. 2017, 130, 2781–2788. [Google Scholar] [CrossRef] [Green Version]
- Cristofani, R.; Piccolella, M.; Crippa, V.; Tedesco, B.; Montagnani Marelli, M.; Poletti, A.; Moretti, R.M. The Role of HSPB8, a Component of the Chaperone-Assisted Selective Autophagy Machinery, in Cancer. Cells 2021, 10, 335. [Google Scholar] [CrossRef]
- Carra, S.; Seguin, S.J.; Landry, J. HspB8 and Bag3: A New Chaperone Complex Targeting Misfolded Proteins to Macroautophagy. Autophagy 2008, 4, 237–239. [Google Scholar] [CrossRef] [Green Version]
- Crippa, V.; Sau, D.; Rusmini, P.; Boncoraglio, A.; Onesto, E.; Bolzoni, E.; Galbiati, M.; Fontana, E.; Marino, M.; Carra, S.; et al. The Small Heat Shock Protein B8 (HspB8) Promotes Autophagic Removal of Misfolded Proteins Involved in Amyotrophic Lateral Sclerosis (ALS). Hum. Mol. Genet. 2010, 19, 3440–3456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, Z.; Brizzee, C.; Johnson, G.V.W. BAG3 Facilitates the Clearance of Endogenous Tau in Primary Neurons. Neurobiol. Aging 2015, 36, 241–248. [Google Scholar] [CrossRef] [Green Version]
- Gamerdinger, M.; Hajieva, P.; Kaya, A.M.; Wolfrum, U.; Hartl, F.U.; Behl, C. Protein Quality Control during Aging Involves Recruitment of the Macroautophagy Pathway by BAG3. EMBO J. 2009, 28, 889–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wattin, M.; Gaweda, L.; Muller, P.; Baritaud, M.; Scholtes, C.; Lozano, C.; Gieseler, K.; Kretz-Remy, C. Modulation of Protein Quality Control and Proteasome to Autophagy Switch in Immortalized Myoblasts from Duchenne Muscular Dystrophy Patients. Int. J. Mol. Sci. 2018, 19, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, T.G.; Myers, V.D.; Dubey, P.; Dubey, S.; Perez, E.; Moravec, C.S.; Willis, M.S.; Feldman, A.M.; Kirk, J.A. Cardiomyocyte Contractile Impairment in Heart Failure Results from Reduced BAG3-Mediated Sarcomeric Protein Turnover. Nat. Commun. 2021, 12, 2942. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, T.; Myers, V.D.; Gordon, J.; Tilley, D.G.; Sharp, T.E.; Wang, J.; Khalili, K.; Cheung, J.Y.; Feldman, A.M. BAG3: A New Player in the Heart Failure Paradigm. Heart Fail. Rev. 2015, 20, 423–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kögel, D.; Linder, B.; Brunschweiger, A.; Chines, S.; Behl, C. At the Crossroads of Apoptosis and Autophagy: Multiple Roles of the Co-Chaperone BAG3 in Stress and Therapy Resistance of Cancer. Cells 2020, 9, 574. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Wang, J.-M.; Yan, J.; Zhang, D.-L.; Liu, B.-Q.; Jiang, J.-Y.; Li, C.; Li, S.; Meng, X.-N.; Wang, H.-Q. BAG3 Promotes Autophagy and Glutaminolysis via Stabilizing Glutaminase. Cell Death Dis. 2019, 10, 284. [Google Scholar] [CrossRef] [Green Version]
- Che, N.; Ng, K.-Y.; Wong, T.-L.; Tong, M.; Kau, P.W.; Chan, L.-H.; Lee, T.K.; Huen, M.S.; Yun, J.-P.; Ma, S. PRMT6 Deficiency Induces Autophagy in Hostile Microenvironments of Hepatocellular Carcinoma Tumors by Regulating BAG5-Associated HSC70 Stability. Cancer Lett. 2021, 501, 247–262. [Google Scholar] [CrossRef]
