The Full Model of the pMHC-TCR-CD3 Complex: A Structural and Dynamical Characterization of Bound and Unbound States
Abstract
:1. Introduction
2. Materials and Methods
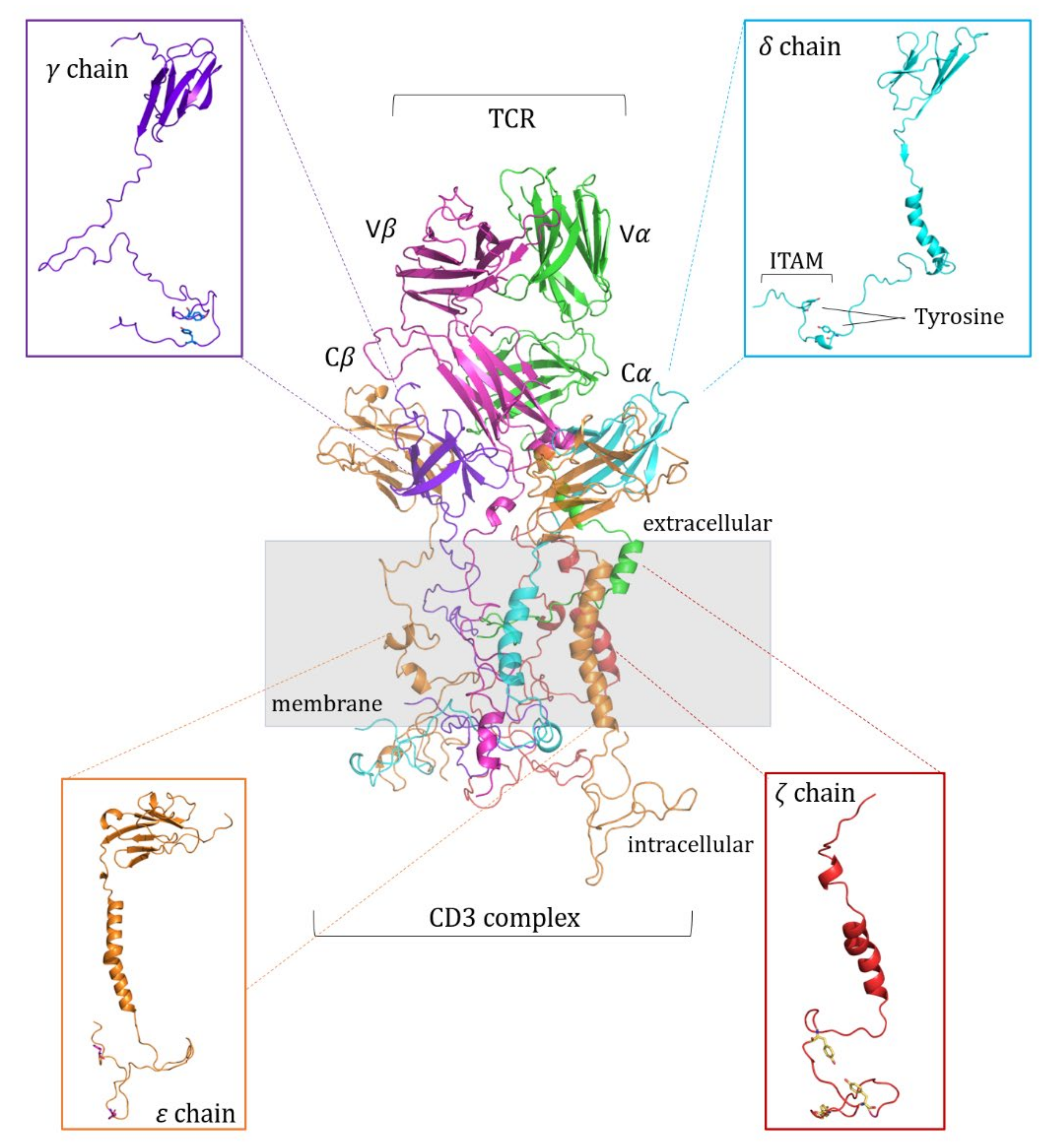
2.1. Modeling of the pMHC:TCR:CD3-Chains Complex
2.2. Molecular Dynamics (MD) Simulations
2.3. Structural Analysis
2.4. Essential Dynamics (ED)
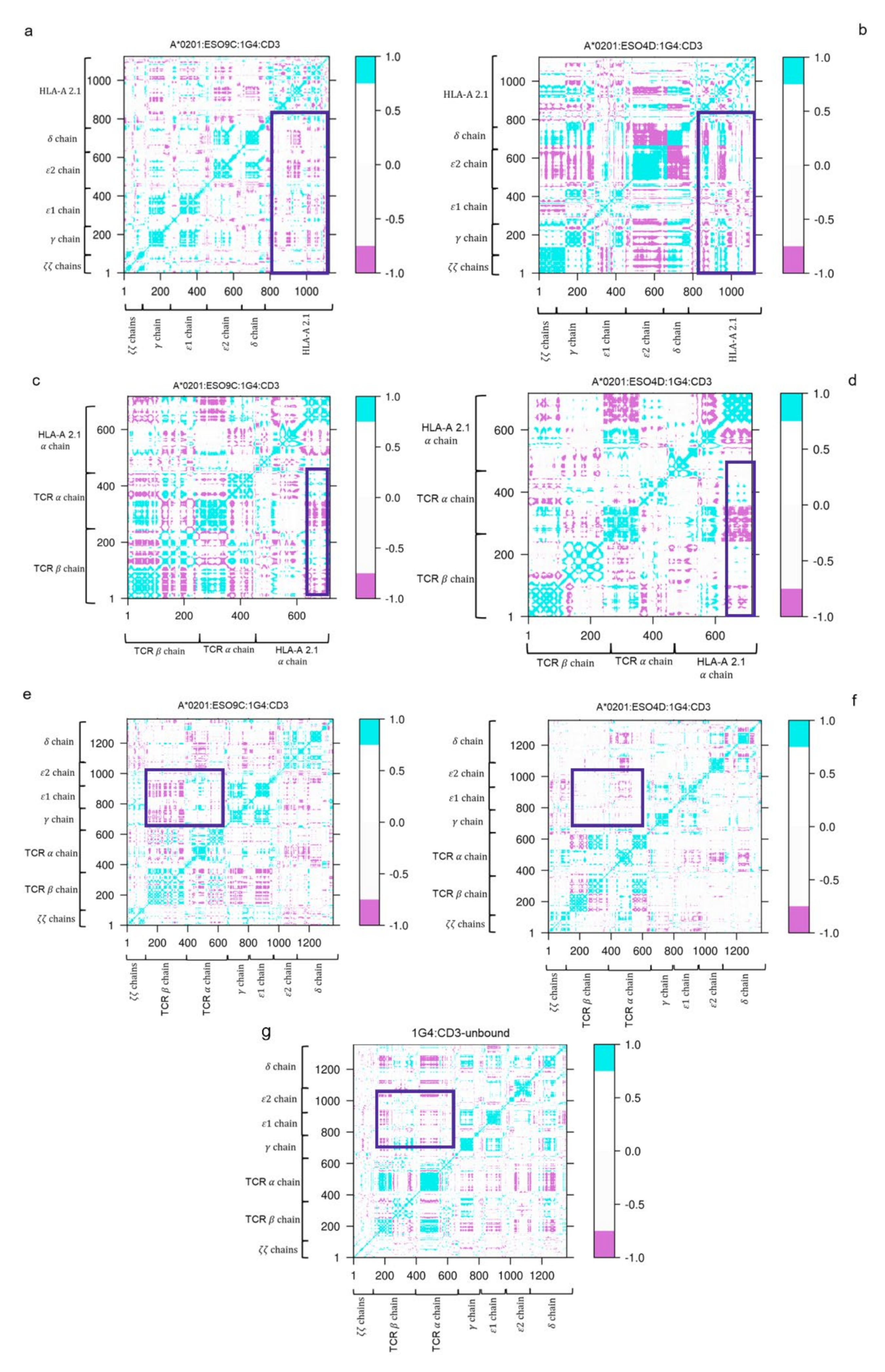
2.5. Cross Correlation Matrix (DCC)
2.6. Hydrogen Bonds and Salt Bridges Interactions
2.7. Cluster Analysis and Binding Free Energy Estimation (MMPBSA)
3. Results
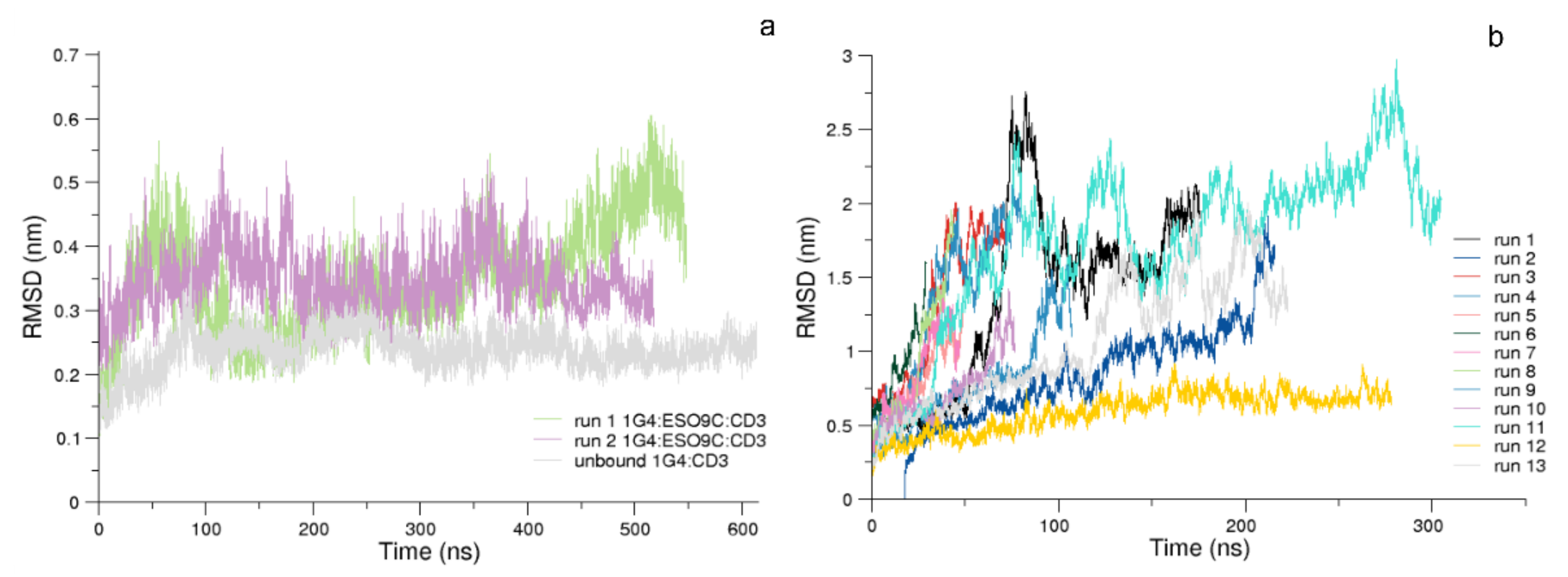
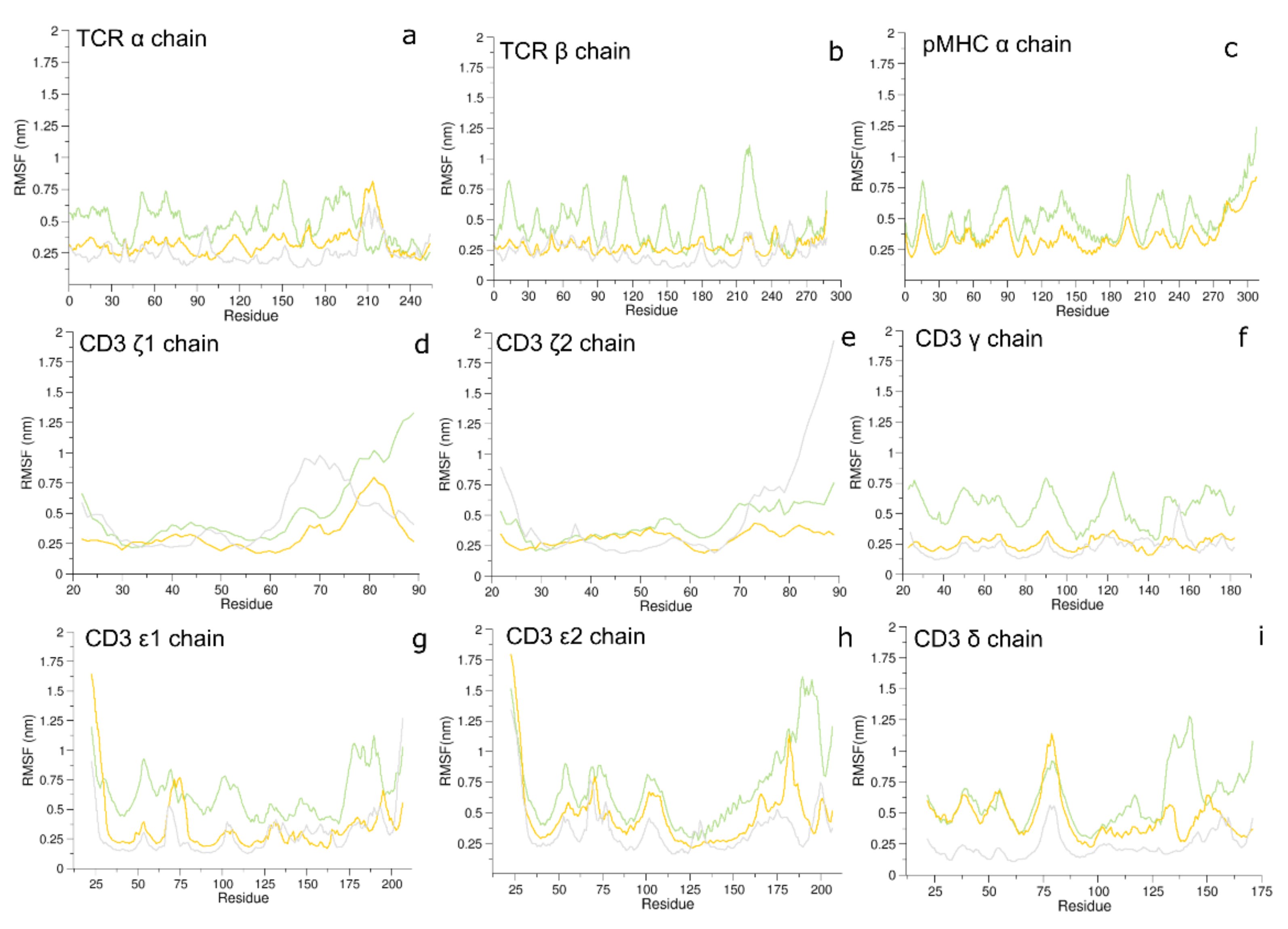
3.1. Conformational Analysis
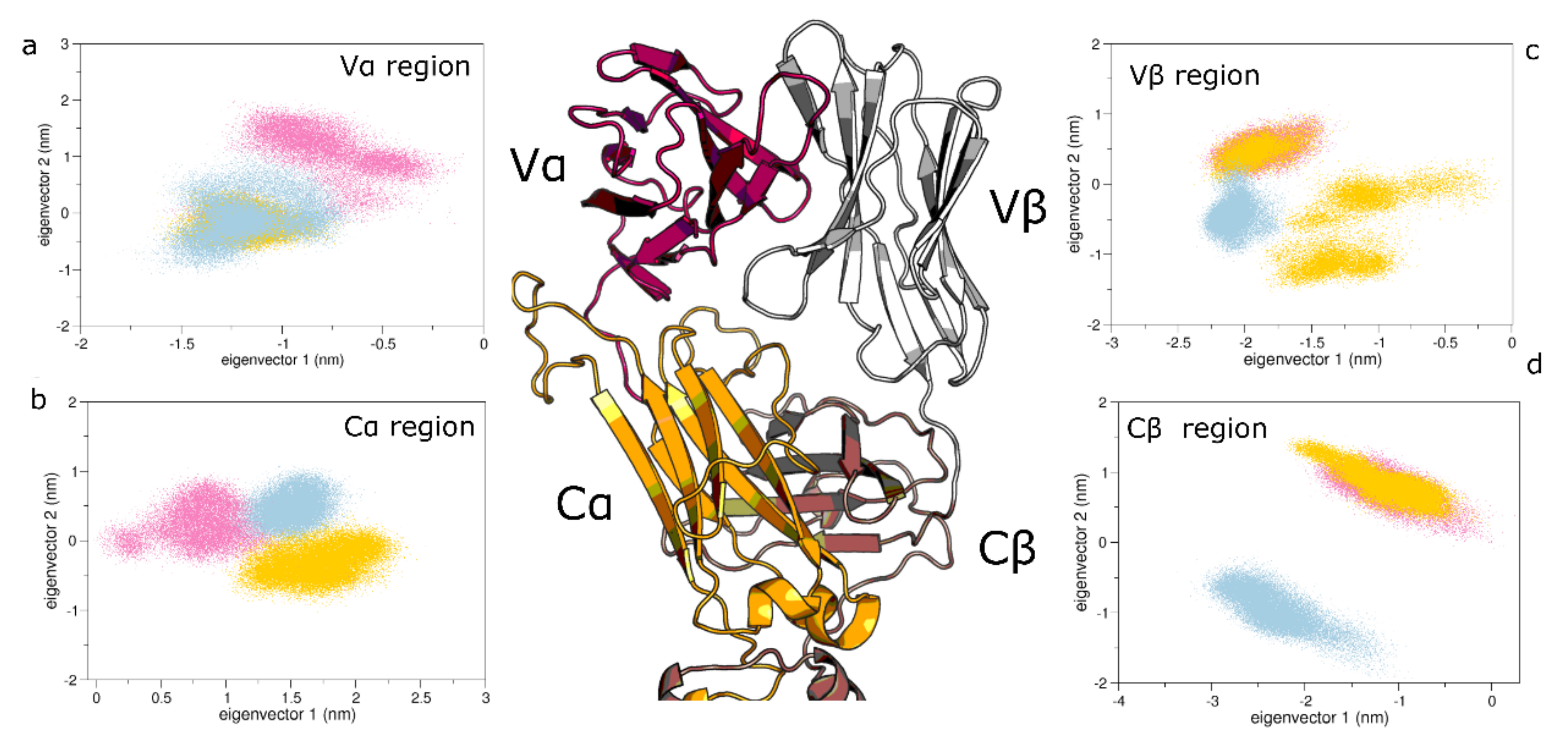
3.2. Collective Motions
3.3. Dynamic Cross-Correlation (DCC) of the Bound States
3.4. Interface Interaction Behavior of the Bound States
3.5. Hydrogen Bonds and Salt Bridges
3.6. Solvent Exposure of the CD3 Chains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Janeway, C.A. The immune system evolved to discriminate infectious non self from noninfectious self. Immunol. Today 1992, 13, 11–16. [Google Scholar] [CrossRef]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major Histocompatibility Complex (MHC) Class I and MHC Class II Proteins: Conformational Plasticity in Antigen Presentation. Front. Immunol. 