- Friesen, E.L.; Zhang, Y.T.; Earnshaw, R.; De Snoo, M.L.; O’Hara, D.M.; Agapova, V.; Chau, H.; Ngana, S.; Chen, K.S.; Kalia, L.V.; et al. BAG5 Promotes Alpha-Synuclein Oligomer Formation and Functionally Interacts With the Autophagy Adaptor Protein P62. Front. Cell Dev. Biol. 2020, 8, 716. [Google Scholar] [CrossRef]
- Sasaki, T.; Gan, E.C.; Wakeham, A.; Kornbluth, S.; Mak, T.W.; Okada, H. HLA-B-Associated Transcript 3 (Bat3)/Scythe Is Essential for P300-Mediated Acetylation of P53. Genes Dev. 2007, 21, 848–861. [Google Scholar] [CrossRef] [Green Version]
- Sebti, S.; Prébois, C.; Pérez-Gracia, E.; Bauvy, C.; Desmots, F.; Pirot, N.; Gongora, C.; Bach, A.-S.; Hubberstey, A.V.; Palissot, V.; et al. BAG6/BAT3 Modulates Autophagy by Affecting EP300/P300 Intracellular Localization. Autophagy 2014, 10, 1341–1342. [Google Scholar] [CrossRef] [Green Version]
- Sebti, S.; Prébois, C.; Pérez-Gracia, E.; Bauvy, C.; Desmots, F.; Pirot, N.; Gongora, C.; Bach, A.-S.; Hubberstey, A.V.; Palissot, V.; et al. BAT3 Modulates P300-Dependent Acetylation of P53 and Autophagy-Related Protein 7 (ATG7) during Autophagy. Proc. Natl. Acad. Sci. USA 2014, 111, 4115–4120. [Google Scholar] [CrossRef] [Green Version]
- Chu, Y.; Dong, X.; Kang, Y.; Liu, J.; Zhang, T.; Yang, C.; Wang, Z.; Shen, W.; Huo, H.; Zhuang, M.; et al. The Chaperone BAG6 Regulates Cellular Homeostasis between Autophagy and Apoptosis by Holding LC3B. iScience 2020, 23, 101708. [Google Scholar] [CrossRef]
- Roca-Portoles, A.; Tait, S.W.G. Mitochondrial Quality Control: From Molecule to Organelle. Cell Mol. Life Sci. 2021, 78, 3853–3866. [Google Scholar] [CrossRef]
- Matsuda, N. Phospho-Ubiquitin: Upending the PINK-Parkin-Ubiquitin Cascade. J. Biochem. 2016, 159, 379–385. [Google Scholar] [CrossRef] [Green Version]
- Navaratnarajah, T.; Anand, R.; Reichert, A.S.; Distelmaier, F. The Relevance of Mitochondrial Morphology for Human Disease. Int. J. Biochem. Cell Biol. 2021, 134, 105951. [Google Scholar] [CrossRef]
- Twig, G.; Elorza, A.; Molina, A.J.A.; Mohamed, H.; Wikstrom, J.D.; Walzer, G.; Stiles, L.; Haigh, S.E.; Katz, S.; Las, G.; et al. Fission and Selective Fusion Govern Mitochondrial Segregation and Elimination by Autophagy. EMBO J. 2008, 27, 433–446. [Google Scholar] [CrossRef] [Green Version]
- Ragimbeau, R.; El Kebriti, L.; Sebti, S.; Fourgous, E.; Boulahtouf, A.; Arena, G.; Espert, L.; Turtoi, A.; Gongora, C.; Houédé, N.; et al. BAG6 Promotes PINK1 Signaling Pathway and Is Essential for Mitophagy. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2021, 35, e21361. [Google Scholar] [CrossRef]
- Saita, S.; Ishihara, T.; Maeda, M.; Iemura, S.-I.; Natsume, T.; Mihara, K.; Ishihara, N. Distinct Types of Protease Systems Are Involved in Homeostasis Regulation of Mitochondrial Morphology via Balanced Fusion and Fission. Genes Cells 2016, 21, 408–424. [Google Scholar] [CrossRef] [Green Version]
- Hayashishita, M.; Kawahara, H.; Yokota, N. BAG6 Deficiency Induces Mis-Distribution of Mitochondrial Clusters under Depolarization. FEBS Open Bio. 2019, 9, 1281–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, X.; Tang, B.; Wang, X.; Chen, D.; Yan, X.; Jiang, H.; Shen, L.; Xu, Q.; Wang, G.; Guo, J. The BAG2 Protein Stabilises PINK1 by Decreasing Its Ubiquitination. Biochem. Biophys. Res. Commun. 2013, 441, 488–492. [Google Scholar] [CrossRef]
- Qu, D.; Hage, A.; Don-Carolis, K.; Huang, E.; Joselin, A.; Safarpour, F.; Marcogliese, P.C.; Rousseaux, M.W.C.; Hewitt, S.J.; Huang, T.; et al. BAG2 Gene-Mediated Regulation of PINK1 Protein Is Critical for Mitochondrial Translocation of PARKIN and Neuronal Survival. J. Biol. Chem. 2015, 290, 30441–30452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.; Kim, J.W.; Heo, H.; Lee, J.; Park, K.Y.; Yoon, J.H.; Chang, J. Identification of BAG2 and Cathepsin D as Plasma Biomarkers for Parkinson’s Disease. Clin. Transl. Sci. 2021, 14, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, J.; Fei, E.; Mu, Y.; He, S.; Che, X.; Tan, J.; Xia, K.; Zhang, Z.; Wang, G.; et al. BAG5 Protects against Mitochondrial Oxidative Damage through Regulating PINK1 Degradation. PLoS ONE 2014, 9, e86276. [Google Scholar] [CrossRef] [Green Version]
- Tsujimoto, T.; Mori, T.; Houri, K.; Onodera, Y.; Takehara, T.; Shigi, K.; Nakao, S.; Teramura, T.; Fukuda, K. MiR-155 Inhibits Mitophagy through Suppression of BAG5, a Partner Protein of PINK1. Biochem. Biophys. Res. Commun. 2020, 523, 707–712. [Google Scholar] [CrossRef]
- Verma, M.; Zhu, J.; Wang, K.Z.Q.; Chu, C.T. Chronic Treatment with the Complex I Inhibitor MPP+ Depletes Endogenous PTEN-Induced Kinase 1 (PINK1) via up-Regulation of Bcl-2-Associated Athanogene 6 (BAG6). J. Biol. Chem. 2020, 295, 7865–7876. [Google Scholar] [CrossRef] [Green Version]
- Tahrir, F.G.; Knezevic, T.; Gupta, M.K.; Gordon, J.; Cheung, J.Y.; Feldman, A.M.; Khalili, K. Evidence for the Role of BAG3 in Mitochondrial Quality Control in Cardiomyocytes. J. Cell. Physiol. 2017, 232, 797–805. [Google Scholar] [CrossRef] [Green Version]
- Schänzer, A.; Rupp, S.; Gräf, S.; Zengeler, D.; Jux, C.; Akintürk, H.; Gulatz, L.; Mazhari, N.; Acker, T.; Van Coster, R.; et al. Dysregulated Autophagy in Restrictive Cardiomyopathy Due to Pro209Leu Mutation in BAG3. Mol. Genet. Metab. 2018, 123, 388–399. [Google Scholar] [CrossRef]
- Kalia, S.K.; Lee, S.; Smith, P.D.; Liu, L.; Crocker, S.J.; Thorarinsdottir, T.E.; Glover, J.R.; Fon, E.A.; Park, D.S.; Lozano, A.M. BAG5 Inhibits Parkin and Enhances Dopaminergic Neuron Degeneration. Neuron 2004, 44, 931–945. [Google Scholar] [CrossRef] [Green Version]