2017, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Birnbaum, M.; Berry, R.; Hsiao, Y.-S.; Chen, Z.; Shingu-Vazquez, M.A.; Yu, X.; Waghray, D.; Fischer, S.; McCluskey, J.; Rossjohn, J.; et al. Molecular architecture of the αβ T cell receptor–CD3 complex. Proc. Natl. Acad. Sci. USA 2014, 111, 17576–17581. [Google Scholar] [CrossRef] [Green Version]
- Bosselut, R. T cell antigen recognition: Evolution-driven affinities. Proc. Natl. Acad. Sci. USA 2019, 116, 21969–21971. [Google Scholar] [CrossRef]
- Li, X.C.; Raghavan, M. Structure and function of major histocompatibility complex class I antigens. Curr. Opin. Organ Transplant. 2010, 15, 499–504. [Google Scholar] [CrossRef]
- Wucherpfennig, K.W.; Gagnon, E.; Call, M.J.; Huseby, E.S. Structural biology of the T-cell receptor: Insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb. Perspect. Biol. 2010, 2, a005140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richman, S.A.; Aggen, D.H.; Dossett, M.L.; Donermeyer, D.L.; Allen, P.M.; Greenberg, P.D.; Kranz, D.M. Structural features of T cell receptor variable regions that enhance domain stability and enable expression as single-chain ValphaVbeta fragments. Mol. Immunol. 2009, 46, 902–916. [Google Scholar] [CrossRef] [Green Version]
- Holland, C.J.; MacLachlan, B.; Bianchi, V.; Hesketh, S.; Morgan, R.; Vickery, O.; Bulek, A.M.; Fuller, A.; Godkin, A.; Sewell, A.; et al. In Silico and Structural Analyses Demonstrate That Intrinsic Protein Motions Guide T Cell Receptor Complementarity Determining Region Loop Flexibility. Front. Immunol. 2018, 9, 674. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Quintero, M.L.; Pomarici, N.D.; Loeffler, J.R.; Seidler, C.A.; Liedl, K.R. T-Cell Receptor CDR3 Loop Conformations in Solution Shift the Relative Vα-VβDomain Distributions. Front. Immunol. 2020, 11, 1440. [Google Scholar] [CrossRef]
- Reiser, J.B.; Darnault, C.; Grégoire, C.; Mosser, T.; Mazza, G.; Kearney, A.; van der Merwe, P.A.; Fontecilla-Camps, J.C.; Housset, D.; Malissen, B. CDR3 loop flexibility contributes to the degeneracy of TCR recognition. Nat. Immunol. 2003, 4, 241–247. [Google Scholar] [CrossRef]
- Wong, W.K.; Leem, J.; Deane, C.M. Comparative Analysis of the CDR Loops of Antigen Receptors. Front. Immunol. 2019, 10, 2454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, K.C.; Adams, E.J. How the T Cell Receptor Sees Antigen—A Structural View. Cell 2005, 122, 333–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Natarajan, K.; McShan, A.C.; Jiang, J.; Kumirov, V.K.; Wang, R.; Zhao, H.; Schuck, P.; Tilahun, M.E.; Boyd, L.F.; Ying, J.; et al. An allosteric site in the T-cell receptor Cβ domain plays a critical signalling role. Nat. Commun. 2017, 8, 15260. [Google Scholar] [CrossRef] [Green Version]
- Beddoe, T.; Chen, Z.; Clements, C.S.; Ely, L.K.; Bushell, S.R.; Vivian, J.P.; Kjer-Nielsen, L.; Pang, S.S.; Dunstone, M.A.; Liu, Y.C.; et al. Antigen Ligation Triggers a Conformational Change within the Constant Domain of the αβ T Cell Receptor. Immunity 2009, 30, 777–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangarajan, S.; He, Y.; Chen, Y.; Kerzic, M.C.; Ma, B.; Gowthaman, R.; Pierce, B.; Nussinov, R.; Mariuzza, R.A.; Orban, J. Peptide–MHC (pMHC) binding to a human antiviral T cell receptor induces long-range allosteric communication between pMHC- and CD3-binding sites. J. Biol. Chem. 2018, 293, 15991–16005. [Google Scholar] [CrossRef] [Green Version]