- De Snoo, M.L.; Friesen, E.L.; Zhang, Y.T.; Earnshaw, R.; Dorval, G.; Kapadia, M.; O’Hara, D.M.; Agapova, V.; Chau, H.; Pellerito, O.; et al. Bcl-2-Associated Athanogene 5 (BAG5) Regulates Parkin-Dependent Mitophagy and Cell Death. Cell Death Dis. 2019, 10, 907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasson, S.A.; Kane, L.A.; Yamano, K.; Huang, C.-H.; Sliter, D.A.; Buehler, E.; Wang, C.; Heman-Ackah, S.M.; Hessa, T.; Guha, R.; et al. High-Content Genome-Wide RNAi Screens Identify Regulators of Parkin Upstream of Mitophagy. Nature 2013, 504, 291–295. [Google Scholar] [CrossRef]
- Onishi, M.; Yamano, K.; Sato, M.; Matsuda, N.; Okamoto, K. Molecular Mechanisms and Physiological Functions of Mitophagy. EMBO J. 2021, 40, e104705. [Google Scholar] [CrossRef]
- Kalvari, I.; Tsompanis, S.; Mulakkal, N.C.; Osgood, R.; Johansen, T.; Nezis, I.P.; Promponas, V.J. ILIR: A Web Resource for Prediction of Atg8-Family Interacting Proteins. Autophagy 2014, 10, 913–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baeken, M.W.; Behl, C. On the Origin of BAG(3) and Its Consequences for an Expansion of BAG3’s Role in Protein Homeostasis. J. Cell Biochem. 2021, 123, 102–114. [Google Scholar] [CrossRef] [PubMed]
- Roperto, S.; Russo, V.; De Falco, F.; Rosati, A.; Catoi, C.; Roperto, F. FUNDC1-Mediated Mitophagy in Bovine Papillomavirus-Infected Urothelial Cells. Vet. Microbiol. 2019, 234, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Roperto, S.; De Falco, F.; Perillo, A.; Catoi, C.; Roperto, F. Mitophagy Mediated by BNIP3 and BNIP3L/NIX in Urothelial Cells of the Urinary Bladder of Cattle Harbouring Bovine Papillomavirus Infection. Vet. Microbiol. 2019, 236, 108396. [Google Scholar] [CrossRef]
- De Falco, F.; Urraro, C.; Cutarelli, A.; Roperto, S. Bovine Papillomavirus E5 Oncoprotein Upregulates Parkin-Dependent Mitophagy in Urothelial Cells of Cattle with Spontaneous Papillomavirus Infection: A Mechanistic Study. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101463. [Google Scholar] [CrossRef] [PubMed]
- Benarroch, R.; Austin, J.M.; Ahmed, F.; Isaacson, R.L. The Roles of Cytosolic Quality Control Proteins, SGTA and the BAG6 Complex, in Disease. Adv. Protein. Chem. Struct. Biol. 2019, 114, 265–313. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pattingre, S.; Turtoi, A. BAG Family Members as Mitophagy Regulators in Mammals. Cells 2022, 11, 681. https://doi.org/10.3390/cells11040681
Pattingre S, Turtoi A. BAG Family Members as Mitophagy Regulators in Mammals. Cells. 2022; 11(4):681. https://doi.org/10.3390/cells11040681
Chicago/Turabian StylePattingre, Sophie, and Andrei Turtoi. 2022. "BAG Family Members as Mitophagy Regulators in Mammals" Cells 11, no. 4: 681. https://doi.org/10.3390/cells11040681
APA StylePattingre, S., & Turtoi, A. (2022). BAG Family Members as Mitophagy Regulators in Mammals. Cells, 11(4), 681. https://doi.org/10.3390/cells11040681