- Gaud, G.; Lesourne, R.; Love, P.E. Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 2018, 18, 485–497. [Google Scholar] [CrossRef]
- Kane, L.P.; Lin, J.; Weiss, A. Signal transduction by the TCR for antigen. Curr. Opin. Immunol. 2000, 12, 242–249. [Google Scholar] [CrossRef]
- Call, M.E.; Pyrdol, J.; Wucherpfennig, K.W. Stoichiometry of the T-cell receptor–CD3 complex and key intermediates assembled in the endoplasmic reticulum. EMBO J. 2004, 23, 2348–2357. [Google Scholar] [CrossRef] [Green Version]
- Love, P.E.; Hayes, S.M. ITAM-mediated signaling by the T-cell antigen receptor. Cold Spring Harb. Perspect. Biol. 2010, 2, a002485. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Guo, X.; Shi, X.; Li, C.; Wu, W.; Yan, C.; Wang, H.; Li, H.; Xu, C. Ionic CD3−Lck interaction regulates the initiation of T-cell receptor signaling. Proc. Natl. Acad. Sci. USA 2017, 114, E5891–E5899. [Google Scholar] [CrossRef] [Green Version]
- Brazin, K.N.; Mallis, R.J.; Boeszoermenyi, A.; Feng, Y.; Yoshizawa, A.; Reche, P.; Kaur, P.; Bi, K.; Hussey, R.E.; Duke-Cohan, J.S.; et al. The T Cell Antigen Receptor α Transmembrane Domain Coordinates Triggering through Regulation of Bilayer Immersion and CD3 Subunit Associations. Immunity 2018, 49, 829–841.e6. [Google Scholar] [CrossRef] [Green Version]
- Mariuzza, R.A.; Agnihotri, P.; Orban, J. The structural basis of T-cell receptor (TCR) activation: An enduring enigma. J. Biol. Chem. 2020, 295, 914–925. [Google Scholar] [CrossRef]
- Davis, S.; van der Merwe, P. The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 2006, 7, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Zheng, L.; Lin, J.; Zhang, B.; Zhu, Y.; Li, N.; Xie, S.; Wang, Y.; Gao, N.; Huang, Z. Structural basis of assembly of the human T cell receptor–CD3 complex. Nature 2019, 573, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Aleksic, M.; Dushek, O.; Zhang, H.; Shenderov, E.; Chen, J.-L.; Cerundolo, V.; Coombs, D.; van der Merwe, P.A. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity 2010, 32, 163–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.-L.; Dunbar, P.R.; Gileadi, U.; Jäger, E.; Gnjatic, S.; Nagata, Y.; Stockert, E.; Panicali, D.L.; Chen, Y.-T.; Knuth, A.; et al. Identification of NY-ESO-1 Peptide Analogues Capable of Improved Stimulation of Tumor-Reactive CTL. J. Immunol. 2000, 165, 948–955. [Google Scholar] [CrossRef]
- Alba, J.; Di Rienzo, L.; Milanetti, E.; Acuto, O.; D’Abramo, M. Molecular Dynamics Simulations Reveal Canonical Conformations in Different pMHC/TCR Interactions. Cells 2020, 9, 942. [Google Scholar] [CrossRef] [Green Version]
- Šali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Martí-Renom, M.A.; Stuart, A.C.; Fiser, A.; Sánchez, R.; Melo, F.; Šali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 291–325. [Google Scholar] [CrossRef] [Green Version]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using Modeller. Curr. Protoc. Bioinform. 2016, 54, 5–6. [Google Scholar] [CrossRef] [Green Version]
- Fiser, A.; Do, R.K.; Sali, A. Modeling of loops in protein structures. Protein Sci. 2000, 9, 1753–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database: Its relevance to human molecular medical research. J. Mol. Med. 1997, 75, 312–316. [Google Scholar] [PubMed]
- Abraham, M.J.; van der Spoel, D.; Lindahl, E.; Hess, B.; Team, D. GROMACS User Manual Version 2016.4 2017. Gromax Web Site. Available online: ftp://ftp.gromacs.org/pub/manual/manual-2016.4.pdf (accessed on 25 January 2020).
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Swamy, M.; Beck-Garcia, K.; Beck-Garcia, E.; Hartl, F.A.; Morath, A.; Yousefi, O.S.; Dopfer, E.P.; Molnár, E.; Schulze, A.K.; Blanco, R.; et al. A Cholesterol-Based Allostery Model of T Cell Receptor Phosphorylation. Immunity 2016, 44, 1091–1101. [Google Scholar] [CrossRef] [Green Version]
- Mark, P.; Nilsson, L. Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef] [Green Version]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.W.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; MacKerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [Green Version]
- Amadei, A.; Linssen, A.B.M.; Berendsen, H.J.C. Essential dynamics of proteins. Proteins Struct. Funct. Bioinform. 1993, 17, 412–425. [Google Scholar] [CrossRef]
- Grant, B.J.; Rodrigues, A.P.; ElSawy, K.M.; McCammon, J.A.; Caves, L.S. Bio3D: An R package for the comparative analysis of protein structures. Bioinformatics 2006, 22, 2695–2696. [Google Scholar] [CrossRef] [Green Version]
- R Studio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015; Available online: http://www.rstudio.com/ (accessed on 16 December 2021).
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1996, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide Folding: When Simulation Meets Experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Valdés-Tresanco, M.S.; Valiente, P.A.; Moreno, E. gmx_MMPBSA: A New Tool to Perform End-State Free Energy Calculations with GROMACS. J. Chem. Theory Comput. 2021, 17, 6281–6291. [Google Scholar] [CrossRef] [PubMed]
- Miller, I.B.R.; McGee, J.T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.R.; Różycki, B.; Lipowsky, R.; Weikl, T.R. Structural variability and concerted motions of the T cell receptor–CD3 complex. eLife 2021, 10, e67195. [Google Scholar] [CrossRef] [PubMed]
- Tedeschi, V.; Alba, J.; Paladini, F.; Paroli, M.; Cauli, A.; Mathieu, A.; Sorrentino, R.; D’Abramo, M.; Fiorillo, M.T. Unusual Placement of an EBV Epitope into the Groove of the Ankylosing Spondylitis-Associated HLA-B27 Allele Allows CD8+ T Cell Activation. Cells 2019, 8, 572. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Complex with HLA-A*0201 | Peptide Sequence | KD (uM) | Koff (s − 1) | t ½ (s) | Kon (M − 1 s − 1) | EC₅₀ (IFN-γ) (μg/mL pMHC) |
|---|---|---|---|---|---|---|
| 1G4-ESO9C | SLLMWITQC | 14 | 0.82 | 0.84 | 57 × 10 + 3 | 115 ± 14 |
| 1G4-ESO4D | SLLDWITQV | 252 | 2.59 | 0.27 | 10 × 10 + 3 | 661 ± 85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alba, J.; D’Abramo, M. The Full Model of the pMHC-TCR-CD3 Complex: A Structural and Dynamical Characterization of Bound and Unbound States. Cells 2022, 11, 668. https://doi.org/10.3390/cells11040668
Alba J, D’Abramo M. The Full Model of the pMHC-TCR-CD3 Complex: A Structural and Dynamical Characterization of Bound and Unbound States. Cells. 2022; 11(4):668. https://doi.org/10.3390/cells11040668
Chicago/Turabian StyleAlba, Josephine, and Marco D’Abramo. 2022. "The Full Model of the pMHC-TCR-CD3 Complex: A Structural and Dynamical Characterization of Bound and Unbound States" Cells 11, no. 4: 668. https://doi.org/10.3390/cells11040668
APA StyleAlba, J., & D’Abramo, M. (2022). The Full Model of the pMHC-TCR-CD3 Complex: A Structural and Dynamical Characterization of Bound and Unbound States. Cells, 11(4), 668. https://doi.org/10.3390/cells11040668






